Abstract
Fluorescence in situ hybridization (FISH) has been used to demonstrate the t(14;18) in up to 100% of follicular lymphoma (FL) cases, however, there is little reproducible data using fixed tissue. The aim was therefore to develop a robust FISH method for the demonstration of translocations in archival tissue. The technique was evaluated by comparison with multiplex polymerase chain reaction (PCR), capable of detecting the majority of known breakpoints. Twenty-eight paired frozen and fixed cases of FL and 20 reactive controls were analyzed. The t(14;18) was detected in 23 of 28 cases using PCR on frozen material and 8 of 20 in paraffin. Using FISH, 24 of 26 frozen and 26 of 28 paraffin cases had a demonstrable translocation. All 20 reactive nodes were negative for the t(14;18) by PCR. Using FISH, one of the reactive cases had occasional cells with a translocation FISH pattern, demonstrable in frozen and paraffin samples. This is consistent with the presence of the t(14;18), which is well described in normal individuals. Both PCR and FISH are highly effective for t(14;18) analysis in unfixed tissue. When only paraffin blocks are available, FISH is the method of choice, and a result was achieved in 100% of cases. The method is applicable to the retrospective analysis of a range of translocations.
Follicular lymphoma (FL) is characterized by the presence of the t(14;18)(q32;q21) chromosomal translocation, which results in the rearrangement and up-regulation of the BCL2 proto-oncogene. The t(14;18) has traditionally been detected using cytogenetic assay or Southern blot analysis, with a reported incidence in follicular lymphoma of around 60 to 80%. 1, 2, 3 More recently, polymerase chain reaction (PCR) has been used, but highly variable assays have resulted in inconsistent results. 4 Between 40 and 70% of breakpoints can be demonstrated by major breakpoint region (MBR) PCR, and 5 to 10% using minor cluster region (mcr) primers. 5, 6, 7, 8, 9, 10 The remaining breakpoints are located 5′ of the BCL2 gene 11 and in the 20-kb region between the MBR and mcr. 12, 13, 14, 15 Long-distance (LD) PCR 6, 14, 15, 16 strategies have been used to identify breakpoints between the MBR and mcr subcluster regions. Positioned 4 kb downstream of the MBR is a further breakpoint region, the 3′MBR subcluster, encompassing a region of 3.8 kb, 12 and 10 kb upstream of the mcr is the 5′ mcr subcluster. 17, 18 LD-PCR techniques are not applicable to routine use, however, for an efficient PCR detection strategy all of these breakpoint regions need to be taken into account. The PCR strategy used in this study is a highly specific multiplex technique capable of detecting the majority of known breakpoints, including MBR, mcr, 3′MBR, and 5′mcr breakpoints and has been validated by the European BIOMED Group. 17, 18
The first reported use of fluorescence in situ hybridization (FISH)-based techniques for the demonstration of the t(14;18), were on cytogenetic samples and involved the demonstration of a break of the signal at 14q32, using chromosome paints, 19 a YAC containing the entire IgH locus, 20 or a dual color IgH break-apart FISH assay. 21 The t(14;18) has been detected in 100% of FLs using a FISH assay based on co-localization of YACs spanning the BCL2 and IgH genes 22 and a BCL2 break-apart interphase FISH assay, validated by comparison with fiber FISH 23, 24 In the present study, the Vysis LSI IGH/BCL2 probe set was used. This has the advantage over alternative FISH strategies in that it utilizes both probe splitting and co-localization, minimizing the risk of false-positives. Using this approach, the t(14;18) has been detected in 25 of 39 (64%) 25 and 63 of 63 (100%) 26 FLs.
For retrospective studies it is vital that molecular techniques used for the detection of translocations are applicable to paraffin-embedded tissue. Although PCR has been used successfully on paraffin-embedded tissue, 6, 27, 28, 29, 30, 31 the detection rate of the t(14;18) is significantly reduced due to poor quality of DNA. The application of FISH techniques for the detection of chromosomal translocations in paraffin tissue has been less well used, and the methodology is not well described and highly variable. The majority of studies have involved either whole chromosome paints or centromeric probes. 32, 33, 34, 35 Locus-specific probes have been used in paraffin material for the demonstration of the Philadelphia chromosome, 36, 37 p53 abnormalities, 38 cERB2 and cMYC amplification in gastric tumors, 39, 40 the t(11;14) in mantle cell lymphoma, 41, 42 the t(14;18) in diffuse large B-cell lymphoma, 43 and more recently FISH on nuclei extracted from cores of tissue taken from paraffin blocks has been used to demonstrate a range of abnormalities. 44
The aim of the study was to develop a relatively simple and reproducible FISH method for the demonstration of chromosomal translocations in archival formalin-fixed, paraffin-embedded tissue. The technique we describe has been evaluated by comparison with paired frozen samples, and with a highly sensitive PCR strategy in the same-paired samples.
Materials and Methods
Twenty-eight histologically defined cases of FL were used in the study. Cases were chosen based on the availability of paired frozen and paraffin-embedded samples. All cases were presentation lymph node biopsies of previously untreated patients. Twenty reactive lymph nodes were used as controls. All paraffin-embedded samples were fixed in 10% formalin and routinely processed.
Multiplex PCR Analysis of the t(14;18)
DNA was extracted from the paired frozen and paraffin-embedded samples in all cases. Amplification of a control gene (β−Globin, 320 bp) was used to assess the quality of the DNA from all frozen and formalin-fixed samples. If samples were not amplifiable initially, the control PCR was repeated on a 1:10 dilution of the DNA sample, to minimize the effect of inhibitors within the sample. Based on previous experience (data not presented), samples (neat or a 1:10 dilution) that failed to amplify to at least 320 bp were considered unsuitable for further PCR analysis. This has also been confirmed by the findings of the Biomed group, who showed that amplification of a 400-bp control gene generally indicated a sample that was suitable for further analysis (work carried out by Dr. H. White 17 ). Twenty-eight of 28 of the frozen samples and 20 of 28 of the fixed samples were suitable for t(14;18) analysis. A single-round multiplex PCR technique was performed on all samples using the European Biomed-2 concerted action t(14;18) primers and protocols. 17, 18 Briefly, two multiplex reactions were used. These included an MBR multiplex strategy and a mcr/3′ MBR/5′ mcr multiplex, used in conjunction with a consensus JH primer. Positive, negative, and no template controls were run in all experiments. The sensitivity of the multiplex reactions was evaluated by amplifying DNA dilutions of the cell lines DoHH2 (MBR +ve), SC-1 (mcr +ve), Oz (5′mcr), and K231 (3′MBR) in normal tonsil DNA. A sensitivity of between 3.3 × 10−2 and 10−3 was consistently achievable using both multiplex reactions (data not shown).
All samples were analyzed on at least two occasions. Amplification was performed in an automated thermal cycler (Geneamp 9700 or 2700 PCR system; PE Biosystems, Foster City, CA) and was identical for each multiplex reaction. Each 50-μl reaction was performed using 100 ng genomic DNA in 1X reaction buffer containing 1.5 mmol/L MgCl2 (Amplitaq Gold Buffer II, PE Biosystems), 0.125 mmol/L dNTP’s (Pharmacia Biotech, Amersham, England), 10 pmols of each primer, and 1 unit of Taq polymerase (Amplitaq Gold, PE Biosystems). Amplification was performed as follows: initial denaturation/enzyme activation at 94°C for 10 minutes; followed by 35 cycles consisting of denaturation at 94°C for 30 seconds, annealing at 60°C for 1 minute, and extension at 72°C for 1 minute; followed by a final extension of 72°C for 10 minutes and rapid cooling to 4°C. Products were resolved on 2% agarose gels and visualized under ultraviolet illumination with ethidium bromide staining.
FISH Analysis
FISH for the t(14;18) was performed on both the frozen and paraffin-embedded samples on all FL and reactive cases using the Vysis LSI IgH Spectrum Green/LSI BCL2 Spectrum Orange probe set (Vysis Inc., No. 32–191018), which includes the probe and the hybridization buffer.
FISH on Touch Preparations from Frozen Tissue
The lymph nodes were taken from the store, allowed to defrost, and a microscope slide was gently touched onto the tissue surface to make an impression. FISH was performed after allowing the slides to air dry.
FISH on Whole Nuclei Extracted from Paraffin-Embedded Tissue
This method was developed by the systematic evaluation of various methodologies, pre-treatments, and enzymes (data not shown). The chosen method, first described for the flow cytometric analysis of ploidy and S-phase, 45 was found to give the most consistent results with a range of paraffin tissue from various sources and with diverse fixation and processing protocols (results not shown).
Between 5 and 10 (depending on the size of the tissue in the block) 35-μm thick paraffin sections were cut, dewaxed in xylene at 37°C, and rehydrated through graded alcohols. Following two changes of distilled water, the sections were incubated in pre-warmed digestion buffer (0.1 mol/L Tris; 0.07 mol/L NaCl; 0.1% NP-40; pH 7.4) at 37°C for 30 minutes. The digestion buffer was replaced with fresh, pre-warmed buffer containing 0.05% Protease XXIV (Sigma P8038). The sections were agitated for 1 hour at 37°C. All processing steps were carried out in 10-ml glass or xylene resistant plastic specimen tubes and the waste solvents were changed by careful pipetting. Digested nuclei were harvested by pipetting the supernatant nuclear suspension from the undigested tissue, and centrifuging at 1800 × g. The harvested nuclei were washed once in phosphate-buffered saline (PBS) and once in 3:1 methanol acetic acid (MAA). Centrifugation at each stage was at 1800 × g. The nuclei were resuspended in MAA, and the nuclear suspension was dropped by pipette onto 3-aminopropyltriethoxy-silane (APES)-coated microscope slides. An optimum concentration resulted in an evenly spread monolayer of nuclei on the slide. The concentration and morphology of the nuclei were checked by staining a slide with May Grunwold Giemsa before commencing FISH.
FISH Method
FISH for the t(14;18) was carried out according to the manufacturer’s instructions. Briefly, the slides were fixed in MAA for at least 30 minutes, and incubated in 2X standard saline citrate (SSC) at 37°C for at least 1 hour. Following dehydration through graded alcohols, the probe was applied in hybridization buffer, and a coverslip was sealed on using rubber cement.
Denaturation/Hybridization
An additional pre-denaturation step was used for the paraffin nuclear preparations. The slides were incubated in hybridization buffer at 85 to 90°C for 2 to 20 minutes, and then allowed to cool back to 37°C. (The pre-denaturation time and temperature can be adjusted accordingly to obtain optimal hybridization efficiency). Using a Vysis Hybrite machine, denaturation was 73°C for 3 minutes and 75°C for 5 minutes for the frozen and paraffin preparations, respectively, and hybridization was at 37°C for 16 to 24 hours.
Post-Hybridization Washes
Post-hybridization washes consisted of 2 × 2 minutes in pre-warmed 0.4XSSC/0.3XNP-40 at 70°C; followed by 5 minutes in 2X SSC/0.1X NP-40 at room temperature. 4′-6-diamino-2-phenylindole (DAPI) was used as the counterstain.
Interpretation of FISH
Representative images were captured using Metasystems ISIS software. Cases were defined as normal if there were two green (IgH) and two red (BCL2) signals (Figure 1a) . A number of cases showed a significant proportion of nuclei in which one each of the signals was co-localizing. In the absence of any extra signals these cases were also classed as normal.
Figure 1.
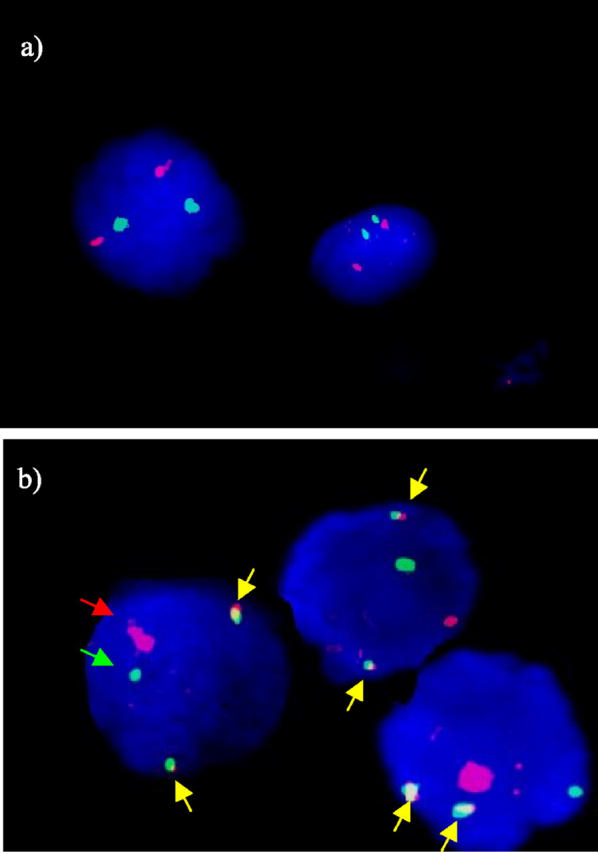
Representative FISH images of the pattern of staining of the BCL2/IgH probe set. a: Normal pattern of staining. Each cell nucleus has two red (BCL2) and two green (IgH) FISH signals, one for each copy of the chromosome. The counterstain is DAPI, which is visualized as blue fluorescence. b: A case with a t(14;18). Each nucleus has three signals with both BCL2 and IgH probe sets, indicating a “split” in one each of the copies of the genes. The reciprocal translocation, t(14;18), is demonstrated by the presence of two fusion signals in each cell (indicated by yellow arrows), along with a residual normal copy of each gene. The counterstain is DAPI.
A t(14;18) was defined when there were extra signals of both the BCL2 and the IgH probes, indicative of a chromosomal break, along with at least two of the extra signals co-localizing to produce a yellow fusion signal (Figure 1b) , as described in the product datasheet. Extra signals of either BCL2 or IgH in the absence of co-localization would be suggestive of either an alternative translocation partner or extra copies of the gene or chromosome.
The entire preparation was analyzed and at least 100 nuclei were formally counted in each case. In general, a cut-off of 5% was used to define a t(14;18)-positive case. If <5% of cells had the classical translocation pattern of signals, up to 500 additional nuclei were counted. Given the stringent criteria used to define the presence of the translocation within a cell, a threshold as low as one cell per preparation is sufficient to class a case as positive, since it is not possible for the translocation pattern of signals to occur by chance.
Results
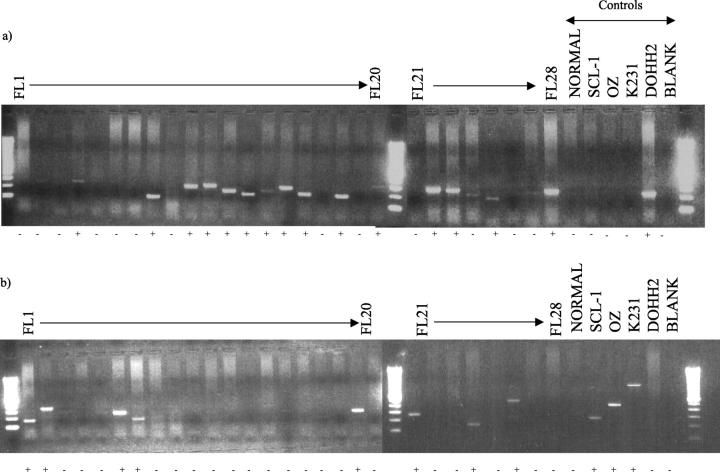
We were able to detect the translocation in 23 of 28 cases using PCR on frozen material, breakpoints were detected at the MBR in 16 of 23 (70%), mcr in 5 of 23 (21%), and 5′mcr in 2 of 23 (9%). No cases were demonstrated with a breakpoint in the 3′MBR region. Figure 2 shows the agarose gel electrophoresis of the MBR and mcr multiplex PCR products from DNA extracted from 28 frozen FL lymph nodes. Eight of 20 (40%) cases with amplifiable DNA were positive using PCR on paraffin material. Using FISH, 24 of 26 cases were positive on the frozen touch preparations, and 26 of 28 were positive on the paraffin nuclei. In five cases the efficiency of hybridization was low, however occasional cells (<5%) were seen with the classical translocation pattern of signals, ie, three red, three green signals with two fusion signals, and a residual red and green signal. All five cases showed concordance between the frozen and paraffin preparations and in 3 of 5 of these cases, the t(14;18) was demonstrated using PCR. These cases were therefore classified as t(14;18)-positive. No cases were positive by PCR but negative by FISH. The results are detailed in Table 1 .
Figure 2.
t(14;18) multiplex PCR analysis on DNA extracted from 28 frozen biopsies of FL. a: MBR multiplex (JH + MBR): 15 of 28 FL cases had a visible band and were classed as positive (+). b: mcr multiplex (JH + mcr, 5′mcr, 3′MBR): 8 of 28 FL cases had a visible band and were classed as positive (+).
Table 1.
Detection of the t(14;18) in 28 Cases of Frozen and Paraffin Samples of FL, Comparing the Effectiveness of PCR to FISH
| Case no. | Amplifable DNA (DNA dilution factor) | PCR for t(14;18) | FISH for t(14;18) | |||
|---|---|---|---|---|---|---|
| Frozen | Paraffin | Frozen | Paraffin | Frozen | Paraffin | |
| 1 | + | + (1/10) | MCR + | − | + | ++ |
| 2 | + | − | 5′ MCR + | NT | Failed | + |
| 3 | + | + (1/10) | − | − | + | + |
| 4 | + | + (1/10) | MBR + | − | +++ | +++ |
| 5 | + | + (1/10) | − | − | − | − |
| 6 | + | + (1/10) | MCR + | + | ++ | ++ |
| 7 | + | + | + | − | ++ | + |
| 8 | + | − | MBR + | NT | ++ | + |
| 9 | + | − | − | NT | − | − |
| 10 | + | + (1/10) | MBR + | + | ++ | + |
| 11 | + | + | MBR + | − | + | ++ |
| 12 | + | + (1/10) | MBR + | + | + | + |
| 13 | + | + | MBR + | + | +++ | +++ |
| 14 | + | + | MBR + | − | ++ | + |
| 15 | + | + | MBR + | + | ++ | ++ |
| 16 | + | + | MBR + | + | + | + |
| 17 | + | + | − | − | ++ | + |
| 18 | + | − | MBR + | NT | ++ | + |
| 19 | + | + (1/10) | MCR + | + | +++ | ++ |
| 20 | + | + | MBR + | − | + | + |
| 21 | + | − | MCR + | NT | ++ | + |
| 22 | + | − | MBR + | NT | ++ | ++ |
| 23 | + | + | MBR + | − | ++ | + |
| 24 | + | + (1/10) | MCR + | + | ++ | ++ |
| 25 | + | − | MBR + | NT | Failed | + |
| 26 | + | + | 5′ MCR + | − | ++ | ++ |
| 27 | + | + | − | − | +* | +* |
| 28 | + | − | MBR + | NT | ++ | ++ |
, Case 27 appeared to have a complex translocation pattern by FISH analysis. Occasional cells were seen with the classical translocation pattern, but additional cells with extra BCL2 signals but no co-localization, and cells with extra IgH signals but without co-localization were also observed.
NT, not tested: no amplifyable DNA; MCR, minor cluster region; MBR, major breakpoint region; +, occasional double fusions; ++, majority of cells double fusions; +++, double and triple fusions.
The concordance of results between the frozen and paraffin-embedded samples was 100% with the FISH method, compared to 60% by PCR. Eight of the 20 cases with amplifiable paraffin-extracted DNA gave a false-negative result by comparison with the PCR on DNA extracted from frozen tissue.
Analysis of Reactive Lymph Nodes
Amplifiable DNA was produced in all reactive lymph nodes in both the frozen and paraffin-embedded paired samples. All cases were negative for the t(14;18) by PCR. Using the FISH method, 1 of 20 of the reactive cases had occasional cells (<5%) with the classical translocation pattern of signals, ie, two co-localized signals along with a residual red and green signal. This was demonstrated in both the frozen touch preparation and the paraffin-extracted nuclei.
Discussion
In this study we have described an effective FISH method, using the Vysis LSI IGH/BCL2 probe set, for the demonstration of the t(14;18) on archival material. The probe set consists of a 1.5 Mb locus-specific IgH probe spanning the entire IgH locus and extending 300 kb beyond the 3′ end, and a 750 kb BCL2 probe spanning the entire BCL2 gene and extending 250 kb both distal and proximal to the gene, labeled Spectrum Green and Spectrum Orange, respectively. This has the advantage over previous FISH strategies in that it utilizes both probe splitting and co-localization. These stringent criteria used to define the translocation have the effect of minimizing false-positives that may occur as a result of random localization of signals, or due to the structural organization of the chromosomes within the nucleus. 46, 47, 48
The method was validated by analysis of the same series of samples using PCR, and by the analysis of paired frozen and paraffin samples using both methods. The PCR strategy used was the method developed and validated by the European Biomed group. 17, 18 The multiplex approach is based on the detailed investigation of breakpoints and primer binding and enables the detection of the majority of known breakpoints, making this PCR method a highly effective technique for the detection of the t(14;18).
The detection rate of these primers has also been extensively studied in paraffin-embedded material 17 and it was shown that amplification of a 400-bp control gene indicated that a sample was suitable for further analysis (work carried out by Dr. H. White 17 ). Amplifiable DNA was obtained in 100% of the frozen lymph nodes and 71% of paraffin cases, which is superior to previous studies that have reported amplifiable DNA in 60% and 50% of cases, respectively. 31
In frozen tissue, all cases produced a result by PCR, however two cases failed using FISH, due to insufficient cells on the touch preparation. The t(14;18) was positive in 82% and 92% of FL cases, respectively. In archival formalin-fixed, paraffin wax-embedded tissue, a result was achieved in 100% of cases using the FISH method, compared to 71% using PCR. In paraffin, the t(14;18) was detected in 93% and 40% of cases, respectively. The reported incidence of the t(14;18) as detected by MBR and mcr PCR is highly variable; reported positive, at best, in 82% of FL patients by analysis of fresh material, 9 and 47% by analysis of DNA extracted from paraffin tissue. 28 In contrast, the t(14;18) has been demonstrated in up to 100% of cases of FL using FISH. 23, 26
Concordance of results comparing FISH and PCR was 88% in frozen tissue and 45% in paraffin tissue. There was a 100% correlation using FISH in the paired frozen and paraffin samples and all cases that were positive using frozen tissue PCR were positive using FISH. In contrast, a significant false-negative result was seen using PCR on archival DNA, with 40% of cases giving a false-negative result compared with PCR on frozen tissue.
Using FISH analysis, a proportion of cases had only occasional cells with the translocation. The low percentage of positive cells is most likely to be due to a high percentage of reactive T and B cells in the lymph node sample. Given the stringent criteria used to define the translocation using FISH, it is unlikely that this low frequency of positive cells was an artifact. In contrast, this suggests that the method is highly sensitive, provided that large numbers of cells are counted.
Using PCR, five cases were translocation-negative in the frozen tissue samples, however only two of these cases were negative using FISH. Of the remaining three cases that were positive by FISH, but negative by PCR, two had only occasional cells positive for the translocation. This highlights a possible sensitivity issue for the PCR technique used in this study, which may help to explain the false-negative results obtained using PCR compared to FISH and in paraffin material compared to the matched frozen tissue. The t(14;18) multiplex developed by the Biomed group is based on the use of a single round PCR amplification, which is sensitive up to 1 in 1000. Previous studies using DNA derived from paraffin-embedded material have used a nested PCR strategy, which has been shown to have an increased sensitivity up to 1000-fold higher than conventional PCR. In addition, PCR requires absolute sequence complementarity that is not as crucial for FISH. In follicular lymphoma, ongoing somatic hypermutation is a key feature and may result in base changes at the primer-binding site, reducing the complementarity. The further case that was positive by FISH but negative by PCR appeared to have a complex translocation pattern. Occasional cells had the classical FISH pattern, but additional cells had extra BCL2 and/or extra IgH signals but without co-localization. This highlights an additional advantage of FISH over PCR for the detection of gene rearrangements, in that PCR will not detect alternative breakpoints that are outside the regions covered by the PCR strategy. Single fusions signals were seen using FISH in a number of cases, and was particularly common in the reactive cases. In the absence of a split of either signal, these cases were classified as translocation-negative. This pattern of signals can be explained simply by the spatial organization of the chromosomes within the nucleus. 46, 47, 48 An alternative explanation is the insertion of the entire BCL2 gene into IgH, which has been reported in FL 24 and would also manifest as a single fusion using FISH, however this possibility was not further investigated. The main disadvantage of this FISH method compared to PCR is that no information regarding the breakpoint can be given. However, due to the mapping of the probes, a positive result was observed by FISH analysis regardless of the breakpoint demonstrated by PCR.
The technique was further validated by analysis of a small series of reactive lymph node biopsies. All cases were PCR-negative and 19 of 20 cases gave a negative FISH result in both the paraffin and frozen material. One reactive case showed a very occasional cell with the classical translocation pattern of signals. This was demonstrated in both the paraffin and the frozen tissue, but was not confirmed by PCR. The presence of the t(14;18) translocation has been well described in reactive tissue by PCR 49, 50, 51, 52 and FISH, 22 and in the peripheral blood of normal individuals. 53, 54, 55 Using TaqMan real-time PCR, 23% of normal individuals were found to be positive for the translocation, with 3% of these at a level of more than 1 in 10,000 cells. 56 As described above, the FISH technique is highly specific for the translocation and is unlikely to producing a false-positive result. The most likely explanation is that the translocation was unable to be detected by PCR for the reasons highlighted above.
Both PCR and FISH are highly effective techniques for the analysis of t(14;18) where unfixed tissue is available. When only paraffin blocks are available, FISH is superior to even this gold standard PCR technique. The main advantage of the method described here is the flexibility it offers for samples that have been fixed and processed using a range of different protocols, which is of particular interest in the clinical trial setting or in a reference laboratory. All cases can be digested using the standard enzyme protocol, and the pre-denaturation time can be adjusted accordingly.
FISH may also have the added advantage over PCR in the analysis of complex translocations. Three cases were demonstrated to have multiple fusion signals by FISH analysis, but there was no difference in these cases by PCR. Although the significance of additional fusion signals is not well understood, it may have prognostic significance. The relative costs of FISH and PCR per test would favor the use of PCR. If cost is a major concern then to minimize expenses, PCR could be used as a “screen” for the presence of the translocation, and FISH only performed on cases that are PCR-negative.
In conclusion, the FISH method described here is a highly sensitive technique for the demonstration of the t(14;18) in archival tissue. The method can be applied to enable the retrospective analysis of a range of translocations, and may be useful in determining the diagnosis in problem cases, for example when FL and marginal zone lymphoma or reactive hyperplasia are the differential diagnoses.
Address reprint requests to Sharon L. Barrans, Haematological Malignancy Diagnostic Service, Academic Unit of Haematology and Oncology, Leeds General Infirmary, Leeds, LS1 3EX, United Kingdom. E-mail: sharonb@hmds.org.uk.
Footnotes
Supported by a grant from the UK Leukaemia Research Fund.
References
- 1.Bloomfield CD, Arthur DC, Frizzera G, Levine EG, Peterson BA, Gajl-Peczalska KJ: Non-random chromosome abnormalities in lymphoma. Cancer Res 1983, 43:2975-2984 [PubMed] [Google Scholar]
- 2.Levine EG, Arthur DC, Frizzera G, Peterson BA, Hurd DD, Bloomfield CD: There are differences in cytogenetic abnormalities among histologic subtypes of the non-Hodgkin’s lymphomas. Blood 1985, 66:1414-1422 [PubMed] [Google Scholar]
- 3.Horsman DE, Gascoyne RD, Coupland RW, Coldman AJ, Adomat SA: Comparison of cytogenetic analysis, Southern analysis, and polymerase chain reaction for the detection of t(14;18) in follicular lymphoma. Am J Clin Pathol 1995, 103:472-478 [DOI] [PubMed] [Google Scholar]
- 4.Johnson PW, Swinbank K, MacLennan S, Colomer D, Debuire B, Diss T, Gabert J, Gupta RK, Haynes A, Kneba M, Lee MS, Macintyre E, Mensink E, Moos M, Morgan GJ, Neri A, Johnson A, Reato G, Salles G, van’t Veer MB, Zehnder JL, Zucca E, Selby PJ, Cotter FE: Variability of polymerase chain reaction detection of the bcl-2-IgH translocation in an international multicentre study. Ann Oncol 1999, 10:1349-1354 [DOI] [PubMed] [Google Scholar]
- 5.Corbally N, Grogan L, Dervan PA, Carney DN: The detection of specific gene rearrangements in non-Hodgkin’s lymphoma using the polymerase chain reaction. Br J Cancer 1992, 66:805-809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pezzella F, Ralfkiaer E, Gatter KC, Mason DY: The 14;18 translocation in European cases of follicular lymphoma: comparison of Southern blotting and the polymerase chain reaction. Br J Haematol 1990, 76:58-64 [DOI] [PubMed] [Google Scholar]
- 7.Said JW, Sassoon AF, Shintaku IP, Corcoran P, Nichols SW: Polymerase chain reaction for bcl-2 in diagnostic lymph node biopsies. Mod Pathol 1990, 3:659-663 [PubMed] [Google Scholar]
- 8.Galoin S, al Saati T, Schlaifer D, Huynh A, Attal M, Delsol G: Oligonucleotide clonospecific probes directed against the junctional sequence of t(14;18): a new tool for the assessment of minimal residual disease in follicular lymphomas. Br J Haematol 1996, 94:676-684 [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Guillermo A, Cabanillas F, McDonnell TI, McLaughlin P, Smith T, Pugh W, Hagemeister F, Rodriguez MA, Romaguera JE, Younes A, Sarris AH, Preti HA, Lee MS: Correlation of bcl-2 rearrangement with clinical characteristics and outcome in indolent follicular lymphoma. Blood 1999, 93:3081-3087 [PubMed] [Google Scholar]
- 10.Aster JC, Longtine JA: Detection of BCL2 rearrangements in follicular lymphoma. Am J Pathol 2002, 160:759-763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsujimoto Y, Bashir MM, Givol I, Cossman J, Jaffe E, Croce CM: DNA rearrangements in human follicular lymphoma can involve the 5′ or the 3′ region of the bcl-2 gene. Proc Natl Acad Sci USA 1987, 84:1329-1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchonnet G, Lenain P, Ruminy P, Lepretre S, Stamatoullas A, Parmentier F, Jardin F, Duval C, Tilly H, Bastard C: Characterisation of BCL2-JH rearrangements in follicular lymphoma: PCR detection of 3′ BCL2 breakpoints and evidence of a new cluster. Leukemia 2000, 14:1563-1569 [DOI] [PubMed] [Google Scholar]
- 13.Wang YL, Addya K, Edwards RH, Rennert H, Dodson L, Leonard DG, Wilson RB: Novel bcl-2 breakpoints in patients with follicular lymphoma. Diagn Mol Pathol 1998, 7:85-89 [DOI] [PubMed] [Google Scholar]
- 14.Willis TG, Jadayel DM, Coignet LJ, Abdul-Rauf M, Treleaven JG, Catovsky D, Dyer MJ: Rapid molecular cloning of rearrangements of the IGHJ locus using long-distance inverse polymerase chain reaction. Blood 1997, 90:2456-2464 [PubMed] [Google Scholar]
- 15.Akasaka T, Akasaka H, Yonetani N, Ohno H, Yamabe H, Fukuhara S, Okuma M: Refinement of the BCL2/immunoglobulin heavy chain fusion gene in t(14;18)(q32;q21) by polymerase chain reaction amplification for long targets. Genes Chromosomes Cancer 1998, 21:17-29 [DOI] [PubMed] [Google Scholar]
- 16.Albinger-Hegyi A, Hochreutener B, Abdou MT, Hegyi I, Dours-Zimmermann MT, Kurrer MO, Heitz PU, Zimmermann DR: High frequency of t(14;18)-translocation breakpoints outside of major breakpoint and minor cluster regions in follicular lymphomas: improved polymerase chain reaction protocols for their detection. Am J Pathol 2002, 160:823-832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Dongen JJ, Langerak AW, Bruggemann M, Evans PAS, Hummel M, Lavender L, Delabesse E, Davi F, Schuuring E, Garzia Sanz R, van Krieken JH, Does J, Gondalez Diaz D, Bastard C, Hodges L, Spaargaren M, San Miguel JF, Parreira A, Smith J, Morgan GJ, Kneba M, Macintyre EA: Design and standardisation of PCR primers and protocols for detection of clonal immunoglobulin and T cell receptor gene rearrangements in suspect lymphoproliferations: report of the BIOMED-2 concerted action BMH4-CT98–3936. Leukemia 2003, in press [DOI] [PubMed]
- 18.van Dongen JJ, Langerak AW, San Miguel JF, Parreira A, Smith JL, Morgan GJ, Kneba M, Macintyre E: PCR-based clonality studies for early diagnosis of lymphoproliferative disorders: report of the BIOMED-2 concerted action. Blood 2001, 98:129a(Abstract) [Google Scholar]
- 19.Dirks W, Nolte M, Werner M, Jager K, Koch C, Drexler HG: Preservation of functional and regulatory domains of expressed bcl-2 genes in non-Hodgkin’s lymphoma. Leukemia 1996, 10:150-158 [PubMed] [Google Scholar]
- 20.Taniwaki M, Matsuda F, Jauch A, Nishida K, Takashima T, Tagawa S, Sugiyama H, Misawa S, Abe T, Kashima K: Detection of 14q32 translocations in B-cell malignancies by in situ hybridization with yeast artificial chromosome clones containing the human IgH gene locus. Blood 1994, 83:2962-2969 [PubMed] [Google Scholar]
- 21.Taniwaki M, Nishida K, Ueda Y, Misawa S, Nagai M, Tagawa S, Yamagami T, Sugiyama H, Abe M, Fukuhara S: Interphase and metaphase detection of the breakpoint of 14q32 translocations in B-cell malignancies by double-color fluorescence in situ hybridization. Blood 1995, 85:3223-3228 [PubMed] [Google Scholar]
- 22.Poetsch M, Weber-Matthiesen K, Plendl HJ, Grote W, Schlegelberger B: Detection of the t(14;18) chromosomal translocation by interphase cytogenetics with yeast-artificial-chromosome probes in follicular lymphoma and non-neoplastic lymphoproliferation. J Clin Oncol 1996, 14:963-969 [DOI] [PubMed] [Google Scholar]
- 23.Vaandrager JW, Schuuring E, Raap T, Philippo K, Kleiverda K, Kluin P: Interphase FISH detection of BCL2 rearrangement in follicular lymphoma using breakpoint-flanking probes. Genes Chromosomes Cancer 2000, 27:85-94 [PubMed] [Google Scholar]
- 24.Vaandrager JW, Schuuring E, Philippo K, Kluin PM: V(D)J recombinase-mediated transposition of the BCL2 gene to the IGH locus in follicular lymphoma. Blood 2000, 96:1947-1952 [PubMed] [Google Scholar]
- 25.Frater JL, Tsiftsakis EK, Hsi ED, Pettay J, Tubbs RR: Use of novel t(11;14) and t(14;18) dual-fusion fluorescence in situ hybridization probes in the differential diagnosis of lymphomas of small lymphocytes. Diagn Mol Pathol 2001, 10:214-222 [DOI] [PubMed] [Google Scholar]
- 26.Godon A, Moreau A, Talmant P, Baranger-Papot L, Genevieve F, Milpied N, Zandecki M, Avet-Loiseau H: Is t(14;18)(q32;q21) a constant finding in follicular lymphoma? An interphase FISH study on 63 patients. Leukemia 2003, 17:255-259 [DOI] [PubMed] [Google Scholar]
- 27.Hickish T, Cunningham D: Detecting t(14;18) in paraffin-embedded lymphoma tissue. Lancet 1989, 2:218-219 [DOI] [PubMed] [Google Scholar]
- 28.Shibata D, Hu E, Weiss LM, Brynes RK, Nathwani BN: Detection of specific t(14;18) chromosomal translocations in fixed tissues. Hum Pathol 1990, 21:199-203 [DOI] [PubMed] [Google Scholar]
- 29.Limpens J, Beelen M, Stad R, Haverkort M, van Krieken JH, van Ommen GJ, Kluin PM: Detection of the t(14;18) translocation in frozen and formalin-fixed tissue. Diagn Mol Pathol 1993, 2:99-107 [PubMed] [Google Scholar]
- 30.Miettinen M, Lasota J: Polymerase chain reaction-based gene rearrangement studies in the diagnosis of follicular lymphoma: performance in formaldehyde-fixed tissue and application in clinical problem cases. Pathol Res Pract 1997, 193:9-19 [DOI] [PubMed] [Google Scholar]
- 31.Liu J, Johnson RM, Traweek ST: Rearrangement of the BCL-2 gene in follicular lymphoma: detection by PCR in both fresh and fixed tissue samples. Diagn Mol Pathol 1993, 2:241-247 [PubMed] [Google Scholar]
- 32.Yoshida H, Nagao K, Ito H, Yamamoto K, Ushigome S: Chromosomal translocations in human soft tissue sarcomas by interphase fluorescence in situ hybridization. Pathol Int 1997, 47:222-229 [DOI] [PubMed] [Google Scholar]
- 33.Aoki T, Hisaoka M, Kouho H, Hashimoto H, Nakata H: Interphase cytogenetic analysis of myxoid soft tissue tumors by fluorescence in situ hybridization and DNA flow cytometry using paraffin-embedded tissue. Cancer 1997, 79:284-293 [DOI] [PubMed] [Google Scholar]
- 34.Nagao K, Ito H, Yoshida H, Minamizaki T, Furuse K, Yoshikawa T, Ushigome S: Chromosomal rearrangement t(11;22) in extraskeletal Ewing’s sarcoma and primitive neuroectodermal tumour analysed by fluorescence in situ hybridization using paraffin-embedded tissue. J Pathol 1997, 181:62-66 [DOI] [PubMed] [Google Scholar]
- 35.Nagao K, Ito H, Yoshida H: Chromosomal translocation t(X;18) in human synovial sarcomas analyzed by fluorescence in situ hybridization using paraffin-embedded tissue. Am J Pathol 1996, 148:601-609 [PMC free article] [PubMed] [Google Scholar]
- 36.Brynes RK, McCourty A, Ho JP, Traweek ST, Snyder DS, Slovak ML: Detection of the Philadelphia chromosome in paraffin-embedded tissue by fluorescence in situ hybridization. Mod Pathol 1994, 7:565-569 [PubMed] [Google Scholar]
- 37.Nolte M, Werner M, Ewig M, von Wasielewski R, Wilkens L, Georgii A: Demonstration of the Philadelphia translocation by fluorescence in situ hybridization (FISH) in paraffin sections and identification of aberrant cells by a combined FISH/immunophenotyping approach. Histopathology 1995, 26:433-437 [DOI] [PubMed] [Google Scholar]
- 38.Li X, Tsuji T, Wen S, Mimura Y, Sasaki K, Shinozaki F: Detection of numeric abnormalities of chromosome 17 and p53 deletions by fluorescence in situ hybridization in pleomorphic adenomas and carcinomas in pleomorphic adenoma: correlation with p53 expression. Cancer 1997, 79:2314-2319 [DOI] [PubMed] [Google Scholar]
- 39.Ooi A, Kobayashi M, Mai M, Nakanishi I: Amplification of c-erbB-2 in gastric cancer: detection in formalin-fixed, paraffin-embedded tissue by fluorescence in situ hybridization. Lab Invest 1998, 78:345-351 [PubMed] [Google Scholar]
- 40.Hara T, Ooi A, Kobayashi M, Mai M, Yanagihara K, Nakanishi I: Amplification of c-myc, K-sam, and c-met in gastric cancers: detection by fluorescence in situ hybridization. Lab Invest 1998, 78:1143-1153 [PubMed] [Google Scholar]
- 41.Li JY, Gaillard F, Moreau A, Harousseau JL, Laboisse C, Milpied N, Bataille R, Avet-Loiseau H: Detection of translocation t(11;14)(q13;q32) in mantle cell lymphoma by fluorescence in situ hybridization. Am J Pathol 1999, 154:1449-1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Remstein ED, Kurtin PJ, Buno I, Bailey RJ, Proffitt J, Wyatt WA, Hanson CA, Dewald GW: Diagnostic utility of fluorescence in situ hybridization in mantle-cell lymphoma. Br J Haematol 2000, 110:856-862 [DOI] [PubMed] [Google Scholar]
- 43.Huang JZ, Sanger WG, Greiner TC, Staudt LM, Weisenburger DD, Pickering DL, Lynch JC, Armitage JO, Warnke RA, Alizadeh AA, Lossos IS, Levy R, Chan WC: The t(14;18) defines a unique subset of diffuse large B-cell lymphoma with a germinal center B-cell gene expression profile. Blood 2002, 99:2285-2290 [DOI] [PubMed] [Google Scholar]
- 44.Paternoster SF, Brockman SR, McClure RF, Remstein ED, Kurtin PJ, Dewald GW: A new method to extract nuclei from paraffin-embedded tissue to study lymphomas using interphase fluorescence in situ hybridization. Am J Pathol 2002, 160:1967-1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heiden T, Wang N, Tribukait B: An improved Hedley method for preparation of paraffin-embedded tissues for flow cytometric analysis of ploidy and S-phase. Cytometry 1991, 12:614-21 [DOI] [PubMed] [Google Scholar]
- 46.Nagele R, Freeman T, McMorrow L, Lee HY: Precise spatial positioning of chromosomes during prometaphase: evidence for chromosomal order. Science 1995, 270:1831-1835 [DOI] [PubMed] [Google Scholar]
- 47.Manuelidis L: A view of interphase chromosomes. Science 1990, 250:1533-1540 [DOI] [PubMed] [Google Scholar]
- 48.Qumsiyeh MB: Impact of rearrangements on function and position of chromosomes in the interphase nucleus and on human genetic disorders. Chromosome Res 1995, 3:455-465 [DOI] [PubMed] [Google Scholar]
- 49.Limpens J, de Jong D, van Krieken JH, Price CG, Young BD, van Ommen GJ, Kluin PM: Bcl-2/JH rearrangements in benign lymphoid tissues with follicular hyperplasia. Oncogene 1991, 6:2271-2276 [PubMed] [Google Scholar]
- 50.Aster JC, Kobayashi Y, Shiota M, Mori S, Sklar J: Detection of the t(14;18) at similar frequencies in hyperplastic lymphoid tissues from American and Japanese patients. Am J Pathol 1992, 141:291-299 [PMC free article] [PubMed] [Google Scholar]
- 51.Ohshima K, Kikuchi M, Kobari S, Masuda Y, Eguchi F, Kimura N: Amplified bcl-2/JH rearrangements in reactive lymphadenopathy. Virchows Arch B Cell Pathol 1993, 63:197-198 [DOI] [PubMed] [Google Scholar]
- 52.Estalilla OC, Medeiros LJ, Manning JT, Jr, Luthra R: 5′->3′ exonuclease-based real-time PCR assays for detecting the t(14;18)(q32;21): a survey of 162 malignant lymphomas and reactive specimens. Mod Pathol 2000, 13:661-666 [DOI] [PubMed] [Google Scholar]
- 53.Limpens J, Stad R, Vos C, de Vlaam C, de Jong D, van Ommen GJ, Schuuring E, Kluin PM: Lymphoma-associated translocation t(14;18) in blood B cells of normal individuals. Blood 1995, 85:2528-2536 [PubMed] [Google Scholar]
- 54.Dolken G, Illerhaus G, Hirt C, Mertelsmann R: BCL-2/JH rearrangements in circulating B cells of healthy blood donors and patients with non-malignant diseases. J Clin Oncol 1996, 14:1333-1344 [DOI] [PubMed] [Google Scholar]
- 55.Rauzy O, Galoin S, Chale JJ, Adoue D, Albarede JL, Delsol G, al Saati T: Detection of t(14;18) carrying cells in bone marrow and peripheral blood from patients affected by non-lymphoid diseases. Mol Pathol 1998, 51:333-338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Summers KE, Goff LK, Wilson AG, Gupta RK, Lister TA, Fitzgibbon J: Frequency of the Bcl-2/IgH rearrangement in normal individuals: implications for the monitoring of disease in patients with follicular lymphoma. J Clin Oncol 2001, 19:420-424 [DOI] [PubMed] [Google Scholar]