Abstract
Rett syndrome is a neurodevelopmental disorder that affects females almost exclusively, and in which eight common point mutations on the X-linked MeCP2 gene are knows to cause over 70% of mutation-positive cases. We explored the use of a novel platform to detect the eight common mutations in Rett syndrome patients to expedite and simplify the process of identification of known genotypes. The Nanogen workstation consists of a two-color assay based on electric hybridization and thermal discrimination, all performed on an electronically active NanoChip. This genotyping platform was tested on 362 samples of a pre-determined genotype, which had been previously identified by a combination of DHPLC (denaturing high performance liquid chromatography) and direct sequencing. This genotyping technique proved to be rapid, facile, and displayed a specificity of 100% with 3% ambiguity. In addition, we present consecutive testing of seven mutations on a single pad of the NanoChip. This was accomplished by tagging down two amplimers together and serially hybridizing for seven different loci, allowing us to genotype samples for seven of the eight common Rett mutations on a single pad. This novel method displayed the same level of specificity and accuracy as the single amplimer reactions, and proved to be faster and more economical.
Rett syndrome is the second most common form of neurodevelopmental disorder in females, behind Down syndrome. 1 Approximately 1:10,000 to 1:22,000 females are affected, with fewer than 20 suspected cases in males. 2, 3 Girls with Rett syndrome typically display normal early development during the neonatal period but present at 6 to 12 months of age with developmental regression and slowed head growth. 4 Regression is associated with a loss of communication and hand movement skills. Stereotypic hand washing or wringing behaviors are typical, as are difficulties with breathing, seizures, and gait apraxia. The clinical course of Rett syndrome usually stabilizes by 5 years of age. 5, 6
The gene that causes Rett syndrome has been identified as the X-linked methyl CpG binding protein 2 (MeCP2) gene on the X chromosome, which is a key mediator in the global transcriptional silencing of genes. 7, 8, 9 Heterozygous MeCP2 mutations are present in 65 to 80% of girls that are diagnosed with classical Rett and in 30 to 50% of atypical cases. 10, 11 The mutations in MeCP2 most likely result in a loss of function of the protein, with downstream gene transcription abnormalities due to lack of methylated DNA regulation. 12, 13
Nearly all Rett cases are caused by new mutations of the MeCP2 gene, often in the father’s germ line. 14 However, studies have shown that there are eight mutations that frequently reoccur (C316T, C397T, C455T, C502T, C763T, C808T, C880T, and C916T) and together these account for over 70% of the mutation-positive cases. 15 Therefore, we sought to develop a rapid, accurate, and cost-effective method to genotype these eight common point mutations as a first screen for patients who are suffering from symptoms common to Rett syndrome.
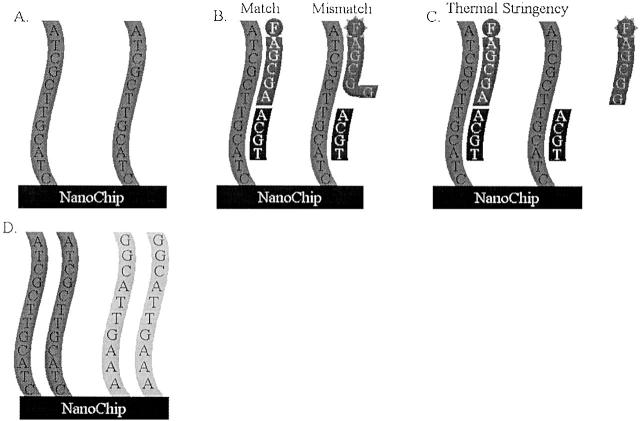
We examined a two-color assay based on thermal discrimination and electronic hybridization as a method to genotype patient samples for these eight point mutations. The NanoChip Molecular Biology Workstation (developed by Nanogen, Inc., San Diego, CA) relies on the binding of biotinylated PCR products of the genomic region of interest from a given sample to a strepavidin-coated matrix of electronic pads 16, 17, 18 (Figure 1) . The NanoChip system has been used to genotype point mutations involved in a number of different disorders, including thrombophilia, hypertension, hemochromatosis, and some forms of cancer (Nanogen Technical Bulletin 143016.1). The 100 distinct streptavidin-coated electronic pads, occupying a 2 mm2 area, allow for concentration of the biotinylated PCR products. After attachment of the PCR amplimers, fluorescent oligonucleotide probes specific to the wild-type (labeled with Cy-3) and mutant (labeled with Cy-5) base are added to the PCR product-loaded pads. In addition, a 27 to 33-mer stabilizer oligo is added to the pad simultaneously to bind to the complementary area directly 5′ of the mutation. The stabilizer allows for base-stacking energy between itself, the probe, and the amplimer. Thermal stringency then strips an improperly binding probe while leaving the correct one bound. Finally, a fluorescence reading is taken of the pad to genotype the sample.
Figure 1.
Schematic diagram of the Nanogen differential hybridization process. A: Attachment of a single set of PCR amplimers on one pad of the NanoChip. B: Fluorescent oligonucleotide probes specific to the wild-type (labeled with Cy-3) and mutant (labeled with Cy-5) base are added to the PCR product-loaded pad. In this case, one probe is a match, the other is mismatched for the base in question. In addition, a 27 to 33-mer stabilizer oligo is added to the pad simultaneously to bind to the complementary area directly 5′ of the mutation. C: Thermal stringency then strips an improperly binding probe while leaving the correct one bound. D: Multiplex binding of two amplimers to the same pad of the NanoChip. The subsequent probing is done serially with distinct probes and stabilizers for one amplimer, followed by thermal stringency and washing, and then the next amplimer is tested.
In this paper, we measure the sensitivity and accuracy of this genotyping method with analysis of 368 patient samples previously identified by a combination of DHPLC and direct sequencing. In addition, we discuss an optimization of a novel multiplex genotyping assay with the Nanogen system, which allowed us to place two separate amplimers on the same pad of the NanoChip. After multiplexing seven heterozygous samples onto seven pads, we serially genotyped them in duplicate for each of seven mutations (a total of 98 genotypings).
Materials and Methods
Sample DNA
Blood samples were sent to Children’s National Medical Center in Washington, D.C. as a component of a previous research study for Rett syndrome. These procedures were reviewed and approved by the Internal Review Board of Children’s National Medical Center (Children’s National Medical Center, IRB no. 2405). Blood was collected at referring sights in Na2EDTA from subjects. Genomic DNA was isolated from blood leukocytes using the Puregene DNA Isolation kit (Gentra Systems). DNA was not quantitated before use, as the subsequent PCR step provided more than enough DNA for tagging down to the NanoChip. All patients had been preciously studied for mutations of the MeCP2 gene using a combination of DHPLC (Transgenomic WAVE) and direct sequencing. The 368 patient samples used were specifically selected for each test so that both heterozygous and homozygous wild-type genotypes were assayed. Unique samples were tested for each mutation locus. For C316T testing: 5 samples were heterozygous (het) for C316T, 20 samples were het at other loci, and 21 were homozygous wild-type (wt); C397T testing: 6 het for C397T, 22 het at other loci, and 18 wt; C455T testing: 2 het for C455T, 15 het for other loci, and 29 wt; C502T testing: 11 het for C502T, 19 het for other loci, and 16 wt; C736T testing: 14 het for C736T, 19 het for other loci, and 13 wt; C808T testing: 10 het for C808T, 11 het for other loci, and 25 wt; C880T testing: 8 het for C880T, 16 het for other loci, and 22 wt; and for C916T testing: 8 het for C916T, 17 het for other loci, and 22 wt.
Loading
PCR
Three separate PCR amplimers, designed to encompass the eight common mutations, were made for each sample. Novel forward and reverse primer sequences (Table 1) were used. Each of the forward primers was labeled on the 5′ end with biotin to facilitate binding of the product to the NanoChip. All samples had PCR reactions done in duplicate.
Table 1.
PCR Primers for the Amplification of the SNPs Located in the MeCP2 Gene
| Mutations | Forward | Reverse |
|---|---|---|
| C316T | GCCCGTGCAGCCATCAGCCC | CCCGACCCACCCTGGGCACAT |
| C397T, C455T, C502T | TTGTCAGAGCGTTGTCACCA | GCTTCCCAGGACTTTTCTCCA |
| C763T, C808T, C880T, C916T | AAGATGCCTTTTCAAACTTCG | CCCAGGGCTCTTACAGGTCT |
The PCR conditions are located in the Materials and Methods section of the paper.
Purification and Preparation
After PCR, samples were de-salted in MultiScreen-PCR 96-well plates (Millipore). For the multiplex reaction both the amplimer for C397T, C455T, and C502T and the amplimer for C763T, C808T, C880T, and C916T were placed in the same well for desalting. The conductivity of the samples was checked with a Twin Cond conductivity meter (Horiba, Japan) before proceeding, to make sure it was <50 μs/cm. Histidine was added to each sample for a final concentration of 50 mmol/L. Sixty μl of each sample was then placed in a 96-well Nunc plate (Nalgene) along with two control samples of 50 mmol/L histidine, a solution of 0.1N NaOH and another solution of 0.3N NaOH. The entire plate was then covered with AlumaSeal (Research Products International Corp.).
Loader Protocol
After the plate was placed in the Nanogen Loader along with a new NanoChip, the loader was primed with 50 mmol/L histidine and water, and the loading protocol was run as follows: 1) the entire chip was bathed in 0.3 mol/L NaOH for 5 minutes; 2) the two control samples of 50 mmol/L histidine were bound to the chip at the appropriate pads in a capture format, meaning each pad was subsequently charged with 2V for 60 seconds; 3) the samples in 50 mmol/L histidine were bound to the chip in a target/amplimer format, meaning that as each sample was loaded, its target pad was charged with 2V for 120 seconds; 4) the samples were denatured by another passive wash, this time with 0.1 mol/L NaOH for 5 minutes.
Reporting
Reporters and Stabilizers
The design of the reporters (Table 2) was such that the 3′ base of each would overlap the locus of the point mutation of interest. For each mutation, both a wild-type and a mutant reporter were made, each possessing the appropriate 3′ base. In addition, each reporter was fluorescently labeled on the 5′ end; wild-type reporters with cy3 and mutant reporters with cy5. The stabilizers were constructed so that the 5′ end annealed against the 3′ end of the reporter. The reporter and stabilizer for the point mutation C502T were constructed so they bound in reverse of the normal positions. The 5′ (rather than 3′) bases of the wild-type and mutant reporters overlapped at the locus of the point mutation, while the 3′ end of the stabilizer annealed directly 5′ of the reporter. All of the reporters and stabilizers, listed 5′ to 3′ are shown in Table 2 .
Table 2.
Stabilizers, Reporters, and Analysis Conditions Used to Genotype MeCP2 SNPs
| Mutation | Stabilizer | WT reporter | Mutant reporter | Contrast temp, °C | Washes |
|---|---|---|---|---|---|
| C316T | GGAAGCTTAAGCAAAGGAAATCTGGCC | GGCTGGACAC | GGCTGGACAT | 38 | 3 |
| C397T | AAAGGCTTTTCCCTGGGGACTGT | CACTTTAGAGCG | CACTTTAGAGCA | 37 | 4 |
| C455T | CCGGGAGGGGCTCCCTCTCCCAGTTACC | GTGAAGTCAAAA | ATGAAGTCAAAA | 39 | 4 |
| C502T | CCGGGAGGGGCTCCCTCTCCCAGTTACC | TCTGCTCTCG | TTCTGCTCTCA | 38 | 3 |
| C736T | CTTCCTGCCGGGGCGTTTGATCACCATGACCTGGG | CTCAGCTTTTCG | CTCAGCTTTTCA | 41 | 2 |
| C808T | GCCCCGTTTCTTGGGAATGGCCTGAGGGTC | CCGGCTTTCG | CCGGCTTTCA | 36 | 4 |
| C880T | GATAGAAGACTCCTTCACGGCTTTCTTTTTGGCCT | CTGCACAGATCG | CTGCACAGATCA | 39 | 4 |
| C916T | CTTCTTGATGGGGAGTACGGTCTCCTGCACAGATC | GGGTCTTGCG | GGGTCTTGCA | 34 | 3 |
Evaluation
After loading was completed, the chip was rinsed twice with High Salt buffer (50 mmol/L sodium phosphate and 500 mmol/L sodium chloride, pH 7.4). For a reading, the chip was bathed for 3 minutes with 100 μl of a cocktail made with 100 pM of wild-type reporter, 100 pM of mutant reporter, and 100 pM of stabilizer in High Salt buffer (50 mmol/L sodium phosphate, 500 mmol/L sodium chloride, pH 7.4). The cocktail was then removed and the chip was again rinsed twice with High Salt buffer. After rinsing, the chip was placed in the reader and the temperature was set to 24°C before alignment and focus. For each probe set an optimum discrimination temperature and number of washes was empirically determined on a small subset of samples before testing. In each case, the chip was raised to the discrimination temperature and washed the appropriate number of times, then scanned at 24°C for both cy3 and cy5 fluorescence. Between each separate point mutation test, the chip was removed from the reader, rinsed twice with dH2O, bathed in 0.1 mol/L NaOH for 5 minutes, rinsed three more times with dH2O and then rinsed five times with High Salt buffer.
Analysis
Initial identification of mutations in the MeCP2 gene in all of the samples had been accomplished by a combination of DHPLC and sequencing as described previously. 15 Briefly, 10 overlapping PCR fragments were amplified and then screened for base changes using a Transgenomic Wave DNA Fragment Analysis system (San Jose, CA) (note that female Rett patients are heterozygous for MeCP2 mutations). Sequencing of heteroduplexes was done using a Thermo Sequenase cycle sequencing kit (Amersham, Arlington Heights, IL) and the reaction products were run on the LiCor automatic sequencer (Gene Reader 4200). To verify that current results recapitulated the previous data, the Nanogen software performed a full analysis on the generated data. Of the heterozygous samples tested, one was randomly chosen to serve as a control for which the laser gain was adjusted by the software so that its cy3 to cy5 fluorescence had a ratio of 1:1. In addition, two-capture format histidine samples were read as a baseline to determine the level of noise inherent in the assay.
Results
Single Amplimer, Multiple Mutations Assay
The Nanogen system relies on the binding of biotin-labeled oligos to the strep-avidin layer of the NanoChip (Figure 1 A–C) . The electric pads embedded in the chip’s surface allow for concentration of the sample of interest in a specific location before binding. This allows a small total amount of sample to reach a very high concentration on the 100 pads within the 2 mm2 area, and thus produce high intensity signals at excellent signal to noise ratios. 19 After the binding step, two different fluorescently labeled (cy3 and cy5) reporter oligos are hybridized to the sample. For detection of point mutations, the two reporters differ at the base in question, one specific to wild-type and the other to the point mutation. When thermal stringency is subsequently applied, the mismatched reporter oligo denatures preferentially from the target, and the signal of the other reporter alone can be detected. In the case of a heterozygous sample, both reporters resist the thermal stringency on the different copies and, consequently, both signals can be detected.
To interpret the two-color signal produced by the binding of the reporters to the sample, genotyping was based on normalized signals rather than raw fluorescence scores. As a baseline, two pads were loaded without sample and fluorescence was measured after binding of reporters. As a precaution, no signal strength was accepted unless it was at least five times that of the baseline signal, which was considered “noise.” In addition, a known control heterozygous sample was loaded and then reported to account for any difference in binding of the wild-type versus mutant reporters. The laser gain was adjusted so that the heterozygous control gives a ratio of 1:1 for cy3 and cy5 fluorescence signal detected.
For each of the eight loci under investigation several different categories of patient sample were used to test the efficacy of the Nanogen system. Previously, all samples had been genotyped using a combination of DHPLC and sequencing to determine the presence or absence of mutation in the exons and intron/exon boarder of the MeCP2 gene. Samples were then chosen non-randomly so that for each locus tested there would be those which were heterozygous mutant for the locus in question, those that were heterozygous for another locus (including polymorphisms), and samples that were homozygous wild-type.
In our first iteration of our assay, individual amplimers were tagged down to each pad. Amplimer 1 was tested for one mutation, amplimer 2 for three mutations and amplimer 3 for four separate point mutations (Table 1) . To test multiple mutations on the same amplimer/NanoChip pad, a single genotype was tested and the fluorescent probes were subsequently removed. Only after removal of the initial probes was another genotype tested by application of a different set of fluorescent probes. This process continued until all of the loci covered by an amplimer were assayed.
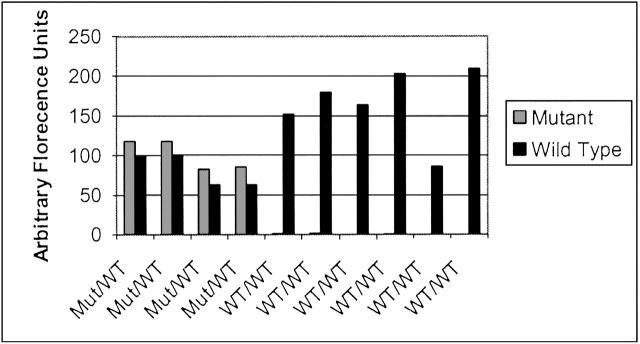
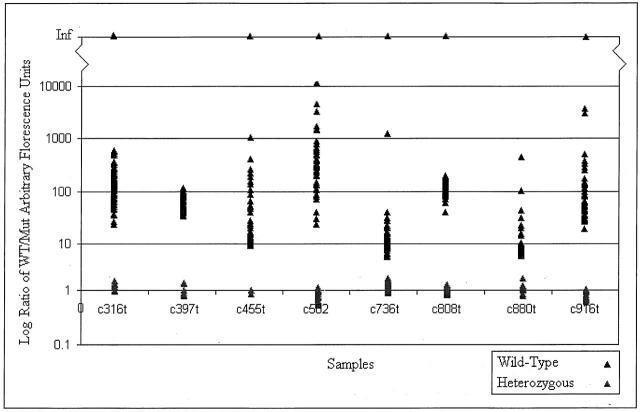
After the fluorescence of both wild-type and mutant probes was taken and the results analyzed, calls were made based on the normalized ratios. Figure 2 shows several samples that are indicative of the results produced for wild-type and mutant signals. Some of the control wild-type samples gave no mutant signal, and so the calculation of a non-infinite ratio (one where mutant signal was not zero) was impossible and those samples were discarded for the purpose of statistical analysis (using a two-tailed, unequal variance t-test) though they are still represented graphically. The comparisons of average heterozygous signal to homozygous wild-type signal for each point mutation can be viewed in Table 3 . Overall, the average ratio of wild-type to mutant signal for those samples possessing a heterozygous genotype was 1.001 ± 0.1139, while the average for all of the samples that were wild-type for the locus in question was 160.96 ± 621.07 (P = 6.7 × 107). Sample ratios of signal data are shown in Figure 3 , demonstrating the clear differentiation between wild-type and heterozygous sample data. Twenty-five of the 736 genotypings gave low signal-to-noise ratio (3% ambiguity), requiring stripping and re-probing. Subsequently, all of the re-probed pads gave correct signal (100% sensitivity and accuracy). In addition, there was no detectable difference in the signal from homozygous wild-type samples and those samples that were heterozygous for a locus other than the one for which the probings were performed.
Figure 2.
Nanogen Reader results for four samples that were heterozygous for MeCP2 mutations and six samples that were homozygous wild-type. After loading onto NanoChips, samples were read by the Nanogen Reader. Arbitrary fluorescence units were measured for both wild-type (Cy-3) and mutant (Cy-5) signals in all samples.
Table 3.
Signal Intensities for the Analysis of MeCP2 Gene SNPs Using the Nanogen System
| Locus | Heterozygous signal | Homozygous wild-type signal |
|---|---|---|
| C316T | 1.09 ± 0.08 | 165 ± 131.5* |
| C397T | 0.988 ± 0.09 | 64.7 ± 19.5* |
| C455T | 0.96 ± 0.05 | 57.8 ± 147.8* |
| C502T | 0.89 ± 0.09 | 778 ± 177* |
| C736T | 1.09 ± 0.09 | 33.5 ± 162* |
| C808T | 0.996 ± 0.06 | 131 ± 35.7* |
| C880T | 1.04 ± 0.09 | 16.6 ± 3.91* |
| C916T | 0.92 ± 0.07 | 206 ± 612* |
Heterozygous signal vs. homozygous wild-type signal, p << 0.001.
Figure 3.
Nanogen system results for 368 tested samples. After detection of fluorescence, wild-type (Cy-3) and mutant (Cy-5) signals were normalized by a control heterozygous sample. The Nanogen software then used the ratio of the normalized data, shown above, to genotype the sample. The generated results were in agreement with those from previous DHPLC and sequencing.
Two Amplimer, Serial Mutation Analysis
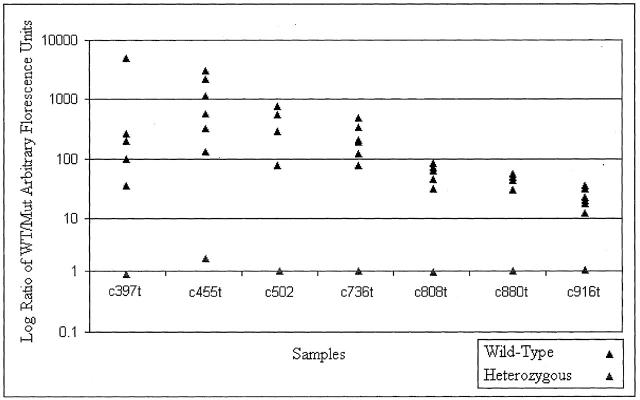
To investigate the optimization of the Nanogen system for analysis for genotyping eight common mutations in Rett syndrome, we tagged down two amplimers (amplimers 2,3; Table 1 ) onto a single pad of the NanoChip and genotyped seven point mutations (C397T, C455T, C502T, C763T, C808T, C880T, and C916T) serially on that pad (Figure 1D) . We analyzed seven individuals, each on with a point mutation at a separate locus. Each sample was probed in duplicate (replicate pads) for all seven mutations and the results are summarized in Figure 4 (a total of 98 genotypings). Across all loci, homozygous samples gave an average wild-type to mutant ratio of arbitrary fluorescence units of 368.3 ± 797.5, which was not significantly lower that the average signal given by assaying point mutations in the single amplimer/pad format (see above). The heterozygous samples for all point mutations gave an average ratio of arbitrary fluorescence units of 1.2 ± 0.63, which was not significantly different from the heterozygous fluorescence ratio given by single amplimers. Separate statistical analysis could not done for each locus individually, as was done the single amplimer reaction, due to the lack of more than one heterozygous data point per locus. As with the single amplimers, there was 100% specificity of calls. There was one call that could not be made due to a low signal-to-noise ratio (2% ambiguity), but subsequent stripping and re-probing led to a correct genotype (100% sensitivity, 100% accuracy). To our knowledge, this is the first report of serial genotyping with multiple products on a single pad of the Nanogen system, or in any other detection assay.
Figure 4.
Nanogen system results for testing of seven samples. For the samples, each point mutation was probed serially. After detection of fluorescence, calls were made in the same manner as non-multiplexed samples and probes were subsequently stripped, readying the samples for the next test. The generated results were in agreement with those from previous DHPLC and sequencing.
Discussion
Electronic hybridization and thermal stringency provides a viable alternative to DHPLC and sequencing. Though the combination of these more classic methods is still invaluable for the location and classification of novel mutations and single nucleotide polymorphisms, for genotyping in certain disorders, the Nanogen system has been shown to be both a swift and accurate surrogate. In a disease like Rett syndrome, where eight different point mutations (C316T, C397T, C455T, C502T, C763T, C808T, C880T, and C916T) in the MeCP2 gene are known to cause the majority of cases, an assay based on that information can be very quick and cost effective. Our results indicate that the Nanogen system is extremely sensitive for the detection of the common Rett syndrome mutations and the cost is significantly less than other mutation detection methods such as DHPLC or sequencing. We tested a total of 362 samples that had previously been screened in our laboratory by DHPLC and sequencing for mutations in the MECP2 gene. All samples heterozygous for the common mutations were accurately detected by the Nanogen system, with no false negatives. This system showed no false positives when samples representing homozygous wild-type or heterozygous for other mutations were tested. We found after one assay there was 100% specificity in mutation detection by the Nanogen system with a 3% ambiguity in calls. After re-testing of ambiguous samples, there was 100% specificity with 100% accuracy. The Nanogen system then proved to have 100% concordance with our precious screening methods of DHPLC and sequencing when taken together with a second NanoChip assay of the ambiguous samples. In our experience, it took only 4 person-hours to run the assay at a cost of $12 per full genotype (all eight mutations). In the future, the cost and time to run will drop due to increased use of the novel multiplexing reaction.
It was exciting to find that the Nanogen system gave a lack of false positives and negatives. Both the assay itself and the attending controls were designed precisely to reduce the chance of an incorrect result. The use of probes specific to either wild-type or mutant afforded a great deal of specificity. Additionally, the base stacking energy provided to the correct complementary probe by the stabilizer oligo meant that then subsequent thermal stringency preferentially removed the incorrect probe. This, in turn, allowed the two-color format of the Nanogen, using a ratio of wild-type to mutant signal, to control for both the innate sample variability and any differences in amplification and preparation of the samples being tested. To further reduce noise due to environment and chip irregularity, two pads were designated as a histidine-only baseline. In addition, one known heterozygous sample was designated as a control and used to normalize both wild-type and mutant signals. Perhaps the best insurance against erroneous calls was the stringent use of a signal-to-noise ratio threshold. If neither the wild-type nor the mutant signal of a sample was over five times the histidine baseline, the sample was considered to have an insufficient signal-to-noise ratio and, consequently, no call was made. Because acceptable signal strength was so high, there was always a clear delineation between homozygous wild-type and heterozygous samples. It should be noted that the wild-type samples showed a large SD in signal strength, both within and across given mutation tests, due to instability of ratios where denominators approach zero. This was not due to any instability within the detection system. While non-specific binding of the mutant and wild-type probes dictates the denominator, and thus the closeness to zero, this has no effect on the interpretation of the resulting data, as the signal-to-noise ratio is still excellent.
The NanoChip Molecular Biology Workstation presents many advantages as a method of identifying single nucleotide polymorphisms and point mutations. The Nanogen system incorporates a number of different control methods; the results have shown that the system has 100% specificity on the eight common Rett mutations with 3% ambiguity on a total of 736 calls. With further testing of ambiguous calls, the sensitivity and accuracy are 100%. The assay itself is inexpensive and further experimentation on multiplexing amplimers on the same chip pad should reduce the price. In addition, future automation of liquid handling and serial point mutation detection would greatly reduce the time currently required for manual handling of chips and fluidics. These factors and general ease of use make the Nanogen system a rapid, uncomplicated, and precise method of genotyping.
Acknowledgments
We thank Dr. Halleh Ahadian of Nanogen Inc., San Diego, for training us on the Nanogen Workstation and her designs for four of the assays, in addition to expert technical advice.
Address reprint requests to Eric P. Hoffman, Research Center for Genetic Medicine, Children’s National Medical Center, 111 Michigan Avenue N.W., Washington, D.C. 20010. E-mail: ehoffman@cnmc.org.
Footnotes
Supported by grants from the National Institutes of Health R01 NS40030 on Rett syndrome to E.P.H.; NCRR PCRC Genetics Core; and NICHD P30HD40677 Molecular Genetics Core.
References
- 1.Kozinetz CA, Skender ML, MacNaughton N, Almes MJ, Schultz RJ, Percy AK, Glaze DG: Epidemiology of Rett syndrome: a population-based registry. Pediatrics 1993, 91:445-450 [PubMed] [Google Scholar]
- 2.Ellaway C, Christodoulou J: Rett syndrome: clinical update and review of recent genetics advances. J Paediatr Child Health 1999, 35:419-426 [DOI] [PubMed] [Google Scholar]
- 3.Jan MM, Dooley JM, Gordon KE: Male Rett syndrome variant: application of diagnostic criteria. Pediatr Neurol 1999, 20:238-240 [DOI] [PubMed] [Google Scholar]
- 4.Naidu S: Rett Syndrome: a disorder affecting early brain growth. Ann Neurol 1997, 42:3-10 [DOI] [PubMed] [Google Scholar]
- 5.Hagberg B, Aicardi J, Dias K, Ramos O: A progressive syndrome of autism, dementia, ataxia, and loss of purposeful hand use in girls: Rett’s syndrome: report of 35 cases. Ann Neurol 1983, 14:471-479 [DOI] [PubMed] [Google Scholar]
- 6.Percy AK: Rett syndrome. Curr Opin Neurol 1995, 8:156-160 [DOI] [PubMed] [Google Scholar]
- 7.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY: Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 1999, 23:185-188 [DOI] [PubMed] [Google Scholar]
- 8.Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A: Purification, sequence, and cellular localization of a novel chromosomal protein that bins to methylated DNA. Cell 1992, 69:905-914 [DOI] [PubMed] [Google Scholar]
- 9.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP: Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 1998, 19:187-191 [DOI] [PubMed] [Google Scholar]
- 10.Bienvenu T, Carrie A, de Roux N, Vinet MC, Jonveaux P, Couvert P, Villard L, Arzimanoglou A, Beldjord C, Fontes M, Tardieu M, Chelly J: MeCP2 mutations account for most cases of typical forms of Rett syndrome. Hum Mol Genet 2000, 9:1377-1384 [DOI] [PubMed] [Google Scholar]
- 11.Xiang F, Buervenich S, Nicolao P, Bailey ME, Zhang Z, Anvret M: Mutations screening in Rett syndrome patients. J Med Genet 2000, 37:250-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A: Transcriptional repression b the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 1998, 6683:386-389 [DOI] [PubMed] [Google Scholar]
- 13.Bird AP, Wolffe AP: Methylation-induced repression: belts, braces, and chromatin. Cell 1999, 99:451-454 [DOI] [PubMed] [Google Scholar]
- 14.Trappe R, Laccone F, Cobilanschi J, Meins M, Huppke P, Hanefeld F, Engel W: MECP2 mutations in sporadic cases of Rett syndrome are almost exclusively of paternal origin. Am J Hum Genet 2001, 68:1093-1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffbuhr K, Devaney JM, LaFleur B, Sirianni N, Scacheri C, Giron J, Schuette J, Innis J, Marino M, Philippart M, Narayanan V, Umansky R, Kronn D, Hoffman EP, Naidu S: MeCP2 mutations in children with and without the phenotype of Rett syndrome. Neurology 2001, 56:1486-1495 [DOI] [PubMed] [Google Scholar]
- 16.Gilles PN, Wu DJ, Foster CB, Dillon PJ, Chanock SJ: Single nucleotide ploymorphic discrimination by an electronic dot blot assay on semiconductor microchips. Nature Biotech 1999, 17:365-370 [DOI] [PubMed] [Google Scholar]
- 17.Radtkey R, Feng L, Muralhidar M, Duhon M, Canter D, DiPierro D, Fallon S, Tu E, McElfresh K, Nerenberg M, Sosnowski R: Rapid, high fidelity analysis of simple sequence repeats on an electronically active DNA microchip. Nucleic Acids Res 2000, 28:E17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forster AH, Krihak M, Swanson PD, Young TC, Ackley DE: A laminated, flex structure for electronic transport and hybridization of DNA. Biosens Bioelectron 2001, 16:187-194 [DOI] [PubMed] [Google Scholar]
- 19.Huang Y, Ewalt KL, Tirado M, Haigis R, Forster A, Ackley D, Heller MJ, O’Connell JP, Krihak M: Electric manipulation of bioparticles and macromolecules on microfabricated electrodes. Anal Chem 2001, 73:1549-1559 [DOI] [PubMed] [Google Scholar]