Abstract
Limited data exist regarding the distribution of gene segments used in T-cell receptor γ gene rearrangements (TCRγGR) in T-cell lymphoproliferative disorders. The reported efficacy of TCRγGR protocols ranges from 60% to greater than 90%. Laboratories reporting a lower detection rate tend to use a limited set of primers. The goal of our study was to provide TCRγGR data to demonstrate the molecular biological basis for needing multiple primer sets targeting all gene segments. Sixty cases with a confirmed histological diagnosis of a T-cell lymphoproliferative disorder and TCRγGR were identified in our lymphoma registry from 1995 to 2001. DNA was obtained from fresh/frozen tissue, cell lysates, or paraffin-embedded tissue. Variable (Vγ) region gene segments were identified using denaturing gradient gel electrophoresis, which was used to select the cases in the study. Capillary electrophoresis using fluorescent-labeled joining (Jγ) region primers was performed to identify Jγ segments. Sixty cases contained a total of 98 TCRγGR, as some cases have more than one rearrangement. The most frequent gene segment combination involved the Vγ1–8 and Jγ1/2 segments. If a single primer set directed at these two segments were used for clinical diagnosis, that pair of primers would only diagnose 67% of cases as positive for TCRγGR. Our gene segment distribution data emphasize the importance of using a comprehensive set of Vγ and Jγ primers for an optimal detection rate of TCRγGR. Protocols with limited numbers of primers should be reconsidered.
Unlike most B-cell lymphomas, T-cell lymphomas are often difficult to diagnose by morphology and immunohistochemistry alone. The neoplastic T-cell infiltrates can be polymorphous and are sometimes difficult to separate from intermixed benign T cells with immunohistochemistry. Demonstrating an aberrant immunophenotype is often not possible, even with flow cytometry. For these reasons, pathologists often resort to molecular methods to demonstrate the presence of a clonal T-cell population.
In most T-cell lymphomas, diagnostic assays in T-cell receptor gene rearrangements targeting the β and γ genes are most useful. Rearrangements of the T-cell receptor γ (TCRγ) chain gene are often analyzed in T-cell lymphoproliferative disorders by polymerase chain reaction (PCR), due to the relative structural simplicity of the gene. The TCRγ chain gene is located on the short arm of chromosome 7 and has 2 constant, 5 joining, and 14 variable region segments. Of the 14 variable region segments, 11 are functional and have been described as rearranged in T-cell lymphoproliferative disorders 1 , 2, 3, 4, 5
There is limited data regarding the distribution of gene segments in rearrangements of the TCRγ chain gene. Two previous studies by Theodorou et al 4 and Födinger et al 6 have demonstrated that most variable region (Vγ) rearrangements occur within the Vγ1–8 subgroup (Group I) and most joining region (Jγ) rearrangements involve the Jγ1/2 segment. This high frequency may have prompted some laboratories to devise TCRγ PCR assays that use only single primer sets for the Vγ1–8 and Jγ1/2 segments. However, both studies have demonstrated that some T-cell lymphoproliferative disorders involve other Vγ and Jγ segments that would not be identified with a single Vγ1–8 and Jγ1/2 primer set. Despite this data, there is a heterogeneous group of primer sets currently used by many laboratories.
A recent multi-center study by Arber et al 7 involving 21 participating laboratories, specifically addressed the use of different primer sets in TCRγPCR testing and compared sensitivity rates. Based on their findings, 25% of laboratories used a single primer set directed at the Vγ1–8 and Jγ1/2 segments. Since the survey was sent only to members of the Association for Molecular Pathology, the percentage of all laboratories using a single primer set is unknown. This study found a 77.9% overall detection rate by TCRγPCR and noted a significant difference in true positive results among laboratories that used multiple primer sets (84%) versus those that used only a single primer set (61.4%) directed against the Vγ1–8 and Jγ1/2 segments. Given the variation in TCRγ primer sets used by laboratories and in the sensitivity results reported by Arber et al, 7 we sought to determine the distribution of involved Vγ and Jγ segments for the purpose of supporting the utilization of complete primer sets in TCRγPCR testing.
Materials and Methods
Case Samples
We identified 60 cases of T-cell lymphoproliferative disorders in our molecular diagnostic laboratory database from 1995 to 2001 that had clonal TCRγGR(s) identified by denaturing gradient gel electrophoresis (DGGE), diagnostic material for morphological review, a T-cell phenotype by immunohistochemical analysis, and adequate DNA amplification on re-analysis. Cases were classified according to the World Health Organization classification system. 8 The cases consisted of 38 peripheral T-cell lymphomas, 10 anaplastic large-cell lymphomas, six cases of mycosis fungoides, three T-cell large granular lymphocyte proliferations, two lymphomatoid papulosis cases, and one hepatosplenic T-cell lymphoma.
Extraction Protocols
DNA from 10 fresh tissue samples, 14 frozen tissue samples, and 36 formalin-fixed, paraffin-embedded tissue samples was used for the analyses. DNA extraction procedures for each sample type have been previously described. 5, 9 Briefly, fresh and frozen tissue samples were homogenized and 0.5 μg DNA was used for PCR following proteinase K (Sigma, St. Louis, MO) digestion, phenol/chloroform (Amresco, Solon, OH) extraction, and ethanol precipitation procedures. For peripheral blood samples, 2.0 to 5.0 μl of cell lysate (estimated 0.5 to 1.0 μg) from a mononuclear cell fraction isolated from a Ficoll-Hypaque (Accurate-Chemical, Westbury, NY) gradient and digested with proteinase K was used for PCR. 10 For formalin-fixed, paraffin-embedded tissue, 5- to 10-μm microtome sections were cut and paraffin was dissolved with xylene (Sigma). The tissue was then washed with 100% ethanol (Sigma) and placed in a dry incubator at 50°C to evaporate residual alcohol. A proteinase K digestion step was performed overnight at 37°C to produce a tissue lysate. Proteinase K was then inactivated by heating to 95°C for 10 minutes before using lysate estimated to contain 0.5 μg of DNA.
Vγ Segment Identification
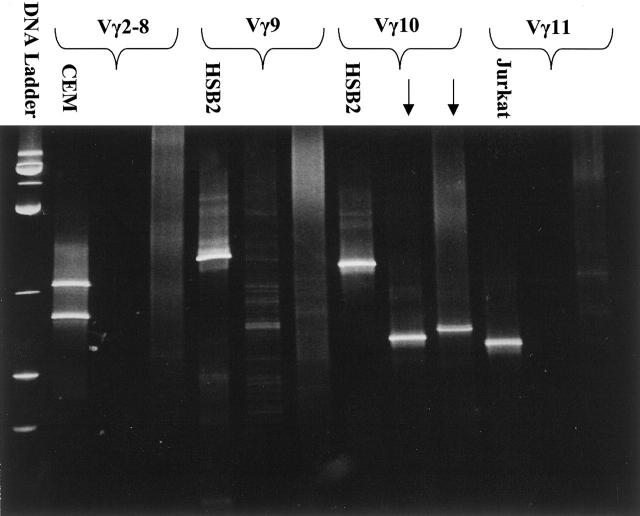
The Vγ gene segment analysis was previously performed by DGGE (CBS Scientific, Del Mar, CA) during clinical testing of the cases using multiple GC-clamped Vγ primers (Vγ2, Vγ9, Vγ10, and Vγ11) and Jγ primers (JγP1/2, Jγ2, and JγP), as described. 5 Primer sequences are listed in Table 1 . Four separate reaction mixes were prepared with each one containing a different GC-clamped Vγ primer to identify the Vγ segment combined with all Jγ primers (Figure 1) . The PCR cycles consisted of a 9-minute denaturing step at 94°C, followed by 45 PCR cycles (94°C for 75 seconds, 66°C for 75 seconds, and 72°C for 10 seconds with a 1-second additive extension per each cycle). A final extension step at 72°C was held for 7 minutes. The GC-clamped products were separated using a modified DGGE procedure using an 8% polyacrylamide gel (Amresco) with a 30 to 60% urea-formamide gradient, and stained with ethidium bromide solution (Fisher, Pittsburgh, PA). 5, 11, 12 DNA from T-cell lines (Jurkat, HSB2, and CEM) served as positive controls and a reaction mix with no DNA was used as a negative control. 5 Only samples with discrete bands and staining intensity similar to the controls were regarded as positive. DNA integrity was assessed using a multiplex β-hemoglobin gene assay. 9, 13
Table 1.
Sequences of the Primers Used in the DGGE PCR Protocol 5
| Primer | Sequence 5′-3′ |
|---|---|
| Vγ2* | GC clamp TAC ATC CAC TGG TAC CTA CAC CAG |
| Vγ9 | GC clamp GAA AGG AAT CTG GCA TTC CGT CAG |
| Vγ10 | GC clamp AAG CAA CAA AGT GGA GGC AAG AAA G |
| Vγ11 | GC clamp AGT AAA AAT GCT CAC ACT TCC ACT TC |
| Jγ2 | TAC CTG TGA CAA CAA GTG TTG TTC |
| JγP | AAG CTT TGT TCC GGG ACC AAA TAC |
| JγP1/JγP2 | GAA GTT ACT ATG AGC T/CTA GTC CCT T |
| GC clamp | CGC CCG CCG CGC CCC GCG CCC GGC CCG |
| CCG CCC CCG CCC G |
, Vγ2 covers all genes in Group I (Vγ1–8).
Figure 1.
Denaturing gradient gel electrophoresis: two cases exhibiting a rearrangement in the Vγ10 segment (arrows). CEM, HSB2, and Jurkat are T-cell lines for positive controls and CEM shows a biallelic rearrangement.
Jγ Segment Identification
Fluorescent-labeled Jγ segment analysis was performed in duplicate assays using capillary electrophoresis (CE) in an ABI™ Prism 310 Genetic Analyzer (Perkin Elmer Applied Biosystems, Foster City, CA). 14, 15 Fluorescent-labeled Jγ primer sets (Jγ1/2, JγP 1/2, and JγP) were coupled with a set of Vγ primers (Vγ2, Vγ3, Vγ9, Vγ10, and Vγ11) in a single multiplex PCR tube using a Hybaid Omnigene thermal cycler (National Labnet). The PCR reagents contained 0.6 μmol/L of fluorescent-labeled Jγ primers, 0.6 μmol/L of Vγ primers, 100 μmol/L of each dNTPs, 1.5 mmol/L MgCl2, 10 mmol/L Tris-HCl, 0.01% gelatin (Perkin Elmer ABI) and 1.25 units of Platinum Taq polymerase (Invitrogen, Carlsbad, CA). Primer sequences are listed in Table 2 . The PCR conditions consisted of a 9-minute denaturing cycle at 94°C, followed by 30 cycles (75 seconds at 94°C, 75 seconds at 60°C, and 10 seconds at 72°C with a 1-second additive extension step per each cycle). A final extension step was performed at 60°C for 45 minutes. A positive control consisting of a total of 1 μg of DNA (2% CEM DNA in peripheral blood DNA) was used and a reaction mix with no DNA served as a negative control. Before the capillary electrophoresis analysis, the amplified PCR products were run on a 2% NuSieve 3:1 agarose gel (Sigma, St. Louis, MO) to estimate the concentration for determining a dilution ratio for capillary electrophoresis. Performance optimized polymer-4 (Perkin Elmer Applied Biosystems) was used for the capillary separation matrix and the samples were electrophoresed at 15,000 volts at 60°C for 24 minutes. The internal size standard GS350 labeled with Rox (Perkin Elmer Applied Biosystems) provided reference standards. The data were analyzed using Gene Scan software (Perkin Elmer Applied Biosystems). Only clonal populations that had a peak height greater than two times the maximum height of the background polyclonal distribution in duplicate assays were interpreted as positive for the capillary electrophoresis method. 14 Dilutional studies and the sensitivity of capillary electrophoresis (2% tumor DNA) were previously described. 14
Table 2.
Primers for TCRγGR for the CE PCR Protocol 14
| Primer | Sequence 5′-3′ |
|---|---|
| Vγ2 | ACTCCAGGGTTGTGTTGGAATCA |
| Vγ3 | CCGCAAGGGATGTGTTGGAATCA |
| Vγ9 | ACGGCACTGTCAGAAAGGAATC |
| Vγ10 | AATCCGCAGCTCGACGCAGCA |
| Vγ11 | GGC TCA AGA TTG CTC AGG TGG |
| Jγ1/Jγ2 | NED-TAC CTG TGA CAA CAA GTG TTG TTC |
| JγP | FAM-AAG CTT TGT TCC GGG ACC AAA TAC |
| JγP1/JγP2 | JOE-GAA GTT ACT ATG AGC T/CTA GTC CCT T |
FAM, JOE and NED are fluorochromes attached to the 5′ end of the Jγ primers. While the Jγ primers are the same as in DGGE, the Vγ primers are different than those in the DGGE assay. Vγ2 has sequence homology with gene segments Vγ1,4–8, and together with Vγ3 amplifies all the Group I Vγ1–8 segments. A specific Vγ3 primer was needed to gain equivalency to the DGGE protocol. 14
Results
We identified a total of 98 TCRγGRs among the 60 cases analyzed, as many cases had biallelic or more rearrangements, a characteristic of TCRγGR. 4, 5, 14, 16 Twenty-five cases had a single clonal rearrangement, 32 cases had two clonal rearrangements, and three cases had three clonal rearrangements. The cases with three rearrangements, which may represent the development of subclones, included two peripheral T-cell lymphomas and a large granular lymphocyte proliferation/leukemia. All cases included had concordant results on the number of TCRγ rearrangements between DGGE and CE.
Analysis of Individual TCRγ Rearrangements
Based on a total of 98 gene rearrangements, we first analyzed the number of individual rearrangements involving each Vγ and Jγ segment (Table 3) . As expected from previous TCRγGR studies, 6, 17 the Group I Vγ1–8 and Jγ1/2 genes were the most commonly rearranged segments. For the Vγ segments, the Vγ1–8 segments were involved in 55 (56%) individual rearrangements, the Vγ9 in 22 (23%) rearrangements, the Vγ10 in 17 (17%) rearrangements and the Vγ11 in 4 (4%) rearrangements. For the Jγ segments, most rearrangements occurred in the Jγ1/2 segments (81 of 98, 83%), followed by the JγP-1/2 segments (14 of 98, 14%), and the JγP segment (3 of 98, 3%).
Table 3.
Number of Individual Rearrangements for Each Possible Combination of Joining Region and Variable Region Primers, Based on a Total of 98 Individual Rearrangements
| Vγ1–8 | Vγ9 | Vγ10 | Vγ11 | Total | |
|---|---|---|---|---|---|
| Jγ1/2 | 46 (47%) | 17 (17%) | 15 (15%) | 3 (3%) | 81 (83%) |
| JγP1/2 | 8 (8%) | 3 (3%) | 2 (2%) | 1 (1%) | 14 (14%) |
| JγP | 1 (1%) | 2 (2%) | 0 (0%) | 0 (0%) | 3 (3%) |
| Total | 55 (56%) | 22 (23%) | 17 (17%) | 4 (4%) | 98 (100%) |
We then determined the number of individual rearrangements for each combination of Vγ and Jγ primers used for analysis. The results of the Vγ and Jγ segment pairs are listed in Table 3 . As expected from previous studies, Vγ1–8 and Jγ1/2 was the most frequently involved segment pair. However, slightly less than half (47%) of all individual TCRγGR involved the Vγ1–8 and Jγ1/2 combination, which supports the conclusion that using only these two primers is insufficient for clinical testing purposes. The data in Table 3 also demonstrates that rearrangements involving other Vγ and Jγ segments are not uncommon, although rearrangements involving either the rare Vγ11 and JγP segments constitute only 7% percent of the total.
Analysis of Case Positivity Detection Rates with Various Primer Sets
To further investigate the efficacy of different primer sets, we determined the case positivity rate for detecting at least one rearrangement (Table 4) . Case positivity, or qualitative sensitivity, is defined as the percentage of cases detected with TCRγGR using a specific set of primers. We first analyzed the case positivity rate if only the Vγ1–8 and Jγ1/2 primers were used, and found only a 67% case positivity detection rate. The sensitivity rate of 61.4% reported by Arber et al 7 for laboratories that only use the Vγ1–8 and Jγ1/2 primer pair is consistent with our TCRγGR distribution data. Distribution data for the segments used in the TCRγGR was not analyzed in the multi-center study. 7 Even if all of the Jγ primers were used with the Vγ1–8 primers, the detection rate only increases to 75% (data not shown), which demonstrates the requirement for using Vγ9, Vγ10, and Vγ11 primers.
Table 4.
Case Positivity Results for Different Primer Sets, Based on 60 Cases Selected with Previously Identified TCRγGR
| Primer set | Positive cases |
|---|---|
| Vγ1–8, Jγ1/2 | 40 (67%) |
| Vγ1–8, Vγ9, Jγ1/2 | 44 (73%) |
| Vγ1–8, Vγ9, Vγ10, Jγ1/2 | 48 (80%) |
| Vγ1–8, Vγ9, Vγ10, Vγ11, Jγ1/2 | 49 (82%) |
| Vγ1–8, Vγ9, Vγ10, Vγ11, Jγ1/2, JPγ1/2 | 58 (97%) |
| Vγ1–8, Vγ9, Vγ10, Vγ11, Jγ1/2, JPγ1/2, JγP | 60 (100%) |
We then determined the detection rate if Vγ primers were added and coupled with a single Jγ1/2 primer. When Vγ9, Vγ10, and Vγ11 primers are cumulatively added to the primer set, the detection rate increases to 73%, 80%, and 82%, respectively. When the entire Vγ primer set covering the Vγ segments (1–11) is used and as additional Jγ primers are added to Jγ1/2, the detection rate increases to 97% (JγP1/2) and 100% (JγP1/2 and JγP). Since only cases with a known TCRγGR were selected in this study, the detection rate with the complete Vγ and Jγ primer set is by definition 100%.
Discussion
Our data demonstrate that TCRγGR protocols that use only primers to the Vγ1–8 and Jγ1/2 gene segments are insufficient for diagnostic testing purposes. In comparison to previous studies that reported TCRγ gene segment distribution data, 4, 6 our larger study of 60 T-cell lymphoproliferative disorders shows a lower overall incidence of Vγ1–8 rearrangements and a higher incidence of Vγ9 and Vγ10 rearrangements. Our Jγ segment data are similar to these previous studies, and together these studies show a very small percentage of rearrangements involve either the Vγ11 or JγP segment. This is the first work that demonstrates the case detection results one can expect with different combinations of TCRγ primers. In addition, we provide the molecular basis that explains why the presence of biallelic TCRγGR provides partial success for protocols with limited primer sets.
If the Vγ and Jγ segments are determined for all of the rearrangements, less than half (47%) of individual rearrangements involved the Vγ1–8 and Jγ1/2 segments. The reason why sensitivity rates for TCRγGR assays which use only a single Vγ1–8 and Jγ1/2 primer set are higher than the frequency of individual gene segments used is due to the frequent occurrence of biallelic rearrangements in T-cell lymphoproliferative disorders. In our study, 53% of cases exhibited biallelic rearrangements and 5% of cases had three rearrangements. Some studies have found higher incidences of biallelic rearrangements. 16, 18 Laboratories that use limited primer sets are actually achieving higher detection rates because biallelic TCRγGR occur, often with one rearrangement involving the Vγ1–8 or Jγ1/2 segments. Among the 33 cases with biallelic or more rearrangements, nearly all (30 of 33, 91%) had a Vγ1–8 and Jγ1/2 in one of the rearrangements. However, cases with a monoallelic and rare biallelic TCRγGR involving the other gene segments will be missed. In a previous study by Födinger et al, 6 eight of 30 (27%) patient samples did not have a TCRγGR involving the Vγ1–8 or Jγ1/2 gene segments. Our study also demonstrates a high false negative (33%) rate would occur if only these two primers are used. Given the importance of identifying a clonal T-cell population in establishing a diagnosis of a T-cell lymphoproliferative disorder, a false negative result could potentially lead to an erroneous diagnosis.
The detection rates for different primer sets in the literature is difficult to evaluate, since case selection methods are variable as is the gold standard used to compare results. Using a single Vγ1–8 and Jγ1/2 primer set, Gutzmer et al 19 found a 59 to 72% detection rate among 22 patients with cutaneous T-cell lymphoma. The detection rate varied due to the different protocols that were used, including using a thermocycler versus a LightCycler for amplification and polyacrylamide gel electrophoresis versus melting curve analysis for detection. In a study by Dippel et al, 20 which included 21 patients with advanced stage cutaneous T-cell lymphoma, a 76% sensitivity rate was reported using primers to the Vγ1–8, Jγ1/2, and JγP1/2 gene segments.
In comparison, Lamberson et al 21 found a 90% sensitivity rate using multiple primer sets in three separate multiplex reactions. The findings by Luo et al 22 and Vega et al 16 also support the trend of a higher detection when complete primer sets are used. They found a 92% and 98% sensitivity rate respectively, in T-cell lymphomas using a large primer set directed at all Vγ and Jγ segments. Since 5 to 10% of T-cell lymphomas will lack a detectable TCRγGR, 4 these are excellent results. Our data and the results of these studies emphasize the need for complete primer sets for an optimal detection rate. Many protocols have been described with complete Vγ and Jγ primer sets 5, 14, 16, 17, 22, 23, 24, 25, 26, 27 that laboratories could choose in their labs. However, since some T-cell lymphomas lack TCRγGR, the use of a TCR-β assay is recommended after a negative TCRγ assay, when lymphoma is still suspected.
Given the literature findings, it is surprising that many laboratories continue to use only a single primer set or limited primer sets. Perhaps one concern may be attributed to a previous report of potential false positive results associated with Vγ9 and JγP primers. 28 TCRγGR that involve the less frequently involved Vγ and Jγ segments should always be interpreted with caution, particularly if the PCR reactions are performed in separate tubes. Amplification with all primers in a single tube helps avoid false positive results with rare Vγ or Jγ segments. If only a small number of normal cells with a particular sized segment are amplified, a true polyclonal distribution may not be apparent and faint bands or small spikes could potentially be interpreted as positive. Combining all primers together in capillary electrophoresis helps in defining the polyclonal background and the significance of small peaks. Information concerning the number of T cells present within the specimen analyzed should be taken into account. In addition, duplicate assays help to identify reproducible peaks and eliminate false positive results.
It is critical to compare the peak intensity with the control samples with a known percentage of clonal cells when making a determination of a clonal population. Capillary electrophoresis is our preferred method due to the decreased turnaround time and labor costs. Using the capillary electrophoresis technique, only well-defined spikes with a peak height that is greater than twice that of the polyclonal background peaks are presently interpreted as positive in our laboratory, as compared to a 2% positive control. 14 Lee et al 29 have described the presence of pseudo-spikes in histologically benign lymphoid tissues, although none of these samples had a maximal peak to polyclonal background ratio that exceeded 1.37 and most were below 1.0. In our experience, we have not identified a significant false positive rate associated with these primers when strict criteria for a true clonal population are used. Similarly, in a comparative study of 21 laboratories by Arber et al, 7 there was no significant difference in false positive results with any Tγ primer group.
In conclusion, pathologists should reassess TCRγ protocols in their laboratories or reference laboratories to ensure that multiple primer sets are used, as many laboratories do not report the primer sets used in the procedure part of their report. Clearly, it is unacceptable to have a false negative rate of up to 33% when protocols can be modified at minimal cost to include primers to the uncommon Tγ gene segments.
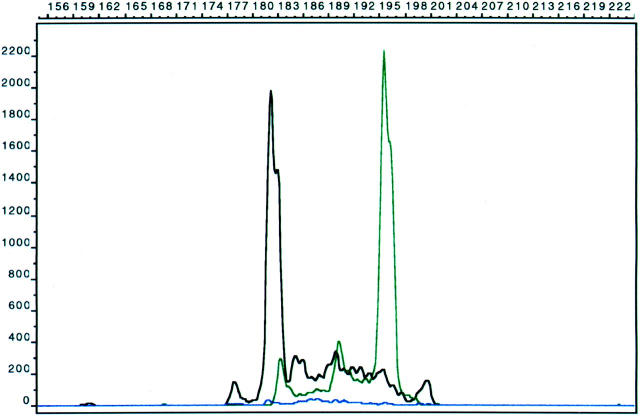
Figure 2.
Capillary electrophoresis method showing biallelic clonal rearrangements with the Jγ1/2 (black) and JγP1/2 (green) primers in a case of peripheral T-cell lymphoma. The heights of the clonal peaks are greater than two times the polyclonal background. x axis, nucleotide length; y axis, intensity of signal.
Acknowledgments
We thank Katrina Matthews for preparing the manuscript.
Address reprint requests to Timothy C. Greiner, M.D., Department of Pathology and Microbiology, University of Nebraska Medical Center, 983135 Nebraska Medical Center, Omaha, NE 68198-3135. E-mail: tgreiner@unmc.edu.
Footnotes
Supported by USPHS grant CA 36727.
Presented at the 2002 United States and Canadian Academy of Pathology Meeting.
References
- 1.Huck S, Lefranc MP: Rearrangements to the JP1, JP, and JP2 segments in the human T-cell rearranging γ gene (TRG γ) locus. FEBS Lett 1987, 224:291-296 [DOI] [PubMed] [Google Scholar]
- 2.LeFranc MP, Forster A, Baer R, Stinson MA, Rabbitts TH: Diversity and rearrangement of the human T cell rearranging γ genes: nine germ-line variable genes belonging to two subgroups. Cell 1986, 45:237-246 [DOI] [PubMed] [Google Scholar]
- 3.Lefranc MP, Forster A, Rabbitts TH: Rearrangement of two distinct T-cell γ-chain variable-region genes in human DNA. Nature 1986, 319:420-422 [DOI] [PubMed] [Google Scholar]
- 4.Theodorou I, Raphael M, Bigorgne C, Fourcade C, Lahet C, Cochet G, Lefranc MP, Gaulard P, Farcet JP: Recombination pattern of the TCR γ locus in human peripheral T-cell lymphomas. J Pathol 1994, 174:233-242 [DOI] [PubMed] [Google Scholar]
- 5.Greiner TC, Raffeld M, Lutz C, Dick F, Jaffe ES: Analysis of T-cell receptor-γ gene rearrangements by denaturing gradient gel electrophoresis of GC-clamped polymerase chain reaction products: correlation with tumor-specific sequences. Am J Pathol 1995, 146:46-55 [PMC free article] [PubMed] [Google Scholar]
- 6.Födinger M, Buchmayer H, Schwarzinger I, Simonitsch I, Winkler K, Jager U, Knobler R, Mannhalter C: Multiplex PCR for rapid detection of T-cell receptor-γ chain gene rearrangements in patients with lymphoproliferative diseases. Br J Haematol 1996, 94:136-139 [DOI] [PubMed] [Google Scholar]
- 7.Arber DA, Braziel RM, Bagg A, Bijwaard KE: Evaluation of T-cell receptor testing in lymphoid neoplasms: results of a multi-center study of 29 extracted DNA and paraffin-embedded samples. J Mol Diagn 2001, 3:133-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaffe ES, Harris NL, Stein H, Vardiman J: Pathology and genetics of tumours of haematopoietic and lymphoid tissues. World Health Organization Classifications of Tumors. Kleihues P Sobin LH eds. 2001:189-230 IARC Press Lyon, France
- 9.Heller MJ, Burgart LJ, TenEyck CJ, Anderson ME, Greiner TC, Robinson RA: An efficient method for the extraction of DNA from formalin-fixed, paraffin-embedded tissue by sonication. Biotechniques 1991, 11:372-374, 376377 [PubMed] [Google Scholar]
- 10.Wan JH, Sykes PJ, Orell SR, Morley AA: Rapid method for detecting monoclonality in B-cell lymphoma in lymph node aspirates using the polymerase chain reaction. J Clin Pathol 1992, 45:420-423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourguin A, Tung R, Galili N, Sklar J: Rapid, nonradioactive detection of clonal T-cell receptor gene rearrangements in lymphoid neoplasms. Proc Natl Acad Sci USA 1990, 87:8536-8540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheffield VC, Cox DR, Lerman LS, Myers RM: Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci USA 1989, 86:232-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA: Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 1988, 239:487-491 [DOI] [PubMed] [Google Scholar]
- 14.Greiner TC, Rubocki RJ: Effectiveness of capillary electrophoresis using fluorescent-labeled primers in detecting T-cell receptor γ gene rearrangements. J Mol Diagn 2002, 4:137-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sprouse JT, Werling R, Hanke D, Lakey C, McDonnel L, Wood BL, Sabath DE: T-cell clonality determination using polymerase chain reaction (PCR) amplification of the T-cell receptor γ-chain gene and capillary electrophoresis of fluorescently labeled PCR products. Am J Clin Pathol 2000, 113:838-850 [DOI] [PubMed] [Google Scholar]
- 16.Vega F, Medeiros LJ, Jones D, Abruzzo LV, Lai R, Manning J, Dunmire V, Luthra R: A novel four-color PCR assay to assess T-cell receptor γ gene rearrangements in lymphoproliferative lesions. Am J Clin Pathol 2001, 116:17-24 [DOI] [PubMed] [Google Scholar]
- 17.Theodorou I, Bigorgne C, Delfau MH, Lahet C, Cochet G, Vidaud M, Raphael M, Gaulard P, Farcet JP: VJ rearrangements of the TCR γ locus in peripheral T-cell lymphomas: analysis by polymerase chain reaction and denaturing gradient gel electrophoresis. J Pathol 1996, 178:303-310 [DOI] [PubMed] [Google Scholar]
- 18.Zhu D, Kadin ME, Samoszuk M: Detection of clonal T-cell receptor-γ gene rearrangement by PCR/temporal temperature gradient gel electrophoresis. Am J Clin Pathol 2001, 116:527-534 [DOI] [PubMed] [Google Scholar]
- 19.Gutzmer R, Mommert S, Kiehl P, Wittmann M, Kapp A, Werfel T: Detection of clonal T-cell receptor γ gene rearrangements in cutaneous T-cell lymphoma by LightCycler-polymerase chain reaction. J Invest Dermatol 2001, 116:926-932 [DOI] [PubMed] [Google Scholar]
- 20.Dippel E, Assaf C, Hummel M, Schrag HJ, Stein H, Goerdt S, Orfanos CE: Clonal T-cell receptor γ-chain gene rearrangement by PCR-based GeneScan analysis in advanced cutaneous T-cell lymphoma: a critical evaluation. J Pathol 1999, 188:146-154 [DOI] [PubMed] [Google Scholar]
- 21.Lamberson C, Hutchison RE, Shrimpton AE: A PCR assay for detecting clonal rearrangement of the TCR-γ gene. Mol Diagn 2001, 6:117-124 [DOI] [PubMed] [Google Scholar]
- 22.Luo V, Lessin SR, Wilson RB, Rennert H, Tozer C, Benoit B, Leonard DG: Detection of clonal T-cell receptor γ gene rearrangements using fluorescent-based PCR and automated high-resolution capillary electrophoresis. Mol Diagn 2001, 6:169-179 [DOI] [PubMed] [Google Scholar]
- 23.Goudie RB, Karim SN, Mills K, Alcorn M, Lee FD: A sensitive method of screening for dominant T-cell clones by amplification of T-cell γ gene rearrangements with the polymerase chain reaction. J Pathol 1990, 162:191-196 [DOI] [PubMed] [Google Scholar]
- 24.Trainor KJ, Brisco MJ, Wan JH, Neoh S, Grist S, Morley AA: Gene rearrangement in B- and T-lymphoproliferative disease detected by the polymerase chain reaction. Blood 1991, 78:192-196 [PubMed] [Google Scholar]
- 25.Lorenzen J, Jux G, Zhao-Hohn M, Klockner A, Fischer R, Hansmann ML: Detection of T-cell clonality in paraffin-embedded tissues. Diagn Mol Pathol 1994, 3:93-99 [DOI] [PubMed] [Google Scholar]
- 26.Delabesse E, Burtin ML, Millien C, Madonik A, Arnulf B, Beldjord K, Valensi F, Macintyre EA: Rapid, multifluorescent TCRG Vγ and Jγ typing: application to T-cell acute lymphoblastic leukemia and to the detection of minor clonal populations. Leukemia 2000, 14:1143-1152 [DOI] [PubMed] [Google Scholar]
- 27.Meier VS, Rufle A, Gudat F: Simultaneous evaluation of T- and B-cell clonality, t(11;14) and t(14;18), in a single reaction by a four-color multiplex polymerase chain reaction assay and automated high-resolution fragment analysis: a method for the rapid molecular diagnosis of lymphoproliferative disorders applicable to fresh frozen and formalin-fixed, paraffin-embedded tissues, blood, and bone marrow aspirates. Am J Pathol 2001, 159:2031-2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delfau MH, Hance AJ, Lecossier D, Vilmer E, Grandchamp B: Restricted diversity of V γ 9-JP rearrangements in unstimulated human γ/δ T lymphocytes. Eur J Immunol 1992, 22:2437-2443 [DOI] [PubMed] [Google Scholar]
- 29.Lee SC, Berg KD, Racke FK, Griffin CA, Eshleman JR: Pseudo-spikes are common in histologically benign lymphoid tissues. J Mol Diagn 2000, 2:145-152 [DOI] [PMC free article] [PubMed] [Google Scholar]