Abstract
Using cDNA microarrays we determined the gene expression patterns in the human acute promyelocytic leukemia (APL) cell line NB4 during all-trans retinoic acid (ATRA)-induced differentiation. We analyzed the expression of 12,288 genes in the NB4 cells after 12 hours, 24 hours, 48 hours, 72 hours, and 96 hours of ATRA exposure. During this time course, we found 168 up-regulated and more than 179 down-regulated genes, most of which have not been reported before. Many of the altered genes encode products that participate in signaling pathways, cell differentiation, programmed cell death, transcription regulation, and production of cytokines and chemokines. Of interest, the CD52 and protein kinase A regulatory subunit α (PKA-Rlα) genes, whose products are being used as therapeutic targets for certain human neoplasias in currently ongoing clinical trials, were among the genes observed to be markedly up-regulated after ATRA treatment. The present study provides valuable data to further understand the mechanism of ATRA-induced APL cell differentiation and suggests potential therapeutic alternatives for this leukemia.
Acute promyelocytic leukemia (APL) is a subtype of acute myeloid leukemia characterized by the accumulation of cells arrested at the promyelocytic stage of myeloid differentiation. This leukemia exhibits a specific chromosomal translocation t(15;17) involving the promyelocytic leukemia (PML) gene locus on chromosome 15, and the retinoid acid receptor α (RARα) locus on chromosome 17. This translocation generates a chimeric fusion gene PML-RARα, which encodes a protein that functions as an aberrant nuclear receptor considered to be the cause of APL. 1, 2
APL cells are extremely sensitive to all-trans retinoic acid (ATRA), which induces APL cell differentiation into mature granulocytes and results in cell apoptosis. 3 Treatment of APL patients with ATRA in addition to chemotherapy yields a high rate of complete remission and long-term survival 4, 5, 6 . Therefore, ATRA had the distinction of being the first “differentiation therapy” drug for cancer.
Almost all APLs are associated with a chromosome translocation involving the rearrangement of the RARα gene. Five different chromosome translocations fusing RARα to distinct partner genes (PML, PLZF, NuMA, NPM, and STAT5b) have been described in APL. 1 However, the impairment of the myeloid differentiation is not a direct result of the translocation. Instead, the disruption of RARα function may alter the expression of a subset of ATRA-inducible genes critical for myeloid differentiation from the promyelocytic stage to the mature granulocytes. In the past decade great efforts have been devoted to the identification of novel gene(s) affected by the t(15;17) chromosome translocation resulting in blocked differentiation at the promyelocytic stage. Several candidates have been proposed, including ATRA-induced genes, RARα target genes, signaling pathway proteins, and transcription factors. 7, 8, 9, 10
It is generally believed that ATRA-induced differentiation of APL cells to a mature state is mediated, at least in part, through the regulation of gene transcription. Therefore, to gain insights into the molecular events associated with cell differentiation, we used the APL cell line NB4 and cDNA microarray technology to elucidate the transcription events occurring in the ATRA-induced cell differentiation process. DNA microarray technology allows the simultaneous analysis of multiple gene expression patterns, 11 an approach that should help elucidate the molecular mechanism of ATRA-induced NB4 cells differentiation. Due to the limitation of gene numbers and types in commercially available DNA arrays, we prepared cDNA chips with selected cDNA clones. The DNA chips used in our microarray analysis were composed of 12,288 genes encoding proteins that are involved in the major signal transduction pathways (ie, cAMP-dependent protein kinase A, protein kinase C, and mitogen activated protein kinase), production of cytokines and chemokines, transcriptional regulation, cell growth and apoptosis, and the interferon system. We also included numerous expressed sequence tags (EST) unknown genes, to identify novel genes that may play important roles in APL cell differentiation. Our studies revealed that a large number of genes were modulated by ATRA, the majority of which have not been observed before. This approach provided a large amount of data that may help improve our understanding of the pathogenesis of APL.
Materials and Methods
Chemicals
All-trans retinoic acid (ATRA), nitroblue tetrazolium (NBT), and phorbol 12-myristate 13-acetate (PMA) were purchased from Sigma (St. Louis, MO).
Cell Culture
NB4 cells were cultured in RPMI 1640 medium (Life Technologies Inc., Grand Island, NY) plus 10% fetal calf serum, 50 units/ml penicillin G, and 50 μg/ml streptomycin. Cells (5 × 106/ml) were incubated at 37°C in the dark with or without 1 μmol/L ATRA for 12, 24, 48, 72, and 96 hours, respectively. At each time point, the cells with or without ATRA treatment were collected and washed with phosphate-buffered saline (PBS) three times and then stored at −70°C for RNA preparation and cDNA microarray analysis. Only ATRA-induced or modulated gene expression were highlighted by either increase or decrease in folds by comparison of treated sample to the corresponding control sample at the same time point. This type of design eliminated non-specific gene expression that occurred spontaneously in the cultured cells without ATRA treatment. Pretreatment cells were not collected because the purpose of the experimental design was to detect differential expression of ATRA-regulated genes.
cDNA Microarray
Total RNA was extracted from NB4 cells using RNeasy kit (Qiagen Inc., Valencia, CA). cDNA was synthesized from 15 μg of each RNA sample and labeled with Cy3 or Cy5 (Amersham Pharmacia Biotech, Uppsala, Sweden). The RNA extracted from untreated NB4 cells was labeled with Cy3 and the RNA from ATRA treated samples with Cy5. Once the reference and experimental probes were created, equal amounts of the two probes were combined and hybridized to our cDNA microarray slides. The microarray slide, which contains 12,288 sequence-verified clones obtained from Research Genetics (Huntsville, AL), were prepared using a MicroGrid TAS II microarrayer (Biorobotics Inc., Cambridge, UK). The hybridization was performed under a coverslip in a specially designed hybridization chamber (Array-It, Inc., Sunnyvale, CA). The hybridization of the probes to the array was allowed to proceed for 16 hours at 60°C. Following hybridization, the slides were washed and spun-dried. For each sample, the hybridization was repeated three times to confirm the results. Detection of probes hybridized to the array was accomplished using the GMS 418 Scanner (Affymetrix, Santa Clara, CA; Genetic Microsystems, Inc). The image data were captured by scanning the slide twice, the first time at 532 nm (Cy3 labeled) and second time at 635 nm (Cy5 labeled). This process generates two 16-bit TIFF image files, one for each wavelength. The two computer images produced from the scanner were combined and the numerical data for each spot was extracted using AnalyzerDG (Molecularware Inc., Cambridge, MA). The intensity data along with background and error measurements were stored into a text file. To determine whether the data quality for each spot was sufficiently good to warrant subsequent analyses, and to eliminate unreliable elements with expression statistically too close to the background, a statistical program was used that identifies (flags) spots with low intensity/background ratio. Reverse labeling of some housekeeping genes was performed to correct any potential artifactual differences in expression levels, which may result from differences in labeling efficiencies. This systemic variation was corrected during the data normalization. Normalization calculation was based on those spots with signal greater than 6 SD of the background distribution in both channels. Genes showing differences in expression between two groups that were statistically significant were clustered with cluster software. 12 Genes with more than twofold increase (>2.0) or decrease (<0.5) in signal intensity ratio were considered as ATRA-modulated genes.
RT-PCR
Total RNA was isolated from NB4 cells treated and untreated with ATRA (1μmol/L). The first-stranded cDNA synthesis was performed in a 20 μl reaction mixture containing 5 μg RNA, 5 mmol/L dNTP, 100 ng of random primers and 200 U of Superscript II reverse transcriptase (Life Technologies, Inc.). The mixture was incubated at 42°C for 50 minutes and the cDNA was amplified for 30 cycles with denaturation at 94°C for 30 seconds, annealing at 56°C for 30 seconds and extension at 72°C for 1 minute. The primers used were MAP4K2 (451 bp), forward: 5′-GAGTGGGAGCCTGCTGCAGTC-3′, reverse 5′-CAGGTTGAGTGTGTAGATGCC-3′. PKA-Rlα (788 bp), forward 5′-CGCAGCCTTCGAGAATGTGAG-3′, reverse 5′-CACTGGTTCCAATGCATCAGC-3′. PKC-δ (461 bp), forward: 5′- TGGAAGTCGACGTTCGATGCC, reverse 5′- CATCTGCCGATGATCTTGTCG-3′. Tissue factor (311 bp), forward: 5′-CGCCAACTGGTAGACATGGAG-3′, reverse 5′-ACATCCTTCACAATCTCGTCG-3′; C/EBP-α (344 bp), forward 5′-AAGGCCAAGAAGTCGGTGGA-3′, reverse 5′-CAAGCCTCGAGATCCGGCGA-3′. The PCR products were separated by electrophoresis in a 2.0% agarose gel.
NBT Reaction
Cells (2 × 105) were added to 200 μl of 1 mg/ml NBT solution contained 3 × 10−7 M PMA in PBS buffer, and incubated at 37°C for 40 minutes. Cytospin preparations were made from 100 μl of the cell suspension and allowed to air dry. The NBT-positive cells were scored by counting at least 200 cells under microscopic examination. 13
Flow Cytometry Analysis of Cell Cycle and CD11b Expression
106 NB4 cells were incubated with an anti-CD11b-phycoerythrin (PE)-conjugated antibody (Serotec Inc., Raleigh, NC) in PBS buffer on ice for 30 minutes. PE-labeled isotype-matched immunoglobulin was used as negative control. The cells were washed three times with PBS and resuspended in 300 μl of PBS. PE fluorescence was measured using a FACScan flow cytometer (Becton Dickinson Biosciences, San Jose, CA) and 104 cells were analyzed per sample as previously described. 14 For cell cycle analysis, the cells were stained with propidium iodide (PI) and analyzed by flow cytometry, as described. 15
Results and Discussion
Gene Expression Profiling of ATRA-Induced NB4 Cell Maturation
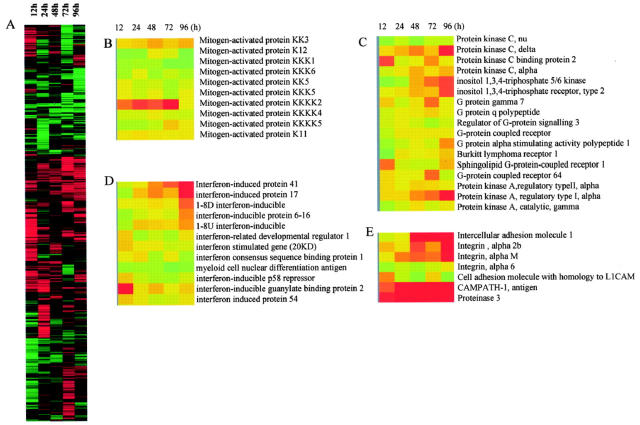
We compared the gene expression profiles of ATRA-treated cells to those of untreated cells to generate the fluorescence intensity ratio of treated versus control signals, as indicated in methods. The data generated from microarray experiments identified that approximately 347 genes, 2.8% of the total 12,288 cDNA elements on the array, were differentially expressed (> twofold up-regulated or down-regulated) in the ATRA treated cells compared to the control cells. It is worth noting that the up-regulated genes at each time point were not always the same ones (Figure 1A) . Table 1 and 2 list the up-regulated and down-regulated known genes induced by ATRA at the indicated time points.
Figure 1.
Expression profiles of 12,288 genes in NB4 cells with or without ATRA (1 μmol/L) treatment. A: Hierarchical clustering of 12,288 genes based on their expression profiles in NB4 cells treated with ATRA at 12 hours, 24 hours, 48 hours, 72 hours, and 96 hours. Each row represents a single gene on the microarray, and each column a separated sample at a different time point. The expression profile of functionally related genes: mitogen activated protein kinase (B), protein kinase C and protein kinase A-related genes (C), IFN-induced genes (D), and neutrophil marker protein genes (E).
Table 1.
List of Genes Up-Regulated in NB4 Cells by ATRA (Fold of Change)
| Name | 12 hour | 24 hour | 48 hour | 72 hour | 96 hour | |
|---|---|---|---|---|---|---|
| DEFA4 | defensin, alpha 4, corticostatin | 1.5 | 7.25 | 38.06 | 41.44 | 152.62 |
| HP-1 | corticostatin/defensin HP-1 protein | 1.94 | 3.86 | 15.09 | 24.7 | 58.55 |
| ICAM1 | intercellular adhesion molecule 1 | 1.06 | 3.06 | 3.12 | 7.2 | 12.38 |
| K6HF | cytokeratin type II | 1.21 | 1.32 | 4.54 | 5.27 | 10.01 |
| PRTN3 | proteinase 3 | 2.08 | 4.61 | 7.43 | 6.53 | 9.56 |
| PDI2 | peptidyl arginine deiminase, type II | 1.98 | 1.98 | 3.56 | 1.85 | 7.26 |
| PENK | proenkephalin | 0.88 | 1.39 | 4.27 | 3.67 | 7.25 |
| MYBL2 | v-myb avian myeloblastosis viral oncogene homolog-like 2 | 1.2 | 0.94 | 2.17 | 1.45 | 6.74 |
| CDW52 | CAMPATH-1 antigen | 1.77 | 3.66 | 5.38 | 3.51 | 6.49 |
| AAD27764 | tyrosine phosphatase homolog | 1.61 | 1.75 | 2.5 | 2.35 | 5.22 |
| CYP26A1 | cytochrome P450, subfamily XXVIA, polypeptide 1 | 1.67 | 1.22 | 2.29 | 1.9 | 5.31 |
| CAPN4 | calpain, small polypeptide | 0.87 | 1.35 | 1.56 | 2.4 | 4.87 |
| CADPS | Ca2+-dependent activator protein for secretion | 0.99 | 1.47 | 2.84 | 2.54 | 4.64 |
| BM040 | bone marrow protein | 2.0 | 2.37 | 3.81 | 2.53 | 3.62 |
| BC002447 | cationic amino acid transporter | 0.74 | 1.29 | 2.05 | 2.41 | 3.6 |
| PSME2 | proteasome activator subunit 2 | 1.56 | 2.65 | 4.26 | 2.37 | 3.6 |
| DUSP6 | dual specificity phosphatase 6 | 1.05 | 1.54 | 1.81 | 2.1 | 3.3 |
| TCIRG1 | T-cell, immune regulator 1 | 1.1 | 1.5 | 2.21 | 2.1 | 3.29 |
| HOXB7 | homeo box B7 | 1.62 | 1.4 | 1.92 | 1.37 | 3.27 |
| ITGA2B | integrin, alpha 2b | 0.95 | 1.16 | 2.17 | 1.69 | 3.23 |
| OAS1 | 2′,5′-oligoadenylate synthetase 1 | 0.92 | 1.12 | 1.51 | 2.01 | 3.18 |
| SH3BGRL | SH3-binding domain glutamic acid-rich protein like | 0.98 | 1.38 | 1.99 | 2.29 | 3.1 |
| XP03817 | hypothetical protein | 1.02 | 1.53 | 1.9 | 2.16 | 3.0 |
| CREG | cellular repressor of E1A-stimulated genes | 1.38 | 1.34 | 1.74 | 1.93 | 2.91 |
| FYB | FYN-binding protein (FYB-120/130) | 1.77 | 1.86 | 2.31 | 2.55 | 2.91 |
| MAP4K2 | mitogen-activated protein kinase kinase kinase kinase 2 | 0.99 | 2.31 | 2.13 | 2.88 | 1.04 |
| ACAA1 | acetyl-coenzyme A acyltransferase 1 | 0.94 | 1.17 | 1.5 | 1.56 | 2.9 |
| HLA-G | HLA-G histocompatibility antigen, class I, G | 1.57 | 1.51 | 2.01 | 1.79 | 2.87 |
| IL5RA | interleukin 5 receptor, alpha | 2.17 | 1.32 | 1.77 | 2.81 | 1.3 |
| KCNC1 | potassium voltage-gated channel, member 1 | 1.29 | 1.05 | 2.04 | 1.66 | 2.84 |
| IFI41 | interferon-induced protein 41, 30kD | 1.47 | 1.11 | 1.71 | 2.05 | 2.76 |
| PRKAR1A | protein kinase A regulatory subunit 1 alpha | 1.26 | 1.29 | 1.71 | 1.92 | 2.75 |
| ITGAM | integrin, alpha M | 1.47 | 1.81 | 2.01 | 1.86 | 2.74 |
| PHTF | putative hemeodomain transcription factor | 1.51 | 1.78 | 1.2 | 1.64 | 3.58 |
| IFI17 | interferon-induced protein 17 | 0.91 | 1.42 | 1.76 | 1.5 | 2.72 |
| BPI | bactericidal/permeability-increasing protein | 0.88 | 0.88 | 1.51 | 2.52 | 2.67 |
| FADSD6 | delta-6 fatty acid desaturase | 2.18 | 2.11 | 2.37 | 1.41 | 2.6 |
| SAT | spermidine/spermine N1-acetyltransferase | 0.85 | 1.34 | 1.95 | 1.62 | 2.57 |
| PTPNS1 | protein tyrosine phosphatase, non-receptor type substrate 1 | 0.88 | 1.1 | 1.64 | 1.71 | 2.57 |
| SCYA20 | small inducible cytokine superfamily A member 20 | 1.41 | 0.87 | 0.98 | 1.38 | 2.0 |
| SCYA14 | small inducible cytokine superfamily A member 14 | 0.77 | 1.17 | 1.17 | 2.52 | 1.37 |
| PRKCD | protein kinase C, delta | 1.28 | 1.53 | 1.81 | 1.43 | 2.56 |
| PTPN22 | protein tyrosine phosphatase, non-receptor type 22 | 0.93 | 1.42 | 1.49 | 1.8 | 2.44 |
| DBI | diazepam binding inhibitor | 1.34 | 1.38 | 2.02 | 1.36 | 2.39 |
| AK3 | adenylate kinase 3 | 1.02 | 1.27 | 1.87 | 1.65 | 2.27 |
| MAFG | v-maf musculoaponeurotic fibrosarcoma, protein G | 1.51 | 1.89 | 2.45 | 1.35 | 2.24 |
| ADD3 | adducin 3 (gamma) | 1.41 | 1.26 | 1.62 | 1.88 | 2.24 |
| MYL4 | myosin, light polypeptide 4 | 1 | 1.55 | 1.91 | 1.54 | 2.23 |
| ITPK1 | inositol 1,3,4-triphosphate 5/6 kinase | 0.8 | 0.72 | 1.58 | 1.9 | 2.22 |
| ITPR2 | inositol 1,4,5-triphosphate receptor, type 2 | 1.02 | 1.21 | 1.52 | 1.44 | 2.21 |
| PPIF F | peptidylprolyl isomerase | 0.83 | 1.38 | 2.03 | 1.19 | 2.2 |
| SUPT3H | suppressor of Ty (S. cerevisiae) 3 homolog | 1.24 | 0.94 | 1.15 | 3.05 | 2.17 |
| CD68 | CD68 antigen | 1.21 | 1.34 | 1.94 | 1.25 | 2.16 |
| MTMR2 | myotubularin related protein 2 | 0.84 | 1.14 | 1.43 | 0.98 | 2.07 |
| PTMA | prothymosin, alpha (gene sequence 28) | 1.22 | 1.55 | 2.44 | 1.42 | 2.07 |
| CYCL | cytochrome c-like antigen | 0.97 | 1.09 | 1.47 | 1.58 | 2.05 |
| SCD | stearoyl-CoA desaturase | 1.23 | 1.58 | 2.6 | 1.44 | 2.05 |
| TCF2 | transcription factor 2 | 0.91 | 1.07 | 1.35 | 1.74 | 2.05 |
| NIPSNAP1 | 4-nitrophenylphosphatase | 0.83 | 1.25 | 1.44 | 1.11 | 2.04 |
| 1-8D | interferon-inducible | 0.71 | 1.09 | 1.22 | 1.12 | 2.03 |
| BIK | BCL-2 iteracting killer | 1.01 | 1.35 | 1.33 | 1.04 | 2.02 |
| PLSCR1 | phospholipid scramblase 1 | 1.03 | 0.95 | 1.71 | 1.78 | 2.02 |
Table 2.
List of Genes Down-Regulated in NB4 Cells by ATRA (Fold of Change)
| Name | 12hr | 24hr | 48hr | 72hr | 96hr | |
|---|---|---|---|---|---|---|
| NYP | neuropeptide Y | 0.9 | 0.72 | 0.49 | 0.72 | 0.19 |
| CFL1 | cofilin 1 (non-muscle) | 1.21 | 1.15 | 0.71 | 0.2 | 0.32 |
| TPS1 | tryptase, alpha | 0.96 | 0.4 | 0.75 | 0.44 | 0.41 |
| CDH1 | cadherin 1, E-cadherin (epithelial) | 0.41 | 0.68 | 0.38 | 0.16 | 0.34 |
| CEP3 | Cdc42 effector protein 3 | 0.66 | 0.58 | 0.35 | 0.18 | 0.09 |
| GTF2H4 | general transcription factor IIH | 1 | 0.59 | 0.41 | 0.56 | 0.52 |
| ASS | argininosuccinate synthetase | 0.34 | 0.33 | 0.21 | 0.23 | 0.24 |
| TP53TG3 | TP53TG3 protein | 0.9 | 0.1 | 0.08 | 0.23 | 0.25 |
| P-B | salivary proline-rich protein | 0.80 | 0.79 | 0.22 | 0.25 | 0.4 |
| PCYT1B | phosphate cytidylyltransferase 1 | 0.8 | 0.9 | 0.33 | 0.29 | 0.04 |
| IGFBP2 | insulin-like growth factor binding protein 2 | 0.36 | 0.51 | 0.42 | 0.29 | 0.4 |
| MNDA | myeloid cell nuclear differentiation antigen | 1.08 | 0.74 | 0.72 | 0.29 | 0.21 |
| MAP3K1 | mitogen-activated protein kinase kinase kinase 1 | 0.65 | 0.76 | 0.79 | 0.31 | 0.38 |
| EBBP | estrogen-responsive B box protein | 0.71 | 0.71 | 0.73 | 0.32 | 0.44 |
| KIAA0266 | KIAA0266 gene product | 0.71 | 0.43 | 0.75 | 0.34 | 0.25 |
| SEMG2 | semenogelin II | 0.88 | 0.65 | 0.24 | 0.36 | 0.59 |
| PTD010 | PTD010 protein | 0.48 | 1.28 | 0.68 | 0.37 | 0.46 |
| CEBPA | CCAAT/EBP, alpha | 1.19 | 0.62 | 0.8 | 0.38 | 0.29 |
| RAD53 | protein kinase Chk2 | 1.33 | 0.75 | 0.87 | 0.61 | 0.19 |
| 20D7-FC4 | hypothetical protein | 0.59 | 0.5 | 0.59 | 0.38 | 0.42 |
| POLH | polymerase eta | 1 | 0.38 | 0.03 | 0.71 | 0.19 |
| SECTM1 | secreted and transmembrane 1 | 1.08 | 1.15 | 1.33 | 0.38 | 0.7 |
| BAI1 | brain-specific angiogenesis inhibitor 1 | 1.05 | 0.68 | 0.33 | 0.87 | 0.32 |
| GABRB3 | gamma-aminobutyric acid A receptor beta 3 | 0.83 | 0.33 | 0.2 | 0.41 | 0.37 |
| BC10 | bladder cancer related protein | 1.02 | 1.08 | 1.04 | 0.78 | 0.38 |
| SPS2 | selenophosphate synthetase 2 | 1.03 | 0.71 | 0.39 | 0.43 | 0.08 |
| MSF | megakaryocyte stimulating factor | 0.82 | 0.12 | 0.24 | 0.43 | 0.45 |
| BCL2 | B-cell CLL/lymphoma 2 | 0.65 | 0.57 | 0.49 | 0.55 | 0.75 |
| TNF | tumor necrosis factor | 1.23 | 1.21 | 0.69 | 0.43 | 0.52 |
| MYO1E | myosin IE | 1.45 | 0.43 | 0.04 | 0.43 | 0.59 |
| KPNB1 | karyopherin (importin) beta 1 | 0.58 | 0.82 | 0.71 | 0.43 | 0.68 |
| UST | uronyl 2-sulfotransferase | 0.49 | 0.67 | 0.32 | 0.44 | 0.52 |
| PCYT2 | phosphate cytidylyltransferase 2, ethanolamine | 0.78 | 0.52 | 0.6 | 0.44 | 0.66 |
| SOX10 | SRY (sex-determining region Y)-box 10 | 0.5 | 0.28 | 0.46 | 0.45 | 0.44 |
| FPGT | fucose-1-phosphate guanylyltransferase | 1.37 | 0.96 | 1.07 | 0.45 | 0.61 |
| PCDH2 | protocadherin 2 (cadherin-like 2) | 0.79 | 0.77 | 0.82 | 0.46 | 0.7 |
| LY9 | lymphocyte antigen | 1.05 | 0.32 | 0.35 | 0.75 | 0.39 |
| INPP4A | inositol polyphosphate-4-phosphatase, type 1, 107kD | 0.75 | 0.4 | 0.3 | 0.5 | 0.47 |
| WSX-1 | class I cytokine receptor | 0.99 | 0.74 | 0.99 | 0.5 | 0.61 |
| PTPN3 | protein tyrosine phosphatase, non-receptor type 3 | 0.58 | 0.72 | 0.48 | 0.5 | 0.69 |
| TF (F3) | tissue factor | 1.03 | 0.75 | 0.69 | 0.52 | 0.46 |
| PLU-1 | putative DNA/chromatin binding motif | 0.67 | 0.53 | 0.52 | 0.51 | 0.5 |
| HFL-EDDG1 | erythroid differentiation and denucleation factor 1 | 1.07 | 0.91 | 0.78 | 0.52 | 0.55 |
| ACVR1 | actin A receptor type 1 | 1.18 | 0.7 | 0.99 | 0.79 | 0.58 |
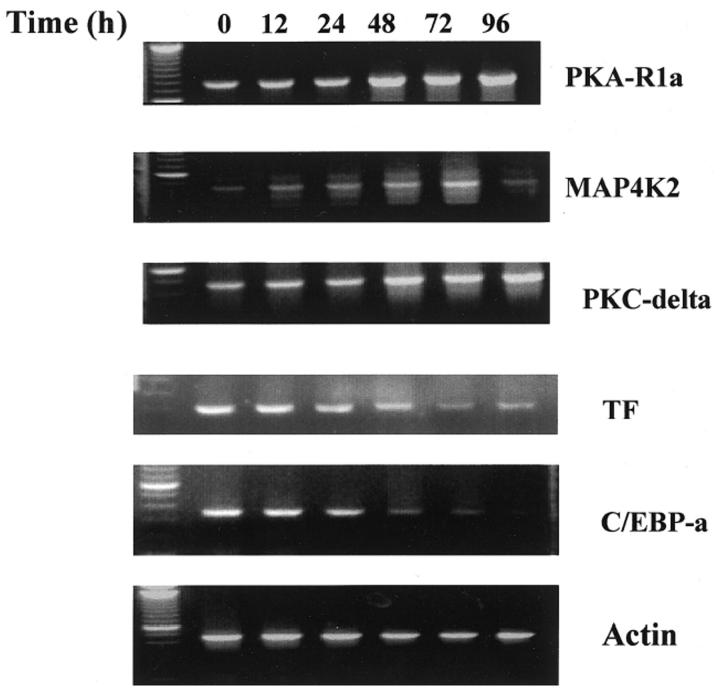
Confirmation of Altered Gene Expression by RT-PCR
We confirmed with RT-PCR the microarray data for ATRA-induced selective gene expression profiles at various time points. The RT-PCR products were analyzed and normalized by using actin as an internal control. As shown in Figure 2 , the expression patterns of three up-regulated genes (PKA-RIα, MAP4K2, and PKC-δ) and two down-regulated genes (TF and C/EBP-α) were consistent with the data obtained from microarray analysis. Our results were entirely consistent with previous findings of ATRA-induced protein(s) reported in the literature by other methods such as differential display, microarray analysis, and Northern blotting. 7, 16 To verify the results of microarray data, we also used neutrophil marker protein cDNAs as internal controls in our array chips, including defensins, proteinase 3, CD11b, ICAM1, CCRL2, and proenkephalin. The array data showed that the levels of gene expression of all of the above-mentioned genes paralleled ATRA-induced NB4 cell maturation (Figure 1E) , supporting the notion that the microarray data reflected changes in gene transcriptional levels during the ATRA-induced myeloid differentiation process.
Figure 2.
RT-PCR analysis of genes in NB4 cells treated with ATRA. cDNA was synthesized from equal amounts of RNA isolated from NB4 cells treated with ATRA at 0, 12, 24, 48, 72, and 96 hours separately. The data are representative examples of three independent experiments that gave similar results. Actin serves as internal control.
Activation of Multi-Signaling Pathways by ATRA
Multiple signaling pathways have been implicated in the ATRA-induced NB4 cells differentiation, including cAMP-dependent protein kinase A (PKA), 17 protein kinase C (PKC), 18 and mitogen-activated protein kinase (MAPK). 19 However, there is relatively little information regarding the effect of ATRA on the transcription level of genes involved in these signaling pathways. We analyzed the transcription level of 10 members of the MAP kinase family, six isoforms of protein kinase C, and two isoforms of cAMP-dependent protein kinase A. Our data revealed that only MAP4K2, one of 10 members of MAP kinases, was up-regulated threefold after treatment with ATRA (Figure 1B) . MAP4K2, also called GC kinase, is a serine/threonine protein kinase, which has been identified in human lymphoid tissue and is activated during the stress response. 20 Recent studies confirmed that MAP4K2 is an upstream activator of the MAP kinase cascade. 21 Activation of MAP4K2 resulted in the activation of Jun N-terminal kinase (JNK) and p38 MAP kinase, but not the extracellular signal-regulated kinase (ERK). It was known that treatment of NB4 cells with ATRA leads to activation of p38 MAP kinase and inactivation of JNK. 22, 23 We observed that ATRA treatment did not affect the gene expression of p38 kinase and JNK, indicating that the modulation of the activity of p38 kinase and JNK by ATRA is mediated by activation of the upstream activator MAP4K2 instead of a change in their gene expression. Our studies were also consistent with the previous observation that ATRA suppressed the transcription of c-jun and c-fos genes. 9
Protein kinase C-δ is the major protein kinase in leukocyte extract that phosphorylates the cytoplasmic domain of CD18 subunit of CD11/CD18 intergrin during cell activation 24 and it also plays an important role in neutrophil activation and apoptosis. 25 Our array results demonstrated that the expression of protein kinase C δ and α, to a lesser extent, increased with ATRA treatment (Figure 1C) , consistent with our previous observation that CD18 expressed on the cell surface in a time-dependent manner following ATRA treatment. 14 These findings also support the previous observations that only the α and δ isoforms of protein kinase C participate in myeloid differentiation 25 and neutrophil apoptosis. 26
cAMP-dependent protein kinase A (PKA) includes two isoforms, PKA-1 and PKA-2. They both share a common catalytic subunit but have distinct regulatory(R) subunits, R1 and R2, respectively. Our results revealed that ATRA only stimulated R1α gene expression, but not the other PKA subunits (Figure 1C) . Expression of R1α of PKA is increased in various human cancers including those of breast, 27 ovary, 28 lung, 29 and colon. 30 Furthermore, the pathogenesis of malignancy and poor prognosis in cancer patients has been reported to correlate with overexpression of Rlα of the PKA gene. 31 Previous experimental evidence has implicated different roles of regulatory subunits of PKA in the regulation of leukemic cell proliferation and differentiation. For instance, treatment of the leukemia cell line HL-60 with antisense oligonucleotide targeted against the mRNA of PKA-R1α caused cell growth inhibition and resulted in cell differentiation, 32 suggesting that PKA-R1α exerted inhibitory effects on HL-60 monocytic cell differentiation. In contrast, the present study demonstrates that ATRA-induced up-regulation of PKA-R1α expression resulted in NB4 cells undergoing granulocytic differentiation, suggesting that PKA-R1α exhibited a stimulatory influence on NB4 cell differentiation. Our results regarding PKA-R1α up-regulation by ATRA have been confirmed independently by a recent similar study using NB4 cells. 33 These conflicting results might be explained by the effects of PKA-R1α being cell-type dependent, due to activation of distinct signal pathways by binding to different regulatory subunits of PKA-R1α. Previous studies have shown that cAMP alone exhibited no differentiation effect on NB4 cells, but it substantially enhanced the maturation effect of ATRA on NB4 cells. 17 It was believed that ATRA “triggers” the cells to become responsive to cAMP and complete the maturation process. Our data have uncovered the molecular mechanism of the phenomenon that stimulation of the expression of R1α gene by ATRA results in NB4 cells being sensitive to cAMP.
Protein tyrosine phosphatase acts in conjunction with protein kinase to regulate the events of tyrosine phosphorylation that have been known to control cell activation and cell differentiation. Of this superfamily, lymphoid-specific protein tyrosine phosphatase non-receptor type 22 (PTPN22) and their substrate type 1 (PTPNS1) were up-regulated by ATRA. The response of protein tyrosine phosphatase to ATRA has not been observed before and the function of this enzyme remains unclear. 34
Up-Regulation of Interferon (IFN) and IFN-Related Genes by ATRA
Previous studies have shown that ATRA is a strong inducer of interferon and interferon-mediated signaling pathways in ATRA-induced NB4 cells differentiation. 35, 36, 37 Our array data showed that among 11 interferon-induced genes contained in our chip, ATRA induced the expression of most IFN-induced genes, such as OSA1, IFI41, IFI17, IFI6–16, 1–8U, and 1–8D, in a time-dependent manner. Three interferon-induced genes, ie, guanylate binding protein 2 (GBP2), interferon-stimulated gene (ISG20), and protein 54 (IFIT2), responded to ATRA immediately and were expressed in NB4 cells within 12 hours of ATRA treatment (Figure 1D) . These early response proteins may be involved in the IFN-induced signaling pathway.
Regulation of the Expression of Cytokines and Chemokines by ATRA
Myeloid cell differentiation and maturation were also accompanied by different gene expression patterns of cytokines and chemokines. 38 The transcription regulation of cytokines and chemokines in ATRA-induced NB4 cells has not been fully explored. Therefore, we also added cDNAs of chemokines and cytokines in our array chips. We found that two chemokines, HCC-1 (SCYA14) and CCL20 (SCYA20) mRNA were increased after ATRA treatment. HCC-1 has been recently described as a human chemokine that is constitutively expressed in various tissues and is present at high concentrations in plasma. 39 However, the major source of circulating HCC-1 is unknown. High expression of HCC-1 in ATRA stimulated NB4 cells suggests that like CCL20, 40 the circulating HCC-1 might be produced by neutrophils. In addition, interleukin-5 receptor α gene (IL-5Rα) was increased up to threefold over untreated cells. Interleukin-5 plays an important role in proliferation and differentiation of eosinophils and basophils. Recent observations by Denburg et al 41 indicated that ATRA could selectively inhibit membrane-bound IL-5 receptor and up-regulate soluble IL-5 receptor transcription in CD34+ cells. The up-regulation of IL-5Rα transcription by ATRA in NB-4 cells suggests that IL-5 might also be involved in granulocytic cell differentiation. An alternative explanation is that the ATRA-induced terminal differentiated NB4 cells do not represent “pure” neutrophils and might exhibit some other kind of granulocytic features. This scenario was supported by our observations that ATRA also induced the expression of CD52 antigen on differentiated NB4 cells (Figure 1E) expression and failed to induce CD16, a mature neutrophil marker expression (data not shown). It has been reported that CD52 is present on the cell membrane of eosinophils, and is not present on neutrophils. 42, 43
Growth Inhibition and Apoptosis Induced by ATRA
We confirmed that ATRA inhibited NB4 cell growth as evidenced by the continuously decrease in cell S-phase and increase in cell apoptosis (data not shown). Our array data showed that ATRA treatment selectively down-regulated the expression of the anti-apoptosis BCL-2 and API3 genes and cell cycle related genes, and up-regulated the pro-apoptosis genes BIK and Daxx. Thus, induction of apoptosis may contribute to the therapeutic value of ATRA.
ATRA-Induced Effects on Transcriptional Factors
Various transcription factors have been associated with hematopoietic cell proliferation and myeloid differentiation. Critical to terminal granulocytic differentiation are the transcription factors PU.1, CCAAT/enhancer binding protein(C/EBP), core-binding factors (CBFs), and homeobox proteins. 44, 45 We closely examined several transcriptional factors affected by ATRA treatment. After cells were exposed to ATRA, there was no change in the expression of CBF-β and C/EBP-γ. The expression of C/EBP-α was down-regulated. This result is consistent with previous studies showing that C/EBP-α plays a role in early myeloid progenitors but its expression is decreased with myeloid differentiation. 46
Human HoxB cluster gene is located in chromosome 17q21.3 47 and the translocation t(15;17) in NB4 cells may affect the HoxB cluster gene expression. The results from our array showed that ATRA stimulated HoxB7 but had no effect on HoxB3, HoxA9 or, HoxA10 gene expression. The putative homeodomain transcriptional factor (PHTF) was originally isolated from human erythroleukemic cells. 48 It is highly homologous with the human homeobox gene family. The biological function of PHTF is still unknown. Our data from the array studies showed that after exposure of cells to ATRA, PHTF gene transcription increased rapidly, suggesting that PHTF may play a role in myeloid cell differentiation.
Relationship between Gene Transcription and Cell Differentiation
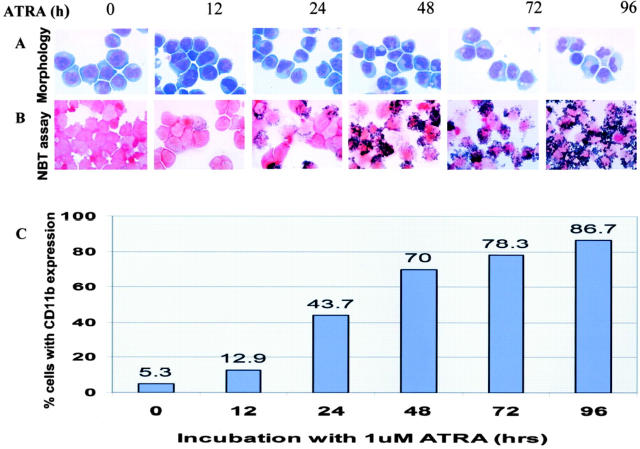
We have also monitored ATRA-mediated NB4 cells differentiation using four classic assays for the study of myeloid cell maturation. They are cell morphology, CD11b/CD18 expression, nitroblue tetrazolium (NBT) reduction test (a measure of granulocytic maturity), and cell cycle analysis at 0, 12, 24, 48, 72, and 96 hours to correlate the biological features with the gene transcription patterns. Using these assays, we observed no obvious morphological change in the NB4 cells during the first 24 hours of ATRA treatment. However, after 24 hours the differentiation markers began to be observed. The ratio of cytoplasm to nucleus increased and the chromatin condensed gradually. CD11b+ expression and NBT- positive cells gradually increased from 24 hours to 96 hours of treatment (Figure 3) . Maximal differentiation (ie, more than 85%) was observed at 96 hours after ATRA treatment. Furthermore, the decrease of the fraction of cells in S-phase paralleled the progression of cell differentiation and programmed cell death. These results indicate that the changes in gene expression observed in the microarray correlate with the degree of differentiation of NB4 cells. It must be emphasized that ATRA-induced granulocytic differentiation of NB4 cells is different from normal myeloid differentiation. NB4 cells strongly express myeloid markers, but they also express some T-cell marker 49 and lack some neutrophil-associated markers. For instance, both CD16 and cathepsin G are neutrophil markers, but they were not expressed in NB4 cells before or after ATRA treatment in our study. The differentiated NB4 cells also expressed some proteins that are not expressed in normal neutrophils, such as CD52 and interleukin 5 receptor. Therefore, the gene expression patterns reported here do not completely represent normal granulocytic differentiation.
Figure 3.
Differentiation of ATRA-induced NB4 cells. A: Morphological features of NB4 cells in response to ATRA treatment at different times. B: Nitroblue tetrazolium enzymatic reduction in ATRA-treated NB4 cells. C: CD11b intergrin expression following ATRA treatment of NB4 cells at each time point as assessed by flow cytometric analysis.
Tissue factor (TF) is known to be an important factor in initiating coagulation cascade. As a result of increased TF expression in APL blast cells, patients with APL are often associated with disseminated intravascular coagulation. We have demonstrated that treatment of APL cells with ATRA significantly reduced TF expression (see Figure 2 , and Table 2 ) from a ratio of 1.02 at 12 hours ATRA treatment to 0.46 at 96 hours ATRA treatment when compared to the untreated cells. This result is consistent with previous study 50 the clinical observation that ATRA treatment improves the APL-associated coagulopathy in patients with APL. 51
Conclusion
In summary, our results demonstrate that ATRA alters the expression of a large number of different genes, most of which have not been reported before. The ATRA-affected genes can be classified into several groups including genes involved in the regulation of signaling pathways, transcription factors, cell differentiation, programmed cell death, and the production of cytokines and chemokines.
Although incorporating ATRA into the treatment of APL has resulted in long-term survival and potential cure in nearly 70% of patients, the disease may relapse and resistance to further ATRA treatment and the retinoic acid syndrome occurred in almost all cases. Therefore, there is need to explore new drugs and new regimens for APL therapy. In this study, we showed that two potential target proteins, R1α subunit of PKA and CD52 antigen, were highly expressed in the ATRA-treated APL cells. Recently, Sandrini et al 52 reported that PKA-R1α acted as a tumor-suppressor gene for sporadic thyroid cancer. The administration of ATRA has been used to treat patients with APL, but ATRA alone is not necessarily sufficient to maintain patients in stable remission. The finding of up-regulation of PKA-1α expression by ATRA support the strategy that combination of ATRA with cyclic AMP analogues synergistically induces APL cell differentiation and possibly reduces both the ATRA-induced side-effects and development of resistance to ATRA treatment. Another important finding in our study is that ATRA can induce CD52 expression on the APL cell surface. CD52 antigen is normally expressed on T and B lymphocytes, natural killer (NK) cells, monocytes, eosinophils and sperm cells, but not on leukemic myeloblasts and neutrophils. 42 CD52 targeted therapy has been used in chronic lymphocytic leukemia and non-Hodgkin’s lymphoma with promising results. 53 Thus, the induced up-regulation of CD52 antigen expression in APL cells could provide a new target for an effective regimen of combination of ATRA and anti-CD52 antibodies for the treatment of this disease.
Acknowledgments
We thank Drs. Raul C. Braylan and Kirsten Madsen for critical evaluation of the manuscript.
Address reprint requests to Lijun Yang, M.D., Department of Pathology, Immunology, and Laboratory Medicine, University of Florida College of Medicine, P.O. Box 100275, Gainesville, FL 32610. E-mail: yanglj@pathology.ufl.edu.
Footnotes
Supported in part by a grant from Stop! Children’s Cancer Foundation, Gainesville, FL (to L.J.Y.)
References
- 1.Benoit G, Roussel M, Pendino F, Segal-Bendirdjian E, Lanotte M: Orchestration of multiple arrays of signal cross-talk and combinatorial interactions for maturation and cell death: another vision of t(15;17) preleukemic blast and APL-cell maturation. Oncogene 2001, 20:7161-7177 [DOI] [PubMed] [Google Scholar]
- 2.Melnick A, Licht JD: Deconstructing a disease: rARα, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood 1999, 93:3167-3215 [PubMed] [Google Scholar]
- 3.Breitman TR, Collins SJ, Keene BR: Terminal differentiation of human promyelocytic leukemic cells in primary culture in response to retinoic acid. Blood 1981, 57:1000-1004 [PubMed] [Google Scholar]
- 4.Huang ME, Ye YC, Chen SR, Chai JR, Lu JX, Zhoa L, Gu LJ, Wang ZY: Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood 1988, 72:567-572 [PubMed] [Google Scholar]
- 5.Warrell RP, Jr, de The H, Wang ZY, Degos L: Acute promyelocytic leukemia. N Engl J Med 1993, 329:177-189 [DOI] [PubMed] [Google Scholar]
- 6.Fenaux P, Chomienne C, Degos L: Acute promyelocytic leukemia: biology and treatment. Semin Oncol 1997, 24:92-102 [PubMed] [Google Scholar]
- 7.Kitareewan S, Pitha-Rowe I, Sekula D, Lowrey CH, Nemeth MJ, Golub TR, Freemantle SJ, Dmitrovsky E: UBE1L is a retinoid target that triggers PML/RARα degradation and apoptosis in acute promyelocytic leukemia. Proc Natl Acad Sci USA 2002, 99:3806-3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshall H, Studer M, Popperl H, Aparicio S, Kuroiwa A, Brenner S, Krumlauf R: A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature 1994, 370:567-571 [DOI] [PubMed] [Google Scholar]
- 9.Han GR, Dohi DF, Lee HY, Rajah R, Walsh GL, Hong WK, Cohen P, Kurie JM: All-trans-retinoic acid increases transforming growth factor-β2 and insulin-like growth factor binding protein-3 expression through a retinoic acid receptor-α-dependent signaling pathway. J Biol Chem 1997, 272:13711-13716 [DOI] [PubMed] [Google Scholar]
- 10.Duprez E, Tong JH, Derre J, Chen SJ, Berger R, Chen Z, Lanotte M: JEM-1, a novel gene encoding a leucine-zipper nuclear factor up-regulated during retinoid-induced maturation of NB4 promyelocytic leukaemia. Oncogene 1997, 14:1563-1570 [DOI] [PubMed] [Google Scholar]
- 11.Schena M, Shalon D, Davis RW, Brown PO: Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270:467-470 [DOI] [PubMed] [Google Scholar]
- 12.Eisen MB, Spellman PT, Brown PO, Botstein D: Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 1998, 95:14863-14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Idres N, Benoit G, Flexor MA, Lanotte M, Chabot GG: Granulocytic differentiation of human NB4 promyelocytic leukemia cells induced by all-trans retinoic acid metabolites. Cancer Res 2001, 61:700-705 [PubMed] [Google Scholar]
- 14.Li SW, Tang D, Ahrens KP, She JX, Braylan RC, Yang L: All-trans-retinoic acid induces CD52 expression in acute promyelocytic leukemia. Blood 2003, 101:1977-1980 [DOI] [PubMed] [Google Scholar]
- 15.Honma Y, Yamamoto-Yamaguchi Y, Kanatani Y: Vesnarinone and glucocorticoids cooperatively induce G1 arrest and have an anti-tumour effect on human non-small cell lung carcinoma cells grown in nude mice. Br J Cancer 1999, 80:96-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Battle TE, Roberson MS, Zhang T, Varvayanis S, Yen A: Retinoic acid-induced blr1 expression requires RARα RXR, and MAPK activation and uses ERK2 but not JNK/SAPK to accelerate cell differentiation. Eur J Cell Biol 2001, 80:59-67 [DOI] [PubMed] [Google Scholar]
- 17.Ruchaud S, Duprez E, Gendron MC, Houge G, Genieser HG, Jastorff B, Doskeland SO, Lanotte M: Two distinctly regulated events, priming and triggering, during retinoid-induced maturation and resistance of NB4 promyelocytic leukemia cell line. Proc Natl Acad Sci USA 1994, 91:8428-8432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Pedrera C, Dobado-Berrios PM, Ros R, Torres A, Garcia-Navarro S, Jardi M, Felez J, Velasco F: Signal transduction pathways underlying the expression of tissue factor and thrombomodulin in promyelocytic cells induced to differentiate by retinoid acid and dibutyryl cAMP. Thromb Haemost 2001, 85:1031-1036 [PubMed] [Google Scholar]
- 19.Yen A, Roberson MS, Varvayanis S: Retinoic acid selectively activates the ERK2 but not JNK/SAPK or p38 MAP kinases when inducing myeloid differentiation. In Vitro Cell Dev Biol Anim 1999, 35:527-532 [DOI] [PubMed] [Google Scholar]
- 20.Katz P, Whalen G, Kehrl JH: Differential expression of a novel protein kinase in human B lymphocytes: preferential localization in the germinal center. J Biol Chem 1994, 269:16802-16809 [PubMed] [Google Scholar]
- 21.Dan I, Watanabe NM, Kusumi A: The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol 2001, 11:220-230 [DOI] [PubMed] [Google Scholar]
- 22.Alsayed Y, Uddin S, Mahmud N, Lekmine F, Kalvakolanu DV, Minucci S, Bokoch G, Platanias LC: Activation of Rac1 and the p38 mitogen-activated protein kinase pathway in response to all-trans-retinoic acid. J Biol Chem 2001, 276:4012-4019 [DOI] [PubMed] [Google Scholar]
- 23.Lee HY, Sueoka N, Hong WK, Mangelsdorf DJ, Claret FX, Kurie JM: All-trans-retinoic acid inhibits Jun N-terminal kinase by increasing dual-specificity phosphatase activity. Mol Cell Biol 1999, 19:1973-1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fagerholm S, Morrice N, Gahmberg CG, Cohen P: Phosphorylation of the cytoplasmic domain of the integrin CD18 chain by protein kinase C isoforms in leukocytes. J Biol Chem 2002, 277:1728-1738 [DOI] [PubMed] [Google Scholar]
- 25.Mischak H, Pierce JH, Goodnight J, Kazanietz MG, Blumberg PM, Mushinski JF: Phorbol ester-induced myeloid differentiation is mediated by protein kinase C-α and -δ and not by protein kinase C-β II, -ε, -ζ, and -η. J Biol Chem 1993, 268:20110-20115 [PubMed] [Google Scholar]
- 26.Webb PR, Wang KQ, Scheel-Toellner D, Pongracz J, Salmon M, Lord JM: Regulation of neutrophil apoptosis: a role for protein kinase C and phosphatidylinositol-3-kinase. Apoptosis 2000, 5:451-458 [DOI] [PubMed] [Google Scholar]
- 27.Gordge PC, Hulme MJ, Clegg RA, Miller WR: Elevation of protein kinase A and protein kinase C activities in malignant as compared with normal human breast tissue. Eur J Cancer 1996, 32A:2120-2126 [DOI] [PubMed] [Google Scholar]
- 28.Simpson BJ, Ramage AD, Hulme MJ, Burns DJ, Katsaros D, Langdon SP, Miller WR: Cyclic adenosine 3′, 5′-monophosphate-binding proteins in human ovarian cancer: correlations with clinicopathological features. Clin Cancer Res 1996, 2:201-206 [PubMed] [Google Scholar]
- 29.Young MR, Montpetit M, Lozano Y, Djordjevic A, Devata S, Matthews JP, Yedavalli S, Chejfec G: Regulation of Lewis lung carcinoma invasion and metastasis by protein kinase A. Int J Cancer 1995, 61:104-109 [DOI] [PubMed] [Google Scholar]
- 30.Bradbury AW, Carter DC, Miller WR, Cho-Chung YS, Clair T: Protein kinase A (PK-A) regulatory subunit expression in colorectal cancer and related mucosa. Br J Cancer 1994, 69:738-742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller WR, Hulme MJ, Bartlett JM, MacCallum J, Dixon JM: Changes in messenger RNA expression of protein kinase A regulatory subunit iα in breast cancer patients treated with tamoxifen. Clin Cancer Res 1997, 3:2399-2404 [PubMed] [Google Scholar]
- 32.Tortora G, Yokozaki H, Pepe S, Clair T, Cho-Chung YS: Differentiation of HL-60 leukemia by type I regulatory subunit antisense oligodeoxynucleotide of cAMP-dependent protein kinase 4. Proc Natl Acad Sci USA 1991, 88:2011-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee KH, Chang MY, Ahn JI, Yu DH, Jung SS, Choi JH, Noh YH, Lee YS, Ahn MJ: Differential gene expression in retinoic acid-induced differentiation of acute promyelocytic leukemia cells, NB4, and HL-60 cells. Biochem Biophys Res Commun 2002, 296:1125-1133 [DOI] [PubMed] [Google Scholar]
- 34.Cohen S, Dadi H, Shaoul E, Sharfe N, Roifman CM: Cloning and characterization of a lymphoid-specific, inducible human protein tyrosine phosphatase, Lyp. Blood 1999, 93:2013-2024 [PubMed] [Google Scholar]
- 35.Pelicano L, Brumpt C, Pitha PM, Chelbi-Alix MK: Retinoic acid resistance in NB4 APL cells is associated with lack of interferon α synthesis Stat1 and p48 induction. Oncogene 1999, 18:3944-3953 [DOI] [PubMed] [Google Scholar]
- 36.Matikainen S, Ronni T, Lehtonen A, Sareneva T, Melen K, Nordling S, Levy DE, Julkunen I: Retinoic acid induces signal transducer and activator of transcription (STAT) 1, STAT2, and p48 expression in myeloid leukemia cells and enhances their responsiveness to interferons. Cell Growth Differ 1997, 8:687-698 [PubMed] [Google Scholar]
- 37.Matikainen S, Ronni T, Hurme M, Pine R, Julkunen I: Retinoic acid activates interferon regulatory factor-1 gene expression in myeloid cells. Blood 1996, 88:114-123 [PubMed] [Google Scholar]
- 38.Scapini P, Lapinet-Vera JA, Gasperini S, Calzetti F, Bazzoni F, Cassatella MA: The neutrophil as a cellular source of chemokines. Immunol Rev 2000, 177:195-203 [DOI] [PubMed] [Google Scholar]
- 39.Vakili J, Standker L, Detheux M, Vassart G, Forssmann WG, Parmentier M: Urokinase plasminogen activator and plasmin efficiently convert hemofiltrate CC chemokine 1 into its active. J Immunol 2001, 167:3406-3413 [DOI] [PubMed] [Google Scholar]
- 40.Scapini P, Laudanna C, Pinardi C, Allavena P, Mantovani A, Sozzani S, Cassatella MA: Neutrophils produce biologically active macrophage inflammatory protein-3α (MIP-3α)/CCL20 and MIP-3β/CCL19. Eur J Immunol 2001, 31:1981-1988 [DOI] [PubMed] [Google Scholar]
- 41.Denburg JA, Sehmi R, Upham J: Regulation of IL-5 receptor on eosinophil progenitors in allergic inflammation: role of retinoic acid. Int Arch Allergy Immunol 2001, 124:246-248 [DOI] [PubMed] [Google Scholar]
- 42.Elsner J, Hochstetter R, Spiekermann K, Kapp A: Surface and mRNA expression of the CD52 antigen by human eosinophils but not by neutrophils. Blood 1996, 88:4684-4693 [PubMed] [Google Scholar]
- 43.Fabian I, Flidel O, Gadish M, Kletter Y, Slavin S, Nagler A: Effects of CAMPATH-1 antibodies on the functional activity of monocytes and polymorphonuclear neutrophils. Exp Hematol 1993, 21:1522-1527 [PubMed] [Google Scholar]
- 44.Khanna-Gupta A, Zibello T, Sun H, Lekstrom-Himes J, Berliner N: C/EBP ε mediates myeloid differentiation and is regulated by the CCAAT displacement protein (CDP/cut). Proc Natl Acad Sci USA 2001, 98:8000-8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lekstrom-Himes JA: The role of C/EBP(ε) in the terminal stages of granulocyte differentiation. Stem Cells 2001, 19:125-133 [DOI] [PubMed] [Google Scholar]
- 46.Scott LM, Civin CI, Rorth P, Friedman AD: A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood 1992, 80:1725-1735 [PubMed] [Google Scholar]
- 47.Chariot A, Senterre-Lesenfants S, Sobel ME, Castronovo V: Molecular cloning of a mutated HOXB7 cDNA encoding a truncated transactivating homeodomain-containing protein. J Cell Biochem 1998, 71:46-54 [PubMed] [Google Scholar]
- 48.Raich N, Mattei MG, Romeo PH, Beaupain D: PHTF, a novel atypical homeobox gene on chromosome 1p13, is evolutionarily conserved. Genomics 1999, 59:108-109 [DOI] [PubMed] [Google Scholar]
- 49.Lanotte M, Martin-Thouvenin V, Najman S, Balerini P, Valensi F, Berger R: NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3). Blood 1991, 77:1080-1086 [PubMed] [Google Scholar]
- 50.Guo W, Wang H, Zhao W: Effects of all-trans retinoic acid and arsenic trioxide on tissue factor expression of acute promyelocytic leukemia cells. Zhonghua Yi Xue Za Zhi 2000, 80:327-331 [PubMed] [Google Scholar]
- 51.Barbui T, Falanga A: Disseminated intravascular coagulation in acute leukemia. Semin Thromb Hemost 2001, 27:593-604 [DOI] [PubMed] [Google Scholar]
- 52.Sandrini F, Matyakhina L, Sarlis NJ, Kirschner LS, Farmakidis C, Gimm O, Stratakis CA: Regulatory subunit type I-α of protein kinase A (PRKAR1A): a tumor-suppressor gene for sporadic thyroid cancer. Genes Chromosomes Cancer 2002, 35:182-192 [DOI] [PubMed] [Google Scholar]
- 53.Flynn JM, Byrd JC: Campath-1H monoclonal antibody therapy. Curr Opin Oncol 2000, 12:574-581 [DOI] [PubMed] [Google Scholar]