Abstract
Synovial sarcomas (SS) are characterized by the t(X;18)(p11;q11) translocation and its resultant fusion gene, SYT-SSX. Two homologues of the SSX gene (ie, SSX1 and SSX2) are involved in the vast majority of SS and the SYT-SSX1 type of fusion has been associated with inferior clinical outcome. Thus, detection of the presence and type of SYT-SSX fusion is critical for diagnosis and prognosis in SS. Identification of SYT-SSX fusion type is typically accomplished by reverse-transcription polymerase chain reaction (RT-PCR) followed by a post-PCR analytic method. As mRNA nucleotide sequences of the SSX1 and SSX2 segments involved in the SYT-SSX fusion are nearly identical, post-PCR methods must be highly discriminatory. We describe a novel method to identify and differentiate these two chimeric transcripts using RT-PCR followed by fluorescent thermostable ligase detection reaction (f-LDR), microparticle bead capture and flow cytometric detection. Evaluation of this unique approach in 11 cases of SS without prior knowledge of SYT-SSX status, six cases of control sarcomas (CS) and three hematopoietic cell lines, revealed that the f-LDR technique was rapid, unambiguous, and highly specific. The f-LDR results were compared to XmnI enzyme digestion patterns and sequencing of PCR products, revealing a 100% concordance for all cases of SS with regards to SYT-SSX transcript type. In addition, there was a strong association of transcript type detected by f-LDR and morphological subclassification of SS, as previously reported. We conclude that this f-LDR method with flow-based detection is a robust approach to post-PCR detection of specific nucleotide sequences in SS and may be more broadly applicable in molecular oncology.
Synovial sarcoma (SS) is the fourth most common soft tissue sarcoma following malignant fibrous histiocytoma, liposarcoma and rhabdomyosarcoma. 1 These tumors predominantly affect adolescents and young adults with a predilection for periarticular regions of the extremities. Morphologically, two patterns of SS are recognized: a monophasic variant, composed entirely of a spindle cell proliferation and a biphasic variant, consisting of glands and spindle cells. Regardless of morphological type, SS is associated with the t(X;18)(p11;q11) translocation in greater than 90% of cases. 2 The presence of the t(X;18) thus constitutes a tumor specific marker for SS and can be helpful when excluding the diagnosis of other spindle cell proliferations. Specifically, the t(X;18) results in fusion of the upstream segment of the SYT gene at 18q11 to sequences of one of two related highly homologous genes at Xp11 (SSX1 or SSX2), forming a chimeric SYT-SSX gene. 3, 4, 5 Evaluation of the gene product of SSX suggests that it may function as a DNA binding factor, which becomes aberrantly functional with fusion to the SYT gene. 5 Although several additional SSX gene family members have been described, 6 the vast majority of SS harbor either the SYT-SSX1 or SYT-SSX2 fusions. A study by Inagaki et al 7 demonstrated a relationship with tumor proliferative index, adverse clinical behavior and the SYT-SSX1 fusion type. These findings are supported by reports documenting a high Ki-67 antigen proliferation rate and a worse clinical outcome for patients harboring the SYT-SSX1 fusion type. 8, 9 Detection of the SYT-SSX fusion type thus not only confirms the diagnosis of SS, but provides valuable prognostic information as well.
A relationship between morphological subtype and SYT-SSX fusion type has been described, with biphasic tumors generally associated with the SYT-SSX1 form and monophasic tumors featuring the SYT-SSX2 transcript, although exceptions to this finding are also known. 5, 10, 11 However, exact determination of the SYT-SSX type in SS is more aptly determined by molecular approaches. As the SYT-SSX fusion gene is actively transcribed, standard reverse-transcriptase polymerase chain reaction (RT-PCR) methods have been developed to specifically detect the chimeric mRNA in tissue biopsies of SS. 7, 8, 9, 10, 11, 12, 13, 14 To delineate the SSX1 and SSX2 fusion products, several approaches have been described. Direct sequencing of the PCR products has been used to distinguish the two types of fusion products (4,7–9,12). Alternatively, some investigators have used allele-specific primers for SSX1 and SSX2 under stringent PCR conditions, or post-PCR hybridization with discriminating oligonucleotide probes (4,7–10,14). The use of restriction enzyme digestion patterns (e.g., XmnI) has also been shown to differentiate the two fusion types. 12 One recent report has suggested the use of quantitative real-time PCR to identify SYT-SSX transcripts. 15 Nearly all of these methods, with the exception of allele-specific or real-time PCR, require some additional post-PCR modification to distinguish between the two SYT-SSX fusion types. The latter point is of significance considering that biopsies of SS are usually not generous and the RNA preparation and RT-PCR is most often performed from paraffin-embedded material. In this setting, the amplicon length for reasonably efficient PCR is limited, in turn producing technical constraints on primer selection for SYT-SSX detection, thus typically requiring some form of post-PCR analysis for confirmation of the PCR product.
We have developed a novel approach for the specific detection of highly-related DNA fragments by using a fluorescent thermostable ligase detection reaction (f-LDR) and flow cytometry following PCR. The specificity afforded by f-LDR ensures correct discrimination of the transcript type and, by modifying the reaction to produce a fluorescent signal, detection is readily accomplished. This general methodology may have applications in a variety of other molecular diagnostic settings as well.
Materials and Methods
Patient Samples
Formalin-fixed paraffin-embedded (FFPE) tissue samples of 11 synovial sarcomas (SS) and six control sarcoma (CS) cases were obtained from the department of Pathology of the University of New Mexico (UNM), the Vancouver Hospital and Health Sciences Center, and the British Columbia Children’s Hospital. Details regarding biopsy site and histological type were obtained. Sections of the FFPE tissue blocks were cut under RNase-free conditions and stored at −20°C in microfuge tubes until use. The research protocol was approved by the UNM Human Research Review Committee.
RNA Extraction
RNA was extracted from 10 μm FFPE sections following the method of Specht et al 16 with minor modifications. In brief, tissue sections were de-paraffinized twice in a 1.5 ml microfuge tube with 1 ml of xylene, then pelleted and washed thoroughly with an equal volume of absolute ethanol. Tissue samples were washed and rehydrated in 90% followed by 70% ethanol/diethyl pyrocarbonate (DEPC)-treated water. All centrifugation steps were performed at 4°C. The pellet was air-dried then resuspended in 200 μl RNA lysis buffer containing 10 mmol/L Tris-HCl (pH 8.0), 0.1 mmol/L ethylenediaminetetraacetic acid (pH 8.0), 2% sodium dodecyl sulfate. 500 μg/ml of proteinase K (Sigma, St. Louis, MO) in DEPC-treated water was added (10 μl of a 10 mg/ml solution) and the sample was digested overnight at 60°C for 16 hours. A second aliquot of proteinase K was added at 16 hours to incompletely digested samples and the incubation was continued until digestion was complete (typically 2 to 3 additional hours). The samples were next extracted in 400 μl acid phenol/chloroform, pH 4.5 (Ambion, Austin, TX) and RNA was precipitated from the aqueous layer in 1/10 volume of 3 mol/L sodium acetate and one volume of isopropanol, in the presence of 1 μl linear acrylamide (Ambion) as carrier. The RNA pellet was washed once with 70% ethanol/DEPC-treated water, air-dried, and resuspended in DEPC-treated water. RNA was stored at −70°C until used.
RT-PCR for Detection of the SYT-SSX Fusion Transcripts
An aliquot (2–5 μl) of RNA from each sample was treated with RNase-free DNase according to the manufacturer’s instructions (DNA-free Kit; Ambion) to eliminate contaminating genomic DNA. The RNA was recovered in a volume of ∼ 10 μl, then reverse transcribed and amplified using the One-Step RT-PCR Kit (Qiagen, Santa Clarita, CA) according to the manufacturer’s instructions. A single set of PCR primers was used to detect both SYT-SSX1 and SYT-SSX2 fusion transcripts. The PCR parameters were as follows (GeneAmp 9700; Applied Biosystems Inc., Foster City CA): RT at 50°C for 30 minutes; PCR at 95°C for 15 minutes, 35 cycles of 95°C for 45 seconds, 60°C for 30 seconds, 72°C for 60 seconds, then 72°C for 5 minutes and cooling to 4°C. The PCR products were analyzed by electrophoresis on 1.5% agarose gel. For each sample, concurrent amplification of a segment of β2-microglobulin was conducted to verify the integrity of the RNA template. All primer sequences are shown in Table 1A . Expected PCR product sizes were: SYT-SSX, 101 bp; β2-microglobulin, 120 bp.
Table 1.
PCR Primers and Ligation Detection Primers
| SYT-SSX and β2-microglobulin RT-PCR |
| SYT: 5′-ACCCCAGCAGAGGCCTTATGG |
| SSX: 5′-TTTGTGGGCCAGATGCTTCTG |
| β2-microglobulin sense: 5′- GAAAAAGATGAGTATGCCTG |
| β2-microglobulin antisense: 5′- ATCTTCAAACCTCCATGATG |
| Ligation detection reactions for SYT-SSX1 and SYT-SSX2 |
| LigSSXcommon: 5′-6FAM- GCCCAAGAAGCCAGCAGAGGA(AC)G |
| LigSSX1specific: 5′- *AAAATGATTCGAAGGGAGTGTCA-BIO |
| LigSSX2specific: 5′- *GAAATGATTCGGAGGAAGTGCCA-BIO |
, phosphorylated nucleotide; BIO, biotin tag; 6FAM, 6-carboxyfluorescein tag; (AC) nucleotides in LigSSXcommon primer indicate 2-base degeneracy; underlined nucleotides in LigSSX1 and LigSSX2 specific primers indicate nucleotide differences between the two respective gene segments. The most 5′ bases in each primer (underlined bold and italic) allow for specific discrimination of the two fusion types when juxtaposed with the LigSSXcommon primer.
Fluorescent Thermostable Ligase Detection Reaction (f-LDR)
Two separate but simultaneous reactions were carried out to distinguish SYT-SSX1 from SYT-SSX2 fusion types in each case studied by relying on single nucleotide differences in the SSX segment of the PCR product. For each reaction, one μl of SYT-SSX PCR product was placed in a total volume of 20 μl containing 5 pmol of the common upstream ligation primer (LigSSXcommon) and 5 pmol of either the LigSSX1-specific or LigSSX2-specific discriminating ligation primers respectively, 2 μl of 10X ligase buffer and 0.5 μl of Taq ligase (New England Biolabs, Beverly, MA). The LigSSXcommon primer was modified at the 5′ end with a fluorescent molecule, 6FAM, whereas both of the LigSSX1 and LigSSX2 specific primers were labeled with a biotin moiety at their 3′ ends. The 5′ nucleotide ends of both LigSSX1 and LigSSX2 specific primers were phosphorylated, enabling specific end-to-end DNA ligation. Sequences of ligation primer oligonucleotides are shown in Table 1B . The ligase detection reactions (LDR) were carried out in a PCR machine (Gene-Amp 9700; Applied Biosystems Inc.) as follows: 95°C for 5 minutes, 20 cycles of 95°C for 30 seconds, 65°C for 2 minutes, then 95°C for 5 minutes and cooling to 4°C.
Bead Capture and Flow Cytometric Analysis
Following LDR, a 10-μl aliquot of ligation product was mixed with 10 μl of 6.0–8.0 μm streptavidin-coated polystyrene microbeads (Spherotech, Inc., Libertyville, IL) in a microfuge tube and incubated at room temperature for 30 minutes, with brief agitation of the tube at 15 minutes. The beads were pelleted in a microcentrifuge for 2 minutes and the supernatant carefully removed and discarded, without disturbing the beads. The beads were then resuspended in 100 μl of 100 mmol/L Tris/300 mmol/L NaCl buffer (pH 8.3) and transferred to a 12 × 75 mm plastic tube for flow cytometric evaluation. Six hundred μl of the 100 mmol/L Tris/300 mmol/L NaCl buffer was added to each tube and flow cytometric analysis was performed on a FACScan flow cytometer (Becton-Dickinson, San Jose, CA) using 488 nm laser excitation. For data analysis, the mean fluorescence intensity values of the f-LDR test samples were compared against negative controls (β2-microglobulin samples and water only samples). A mean fluorescence shift above 101 (log scale) was considered positive and below this value, negative.
XmnI Enzyme Digestion Studies
A 15-μl aliquot of SYT-SSX PCR product from each SS case was incubated for 3 hours with XmnI and supplied buffer (New England Biolabs) according to manufacturer’s directions. Postdigest products were evaluated by electrophoresis on a 4% NuSieve gel (Cambrex, Rockland, ME, USA). Each XmnI-digested PCR product was electrophoresed adjacent to an identical undigested PCR product for comparison. Enzyme digestion results were assessed by ethidium bromide staining and UV illumination. A positive digestion reaction resulted in production of two predicted smaller fragments (69 bp and 32 bp), indicative of the SSX2 nucleotide sequence in the PCR product. 12
Sequencing of SYT-SSX PCR Products
PCR products from SS cases were cloned into pCR 2.1 plasmid using the TopoTA system (Invitrogen, Carlsbad, CA) according to manufacturer’s directions. After transfection and propagation in supplied competent bacteria, colonies representing both SYT-SSX fusion types were selected and minipreparations of purified plasmid were obtained using the Qiaprep Miniprep system (Qiagen, Valencia, CA). The insert regions were sequenced in both directions on an automated fluorescent capillary sequencer (ABI 3100; Applied Biosystems) using M13 primers. Sequence data were compared to known gene database sequence for SYT-SSX2 fusion mRNA (GenBank Accession no. AF230662) and published data of the bp differences between SSX1 and SSX2 regions, 12 to determine the SYT-SSX fusion type in the PCR amplicons.
Results
Patient Samples, RNA Extraction, and RT-PCR for SYT-SSX Fusions
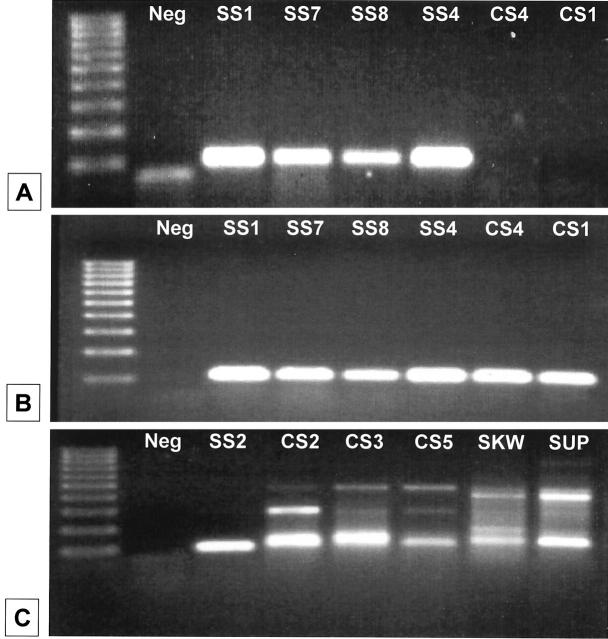
Details of biopsy sites and histological diagnoses of the 11 SS cases, 6 control sarcoma (CS) cases and 3 hematopoietic cell lines are presented in Table 2 . RNA was successfully obtained from paraffinized material in all cases of SS and CS and from cultured hematopoietic cell lines, followed by DNase digestion before RT-PCR. To amplify specific targets from paraffin-embedded material, the amplicons for both SYT-SSX and β2-microglobulin were kept to a minimal size. A PCR product of correct size (101 bp) for the SYT-SSX fusion was clearly evident in 10/11 cases of SS (Figure 1A) . In one remaining case (SS3, Table 2 ), no PCR product was generated, despite adequate and intact RNA. Pathological review of this latter sample revealed that it was in fact a sclerosing fibrosarcoma, errantly included among the group of SS. In all samples evaluated, the integrity of extracted RNA was confirmed by concurrent amplification of a segment of β2-microglobulin (Figure 1B) . Although some of the six CS cases lacked an amplified product by gel electrophoresis (Figure 1A) , others, as well as the hematopoietic cell line negative controls, generated nonspecific PCR products similar to the expected size for the SYT-SSX amplicon (Figure 1C) .
Table 2.
Patient Cases, Tumor Location, and Diagnosis
| Case no. | Biopsy site | Diagnosis |
|---|---|---|
| SS1 | Gluteus maximus | Synovial sarcoma-MP |
| SS2 | Chest wall | Synovial sarcoma-MP |
| SS3* | Lower abdominal wall | Synovial sarcoma-MP→ Sclerosing fibrosarcoma |
| SS4 | Right groin | Synovial sarcoma-MP |
| SS5 | Right knee mass | Synovial sarcoma-MP |
| SS6 | Left knee joint | Synovial sarcoma-BP |
| SS7 | Left chest wall | Synovial sarcoma-BP |
| SS8 | Right thigh | Synovial sarcoma-BP |
| SS9 | Right thigh | Synovial sarcoma-BP |
| SS10 | Left popliteal fossa | Synovial sarcoma-MP |
| SS11 | Left lower extremity | Synovial sarcoma-BP |
| CS1 | Left shoulder | High-grade sarcoma |
| CS2 | Left shoulder | High-grade sarcoma |
| CS3 | Right thigh | Malignant fibrous histiocytoma |
| CS4 | Cervix | Embryonal rhabdomyosarcoma |
| CS5 | Retroperitoneal mass | Leiomyosarcoma |
| CS6 | Skin right groin | Dermatofibrosarcoma protruberans |
| MHH-CALL2† | N/A | Leukemia cell line |
| SKW3† | N/A | Leukemia cell line |
| SUP-T1† | N/A | Leukemia cell line |
This case subsequently re-diagnosed as a sclerosing fibrosarcoma.
RNA isolated from cultured cells.
SS, synovial sarcoma; MP, monophasic; BP, biphasic; CS, control sarcoma.
Figure 1.
Results of RT-PCR for SYT-SSX and β2-microglobulin products. Representative results of SYT-SSX RT-PCR are shown. Far left lane in each panel shows 100-bp molecular size standard (lowest marker represents 100 bp). Cases are designated as “SS” for synovial sarcoma and “CS” for control sarcoma. “SKW” and “SUP” are hematopoietic cell line negative controls. “Neg” is a water-only (no template) blank (primer artifact is present in A). A: SYT-SSX PCR products (expected size 101 bp). B: β2-microglobulin internal control PCR products for cases shown in A (expected size 120 bp). C: Illustration of non-specific, but similarly sized PCR products in CS cases and hematopoietic cell line controls. Note lower PCR band in CS2, CS3, CS5, SKW, and SUP sample lanes, very close to the size of positive SS case SS2. However, none of the control cases gave positive fluorescent LDR results (see Figure 3 and text). Details of cases are given in Table 2 .
f-LDR and Detection of SYT-SSX Fusion Type by Flow Cytometry
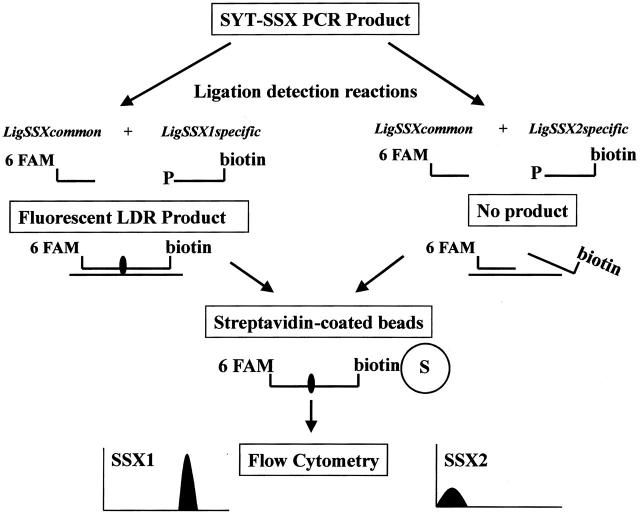
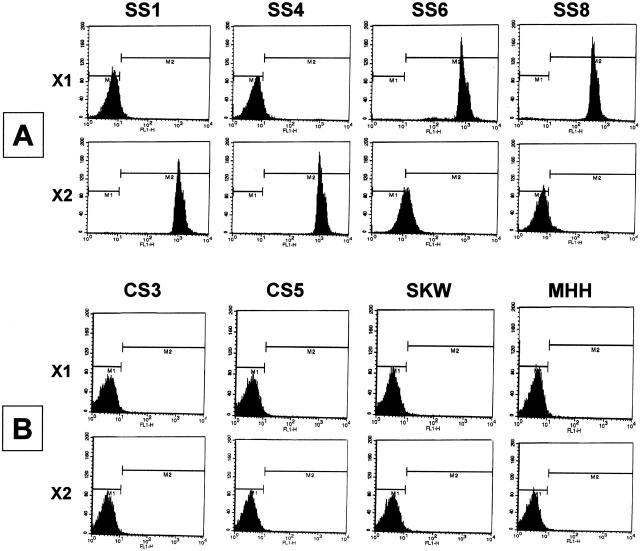
Given the common SYT segment and the close homology between SSX1 and SSX2 regions, a fluorescent ligase detection reaction (f-LDR) technique was developed to discriminate between the latter two sequences (see schematic, Figure 2 ). This approach takes advantage of a small number of coding nucleotide differences between the two highly homologous SSX genes, just downstream of the fusion site with SYT in the chimeric mRNA. By using a common 5′ ligation primer (LigSSXcommon) and two discriminating 3′ ligation primers (either LigSSX1specific or LigSSX2specific), the two SSX genes can be uniquely identified. f-LDR for both SSX1 and SSX2 sequences in the RT-PCR products was accomplished in a thermocycler using a thermostable DNA ligase. To enable detection of the products, the LigSSXcommon primer was labeled with a fluorescent tag (6FAM) and each of the LigSSX1specific and LigSSX2specific primers was modified with a 5′ phosphate and 3′ biotin end-label. If a specific ligation product was subsequently obtained, the resultant fluorescent probe was captured onto streptavidin-coated polystyrene microbeads and fluorescence emission was then detected by flow cytometry. The absence of a ligation product would thus result in lack of a fluorescent signal in the assay. Figure 3A and B demonstrate representative results obtained using the f-LDR method in SS, CS and hematopoietic cell line negative control samples. Intense positive fluorescent signals were obtained in every SS specimen, but none of the CS cases or hematopoietic cell lines, thus demonstrating 100% sensitivity and specificity. The case of sclerosing fibrosarcoma (SS3) correspondingly did not show positivity in the f-LDR assay (data not shown). Among the 10 morphologically diagnosed SS cases, 5 were found to have the SYT-SSX1 fusion and 5 others the SYT-SSX2 transcript type (Table 3) . To further verify the specificity of the f-LDR, control β2-microglobulin PCR products of SS and CS cases were also subjected to the assay, as well as water-only (no RNA template) blank reactions. No fluorescence signal above baseline was obtained in any of these latter samples by flow cytometry (data not shown).
Figure 2.
Schematic diagram of fluorescent ligase detection reaction (f-LDR) method. A 1-μl aliquot of SYT-SSX PCR product is subjected to two separate fluorescent ligase detection reactions (f-LDR) using a common upstream ligation primer (LigSSXcommon) and one of two discriminating primers (LigSSX1specific or LigSSX2specific), shown as left and right branches respectively. The ligation primers are situated in the SSX segment of the fusion transcript amplicon and the discriminating primers are situated to take advantage of single nucleotide differences between SSX1 and SSX2 regions. The LigSSXcommon primer is 5′ fluorescently labeled (with 6-FAM), whereas each of the specific primers is modified by 5′phosphorylation and 3′ biotinylation. In the case of specific primer binding and alignment on the correct PCR product template (in this case shown for SYT-SSX1, left branch), the f-LDR will generate a fluorescent and biotinylated nucleotide fragment. If correct sequence is not present for the specific primer interaction, no product will be formed (ie, right branch with SSX2 primer reaction). Each f-LDR sample is then incubated briefly with a 10 μl aliquot of streptavidin-coated polystyrene microbeads and analyzed by flow cytometry for the presence or absence of a fluorescent signal.
Figure 3.
Results of fluorescent ligase detection reactions (f-LDR) detected by flow cytometry. A demonstrates representative results for synovial sarcoma cases (SS) whereas B illustrates control sarcoma (CS) and hematopoietic cell line results (case details in Table 2 ). Left panel designation “X1” refers to discriminating f-LDR primer set for the SSX1 segment of the SYT-SSX1 fusion; “X2” refers to discriminating f-LDR primer set for the SSX2 segment of the SYT-SSX2 fusion. Relative fluorescence intensity is shown on x axis (negative result indicated as below 101 intensity). Samples SS1 and SS4 demonstrate SYT-SSX2 fusion type and samples SS6 and SS8 reveal SYT-SSX1 fusion type. Note that the alternate f-LDR assay for a given SS sample (for example, sample SS1 with the X1 primer set) does not generate false positivity despite the use of highly homologous primers. CS cases and non-sarcoma hematopoietic cell line control samples are unequivocally negative in both f-LDR primer set assays, despite the presence of apparent PCR products on agarose gel for some of these samples (Figure 1C) . The f-LDR data for SS cases are summarized in Table 3 .
Table 3.
Comparison of f-LDR Results, Xmn Digestion and Histologic Features for SYT-SSX Fusion Type Determination in SS Cases
| Case no.* | Fusion type by f-LDR† | XmnI digest | Histologic SS type |
|---|---|---|---|
| SS1 | SSX2 | Positive | Monophasic |
| SS2 | SSX2 | Positive | Monophasic |
| SS3 | Negative | Negative | Sclerosing fibrosarcoma |
| SS4 | SSX2 | Positive | Monophasic |
| SS5 | SSX2 | Positive | Monophasic |
| SS6 | SSX1 | Negative | Biphasic |
| SS7 | SSX1 | Negative | Biphasic |
| SS8 | SSX1 | Negative | Biphasic |
| SS9 | SSX1 | Negative | Biphasic |
| SS10 | SSX2 | Positive | Monophasic |
| SS11 | SSX1 | Negative | Biphasic |
Case SS3 was subsequently re-diagnosed as a sclerosing fibrosarcoma.
There is 100% concordance between f-LDR results, Xmn digestion and PCR product sequence data.
Correlation of f-LDR Results with XmnI Digestion of PCR Products and with Morphological Features of SS
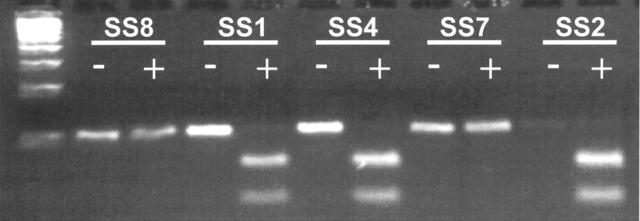
To verify the specificity of the f-LDR method, all SS sample PCR products were subjected to restriction enzymatic digestion with XmnI. This enzyme cleaves the SSX2 segment within the corresponding SYT-SSX2 PCR amplicon, but does not have a consensus recognition site in the homologous SYT-SSX1 DNA sequence. 12 Representative results of XmnI digestion of SYT-SSX PCR products are shown in Figure 4 . In all cases of true SS, the XmnI digestion pattern correlated with the results of f-LDR (Table 3) . In occasional cases, incomplete digestion products were encountered, requiring repeat of the enzymatic procedure with longer digestion times. Notably, the restriction digestion technique using the protocol described here required more laboratory time than completing the f-LDR analysis. Finally, as summarized in Table 3 , the specific results of SYT-SSX fusion type in the SS cases, as determined by f-LDR, were highly correlated with morphological subclassification as monophasic or biphasic tumors (Table 3) .
Figure 4.
Results of XmnI digestion for SYT-SSX type. Results of XmnI enzyme digestion is demonstrated for representative SS cases. XmnI recognition site is present in the SSX2, but not the SSX1 sequence of the SYT-SSX PCR product. Far left lane is a 100 bp ladder. SS case numbers are shown above gel bands. Below each SS case designation, the undigested (no enzyme added) PCR product is indicated on the left (-) and the digested product (enzyme added) is indicated immediately adjacent on the right (+). A positive XmnI digestion is evidenced by the production of two smaller fragments of predicted sizes 69 bp and 32 bp. Cases SS1, SS4 and SS2 are interpreted as showing complete digestion with XmnI (SYT-SSX2 fusion), whereas SS8 and SS7 do not show digestion byproducts (SYT-SSX1 fusion). These results were 100% concordant with f-LDR data (see Figure 3 and Table 3 ) and were further confirmed by PCR product sequencing (see Figure 5 ).
Sequencing of PCR Amplicons in SS Cases
PCR products of SS representing both types of SYT-SSX fusion (as determined by f-LDR method and XmnI digestion) were cloned and sequenced in both directions. The sequence data obtained were compared with published sequence information and demonstrated complete concordance with the f-LDR results for SYT-SSX1 or SYT-SSX2 fusion types (Figure 5A and B) .
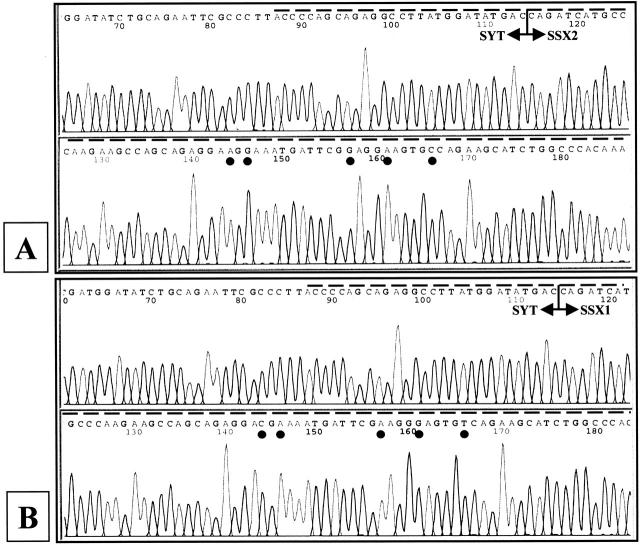
Figure 5.
Sequence data for SYT-SSX fusions. Shown are the representative automated fluorescent sequencing data derived from two cases, SS1 and SS7. A: Case SS1 demonstrates SYT-SSX2 fusion; B: Case SS7 demonstrates SYT-SSX1 fusion. Sequences are shown in 5′-3′ sense orientation. Dashed line above each tracing represents insert sequence of SYT-SSX PCR product. T-bars with arrows indicate location of SYT and SSX junction in mRNAs. Filled circles below nucleotide symbols in each panel show single-base differences in SSX2 and SSX1 regions of the respective fusion transcripts. Sequence data were completely concordant with f-LDR results (Figure 3 and Table 3 ).
Discussion
Specific DNA sequence detection by enzymatic ligation methods was initially described by Landegren 17 and co-workers and Whiteley et al. 18 The production of thermostable DNA ligase subsequently enabled specific ligation assays at higher temperatures (thus with greater stringency), as well as the development of ligation amplification (ligase chain reaction, LCR) to generate large amounts of DNA product. 19, 20 The requirement for precise sequence alignment of an adjacent set of oligonucleotides on a complementary target DNA template underlies the great specificity of DNA ligase analysis. If two pairs of complementary ligation primers are used at a given target DNA site (ie, specific for sequences on both sense and antisense DNA strands), exponential production of ligated DNA molecules can be generated in theory. LCR has been used in research applications for detecting point mutations in DNA with high specificity and sensitivity. 21, 22 In the clinical setting, LCR has been used in the detection of infectious disease pathogens, including Chlamydia trachomatis and Mycobacterium tuberculosis. 23, 24, 25, 26 Eggerding and co-workers 27, 28 pioneered the use of a multiplex, one-sided (one-target strand) oligonucleotide ligation assay (OLA), either simultaneous with, or following PCR to identify CFTR and RAS gene mutations. These studies also used fluorescently-labeled oligonucleotides to detect specific wild-type or mutant sequences by differences in size and fluorescence signals on a DNA sequencer. Fluorescently-modified oligoprobes have also been used in single nucleotide polymorphism (SNP) analysis using OLA, followed by complementary binding of the ligated products to “capture” tags on dye-impregnated microsphere beads and flow cytometric analysis. 29 We wished to extend our recent experience using simple fluorescently-labeled oligonucleotide probes and flow cytometry in the detection of leukemia-associated fusion gene RT-PCR products, 30 by developing a one-sided post-PCR fluorescent ligation assay, here termed f-LDR. Synovial sarcoma provided an ideal model to demonstrate the discriminatory ability of the new assay, given that the two major fusion SYT-SSX products are closely homologous, but nevertheless amenable to specific detection by oligoprobe ligation technique. In addition, the type of SYT-SSX fusion product in SS appears to be clinically significant and there exists a variety of analytic methods described to delineate these fusion types, with various limitations.
In our series of eleven cases of morphological SS, ten were found to have RT-PCR products of correct size using SYT and SSX region primers. The one negative case was found to be a sclerosing fibrosarcoma erroneously included with this group. In contrast, 6 cases designated control sarcomas and 3 hematopoietic cell lines did not show PCR positivity, or demonstrated nonspecific banding patterns that were not identical to SS cases. Nonetheless, given the small PCR product size range, further post-PCR evaluation is required to determine whether a true SYT-SSX fusion is present and if so, the type of SYT-SSX fusion. The f-LDR method we describe was able to unequivocally identify the type of SYT-SSX fusion in all 10 true SS cases, with 5 demonstrating the SYT-SSX1 chimeric product and 5 showing the SYT-SSX2 transcript type. These results were validated by XmnI enzymatic digestion studies, as well as sequencing of the PCR products. None of the CS cases or hematopoietic cell line controls was positive by f-LDR. Furthermore, performing the assay with additional non-specific PCR products (eg, β2-microglobulin internal control) did not generate false positive results. As further evidence of the accuracy of this method, the f-LDR data for SS were highly correlated with morphological subtype (monophasic versus biphasic), as has been previously reported. 10
In this technical approach, the fluorescent ligated products are generated from the SSX segment of the SYT-SSX PCR amplicons. The 100% specificity of this approach is determined by several factors: DNase digestion of the extracted RNA samples, generation of SYT-SSX product by PCR as the template for f-LDR, and use of one-sided ligation reaction to reduce the possibility of nonspecific primer interactions. As paraffin-embedded tissues were required for this work, the importance of creating short PCR amplicons and avoidance of DNA contamination were important considerations. In addition to detecting the SYT-SSX anomaly as a diagnostic marker for SS, delineation of SYT-SSX1 from SYT-SSX2 fusion types has been implicated as prognostically important in this cancer, as patients with SS harboring the former transcript may have an inferior outcome. 7, 8, 9 Investigators have used a variety of methods for this purpose, including restriction enzyme digestion, stringent SSX allele-specific PCR, PCR product sequencing, discriminating probe hybridization and real-time quantitative PCR. 4, 7, 8, 9, 10, 11, 12, 13, 14, 15 Each of these approaches has merits and shortcomings. Enzymatic digestion, for example, though easily performed, is prone to subjective visual interpretation. Sequencing of PCR products is highly specific, but relatively time-consuming and expensive as a routine diagnostic test. Similarly, real-time PCR methods require relatively costly equipment and supplies for routine molecular laboratory detection purposes. Oligonucleotide probe hybridization requires multiple additional post-PCR manipulations.
With the f-LDR method, we describe a highly specific and easily interpreted assay, which can be completed in approximately 2 hours following PCR. Furthermore, the methodology is cost-effective, since only small amounts of fluorescent and biotin-labeled primers are used and the respective stock primer solutions can be stably stored in a minus 80°C freezer for many months without significant degradation. The initial cost of purchasing biotinylated and fluorescently modified oligonucleotides is offset by the reduced need for gel electrophoresis and additional post-PCR manipulations, many of which involve increased technical time. Given the ability to conjugate an increasing number of fluorophores to nucleotides, the development of two- or three-color multiplex assays for detecting SS fusion types and control amplification products simultaneously can be envisioned. The latter point is of significance given the recent descriptions of rarely occurring, alternate forms of the SYT-SSX fusion. 31, 32 Finally, the use of flow cytometry to detect the presence of specific nucleotide species is considered a novel approach to post-PCR evaluation in the clinical molecular diagnostic setting. Conceivably, this approach of combined molecular methodology and flow cytometry could be used in a diverse set of applications, including mutation detection, or screening for other tumor-associated genetic anomalies.
Acknowledgments
We thank Terry Mulcahy and the Center for Genetics in Medicine (Departments of Biochemistry and Molecular Biology, University of New Mexico) for providing DNA sequencing services, and Michael Grady (Department of Pathology, UNM) for assistance with preparation of figures.
Address reprint requests to David S. Viswanatha, M.D., Rm 337-C, BRF Building, University of New Mexico HSC, 915 Camino de Salud NE, Albuquerque, NM 87131. E-mail: dviswanatha@salud.unm.edu.
References
- 1.Malignant soft tissue tumors of uncertain type. Enzinger FM Weiss SW eds. Soft tissue tumors 2001:1483-1509 Mosby St Louis, MO
- 2.Ladanyi M: The emerging molecular genetics of sarcoma translocation. Diagn Mol Pathol 1995, 4:162-173 [DOI] [PubMed] [Google Scholar]
- 3.Clark J, Philippe J, Crew AJ, Gill S, Shipley J, Chan A, Gusterson AB, Cooper CS: Identification of novel genes, SYT and SSX, involved in the t(X;18)(p112;q112) translocation found in human synovial sarcoma. Nat Genet 1994, 7:502-508 [DOI] [PubMed] [Google Scholar]
- 4.Leeuw B, Balemans M, Olde Weghuis D, Geurts van Kessel A: Identification of two alternative fusion genes, SYT-SSX1 and SYT-SSX2, in t(X;18)(p112)(q112)-positive synovial sarcomas. Hum Mol Genet 1995, 4:1097-1099 [DOI] [PubMed] [Google Scholar]
- 5.Crew AJ, Clark J, Fisher C, Gill S, Grimer R, Chand A, Shipley J, Gusterson BA, Cooper CS: Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. EMBO J 1995, 14:2333-2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gure AO, Wei IJ, Old LJ, Chen YT: The SSX gene family: characterization of 9 complete genes. Int J Cancer 2002, 101:448-453 [DOI] [PubMed] [Google Scholar]
- 7.Inagaki H, Nagasaka T, Otsuka T, Sugiura E, Nakashima N, Eimoto T: Association of SYT-SSX fusion types with proliferative activity and prognosis in synovial sarcoma. Mod Pathol 2000, 13:482-488 [DOI] [PubMed] [Google Scholar]
- 8.Kawai A, Woodruff J, Healey JH, Brennan MF, Antonescu CR, Ladanyi M: SYT-SSX gene fusion as a determinant of morphology and prognosis in synovial sarcoma. N Engl J Med 1998, 338:153-160 [DOI] [PubMed] [Google Scholar]
- 9.Nilsson F, Skytting B, Xie Y, Brodin B, Perfekt R, Mandahl N, Lundeberg J, Uhlen M, Larsson O: The SYT-SSX variant of synovial sarcoma is associated with a high rate of tumor cell proliferation and poor clinical outcome. Cancer Res 1999, 59:3180-3184 [PubMed] [Google Scholar]
- 10.Antonescu CR, Kawai A, Leung DH, Lonardo F, Woodruff JM, Healey JH, Ladanyi M: Strong association of SYT-SSX fusion type and morphologic epithelial differentiation in synovial sarcoma. Diagn Mol Pathol 2000, 9:1-8 [DOI] [PubMed] [Google Scholar]
- 11.Tsuji S, Hisaoka M, Morimitsu Y, Hashimoto H, Shimajiri S, Komiya S, Ushijima M, Nakamura T: Detection of SYT-SSX fusion transcripts in synovial sarcoma by reverse transcription-polymerase chain reaction using archival paraffin-embedded tissues. Am J Pathol 1998, 153:1807-1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lasota J, Jasinski M, Debiec-Rychter M, Szadowska A, Limon J, Miettinen M: Detection of the SYT-SSX fusion transcripts in formaldehyde-fixed, paraffin-embedded tissue: a reverse transcription polymerase chain reaction amplification assay useful in the diagnosis of synovial sarcoma. Mod Pathol 1998, 11(7):626-633 [PubMed] [Google Scholar]
- 13.Argani P, Zakowski MF, Klimstra DS, Rosai J, Ladanyi M: Detection of the SYT-SSX chimeric RNA of synovial sarcoma in paraffin-embedded tissue and its application in problematic cases. Mod Pathol 1998, 11:65-71 [PubMed] [Google Scholar]
- 14.van de Rijn M, Barr FG, Collins MH, Xiong QB, Fisher C: Absence of SYT-SSX fusion products in soft tissue tumors other than synovial sarcoma. Am J Clin Path 1999, 112:43-49 [DOI] [PubMed] [Google Scholar]
- 15.Bijwaard KE, Fetsch JF, Przygodzki R, Taubenberger JK, Lichy JH: Detection of SYT-SSX fusion transcripts in archival synovial sarcomas by real-time reverse transcriptase-polymerase chain reaction. J Mol Diagn 2002, 4:59-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Specht K, Richter T, Muller U, Walch A, Werner M, Hofler H: Quantitative gene expression analysis in microdissected archival formalin-fixed and paraffin-embedded tumor tissue. Am J Pathol 2001, 158:419-429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landegren U, Kaiser U, Sanders J, Hood L: A ligase-mediated gene detection technique. Science 1988, 241:1077-1080 [DOI] [PubMed] [Google Scholar]
- 18.Whiteley NM, Hunkapiller MW, Galzer AN: Detection of specific sequences in nucleic acids. U. S. Patent no. 4,885,750
- 19.Barany F: Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc Natl Acad Sci USA 1991, 88:189-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiedmann M, Barany F, Batt CA: Dieffenbach C Dveksler G eds. Ligase chain reaction: a PCR primer: a laboratory manual. 1995:631-652 Cold Spring Harbor Laboratory Press Cold Spring Harbor, NY
- 21.Kalin I, Shephard S, Candrian U: Evaluation of the ligase chain reaction (LCR) for the detection of point mutations. Mutat Res 1992, 283:119-123 [DOI] [PubMed] [Google Scholar]
- 22.Wilson VL, Wei Q, Wade KR, Chisa M, Bailey D, Kanstrup CM, Yin X, Jackson CM, Thompson B, Lee WR: Needle-in-a haystack detection and identification of base substitution mutations in human tissues. Mutat Res 1999, 406:79-100 [DOI] [PubMed] [Google Scholar]
- 23.Kissin DM, Holman S, Minkoff HL, DeMeo L, McCormack WM, DeHovitz JA: Epidemiology and natural history of ligase chain reaction detected chlamydial and gonococcal infections. Sex Transm Infect 2002, 78:208-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battle TJ, Golden MR, Suchland KL, Counts JM, Hughes JP, Stamm WE, Holmes KK: Evaluation of laboratory testing methods for Chlamydia trachomatis infection in the era of nucleic acid amplification. J Clin Microbiol 2001, 39:2924-2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang SX, Tay L: Early identification of Mycobacerium tuberculosis complex in BACTEC cultures by ligase chain reaction. J Med Microbiol 2002, 51:710-712 [DOI] [PubMed] [Google Scholar]
- 26.Ivannisci DM, Winn-Deen ES: Ligation amplification and fluorescence detection of Mycobacterium tuberculosis DNA. Mol Cell Probes 1993, 7:35-43 [DOI] [PubMed] [Google Scholar]
- 27.Eggerding FA: Fluorescent Oligonucleotide Ligation Technology for Identification of ras Oncogene Mutations. Mol Biotechnol 2000, 14:223-233 [DOI] [PubMed] [Google Scholar]
- 28.Eggerding FA, Iovannisci DM, Brinson E, Grossman P, Winn-Deen ES: Fluorescence-based oligonucleotide ligation assay for analysis of cystic fibrosis transmembrane conductance regulator gene mutations. Hum Mutat 1995, 5:153-65 [DOI] [PubMed] [Google Scholar]
- 29.Iannone MA, Taylor JD, Chen J, Li MS, Rivers P, Slentz-Kesler KA, Weiner MP: Multiplexed single nucleotide polymorphism genotyping by oligonucleotide ligation and flow cytometry. Cytometry 2000, 39:131-140 [PubMed] [Google Scholar]
- 30.Zhang Q-Y, Garner K, Viswanatha DS: Rapid detection of leukemia-associated translocation fusion genes using a novel combined RT-PCR and flow cytometric method. Leukemia 2002, 16:144-149 [DOI] [PubMed] [Google Scholar]
- 31.Tornkvist M, Brodin B, Bartolazzi A, Larsson O: A novel type of SYT/SSX fusion: methodological and biologic implications. Mod Pathol 2002, 15:679-685 [DOI] [PubMed] [Google Scholar]
- 32.O’Sullivan MJ, Humphrey PA, Dehner LP, Pfeifer JD: T(X;18) reverse transcriptase-polymerase chain reaction demonstrating a variant transcript. J Mol Diagn 2002, 4:178-180 [DOI] [PMC free article] [PubMed] [Google Scholar]