Abstract
Adenomatous polyposis coli (APC) is a tumor suppressor gene important in colorectal tumorigenesis. A genetic variant of APC, I1307K, results from a T-to-A transversion at nucleotide 3920 which converts the wild-type sequence to a homopolymer tract (A8). The I1307K alteration is not itself oncogenic, but creates a hypermutable region (A8) that is prone to frame-shift mutations. The APC I1307K variant occurs in approximately 6% of the Ashkenazi Jewish population and is reported to approximately double an individual’s risk for colorectal cancer. Here we describe a single nucleotide primer extension assay for the detection of the APC I1307K mutation. Following PCR amplification, nucleotide 3920 of the APC gene is directly sequenced using single nucleotide primer extension technology. The assay is in a multiplex format allowing simultaneous forward and reverse sequencing of the I1307K variant, which provides an internal, independent confirmation of each testing result. The assay was validated against 60 samples previously characterized by an allele-specific oligonucleotide (ASO) hybridization assay, with 100% concordance of results. Compared to the ASO assay, this single nucleotide primer extension assay requires significantly less technical time to perform, and has a greatly increased throughput capacity. The single nucleotide extension assay provides a highly sensitive and specific assay to identify individuals with the APC I1307K gene variant who may benefit from increased colorectal screening.
Colorectal cancer (CRC) is the second leading cause of cancer death in the United States. 1 Approximately 15 to 20% of cases occur in familial aggregations and are believed to be associated with some form of inherited susceptibility. 2, 3 Genetic predisposition to CRC includes dominantly inherited diseases such as familial adenomatous polyposis (FAP) and hereditary non-polyposis colorectal cancer (HNPCC). 4 FAP is caused by germ-line truncating mutations of the tumor suppressor gene Adenomatous polyposis coli (APC) located on chromosome 5q21. In addition to the critical role of APC in FAP, the APC gene also plays an important role in sporadic colorectal tumorigenesis. Somatic mutation of the APC gene is found in >80% of sporadic CRC and is considered to be one of the earliest common genetic events leading to colorectal adenomatous lesions. 4, 5
In addition to the truncating mutations of APC, a germ-line genetic variant of APC, I1307K, has been identified, which results from a T-to-A transversion at nucleotide position 3920. 6 The resulting lysine for isoleucine amino acid substitution does not itself significantly alter the function of the protein, but rather the variant creates a homopolymer tract (A8) which is prone to frame-shift mutations resulting in subsequent somatic inactivation of the APC gene product. 6, 7 This type of genetic alteration represents a novel carcinogenic mechanism in which the alteration itself does not alter the function of the encoded protein, but rather predisposes the gene to further alterations which promote tumorgenesis. Several studies have demonstrated that the APC I1307K gene variant is associated with an estimated relative risk for developing CRC of 1.5 to 2.0. 6, 8, 9 The variant has been associated with increased adenomatous polyp formation 10 and may also be associated with an increased risk of transition from adenoma to invasive cancer. 11 Carriers of the APC I1307K variant have also been reported to be at a somewhat increased risk for other cancers including carcinoma of the breast. 8, 12 The APC I1307K variant has been shown to occur in approximately 6% of the Ashkenazi Jewish population and in approximately 10% of Ashkenazim with CRC. The variant is also seen in approximately 1% of the Jewish, non-Ashkenazi, population 13 and has been reported to occur rarely in non-Jewish individuals. 14, 15 Jewish I1307K mutation carriers share a common allelic pattern with APC-linked markers supporting the notion of a founder effect for the I1307K variant.
Identification of APC I1307K carriers is clinically important because these individuals may benefit from increased colorectal surveillance and early eradication of pre-cancerous polyps. Here we describe the design and validation of a liquid-phase single nucleotide primer extension assay (also called SNaPshot, SNuPE, or minisequencing) to detect the APC I1307K mutation. Single nucleotide extension assays require PCR amplification of the gene region of interest followed by thermal cycling with an unlabeled primer that anneals to the PCR product template immediately 5′ to the position of interest on the template. The reaction contains only dideoxynucleotide (ddNTP) terminators (no dNTPs) such that during thermal cycling, the DNA polymerase can extend the primers by only one base (ddNTP), which is complimentary to the template strand at the site of interest. In this case, the primer is extended by a fluorescently labeled ddNTP terminator so that the products can be separated, sized, and detected by capillary electrophoresis (CE). In this report we have designed an assay in which both PCR strands are interrogated simultaneously in a single multiplex reaction, allowing internal confirmation of each test result. Validation of the single nucleotide primer extension assay against a published allele-specific oligonucleotide (ASO) hybridization assay is described and the assay characteristics are discussed.
Materials and Methods
Samples
DNA was extracted from patient peripheral blood samples using the QIAmp DNA kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Appropriate Johns Hopkins University Institutional Review Board approval was obtained to perform this study.
ASO Assay
PCR reactions consisted of 1X PCR buffer (Applied Biosystems, Foster City, CA), 200 μmol/L each dNTP (Applied Biosystems), 0.01% gelatin, 2.5 U Taq (Applied Biosystems), 0.4 μmol/L each primer in a 50 μl reaction volume. Primers, 1285F 5′-GATGAAATAGGATGTAATCAGACG-3′ and 1316R 5′-CTTCGCTCACAGGATCTTCAGC-3′ were synthesized by Oligos Etc., Wilsonville, OR. Reactions were cycled at 95°C for 2 minutes followed by 35 cycles of 95°C for 1 minute, 55°C for 1 minute, and 72°C for 1 minute. PCR products were slot-blotted twice onto nylon filters and each was hybridized with 32P-labeled oligonucleotides 5′-AATAGCAGAAATAAAAGAAAAGAT-3′(wild-type) or 5′-AAATAGCAGAAAAAAAAGAAAGAT-3′ (mutant). Following hybridization at room temperature for 1 hour, the filters were washed for 10 minutes in 2X saline sodium citrate (SSC), 0.1% sodium dodeycl (lauryl) sulfate (SDS) at room temperature, then washed for 3 minutes at 56°C in 0.2X SSC, 0.1% SDS, and subjected to autoradiography. 6
SNaPshot (Single Nucleotide Primer Extension) Assay
PCR reactions consisted of 1X buffer (Applied Biosystems), 167 μmol/L each dNTP (Applied Biosystems), 0.67 μmol/L each primer (Oligos Etc.) and 2.5 U AmpliTaq Gold (Applied Biosystems) in a 15 μl reaction volume. Primers were 5′-GATGAAATAGGATGTAATCAGACG-3′ (forward) and 5′-CTGCTGGAACTTCGCTCACA-3 (reverse) (Oligos Etc). Thermal cycling conditions were the same as for the PCR for the ASO assay. Following amplification, 6 μl of ExoSAP-IT (USB, Cleveland, OH) was added to each PCR reaction and incubated at 37°C for 1 hour, followed by heat inactivation at 75°C for 15 minutes. The single nucleotide primer extension assay consisted of 5 μl SNaPshot Reaction Mix (Applied Biosystems), 3 μl PCR product, and 0.2 μmol/L of each primer in a 10 μl final reaction volume. Primers 5′-ATACCCTGCAAATAGCAGAAA-3′ and 5′-GACCTAGTTCCAATCTTTTCTTTT-3′ were synthesized by Oligos Etc. Reactions were subjected to 25 cycles of 96°C for 10 seconds, 50°C for 5 seconds, and 60°C for 30 seconds. One μl of SAP (USB) was then added to each reaction and incubated at 37°C for 1 hour followed by heat inactivation at 75°C for 15 minutes. One μl of the SNaPshot products was mixed with deionized formamide and LIZ120 (Applied Biosystems) size standard per the manufacturer’s protocol, heated to 95°C for 2 minutes, and placed on ice for at least 1 minute before injection to the ABI Prism 3100 capillary electrophoresis instrument (Applied Biosystems).
TaqMan Allelic Discrimination Assay
Primers 5′-CTGCTGGAACTTCGCTCACA-3′ (forward), 5′-ACGACACAGGAAGCAGATTCTG-3′ (reverse) and probes 5′-6FAM-TCTTTTCTTTTATTTCTGCTATT-MGBNFQ (wild-type), 5′-VIC-CTTTTCTTTTTTTTCTGCTATT-MGBNFQ (mutant) were synthesized by Applied Biosystems. Reactions consisted of 1X Universal Master Mix (Applied Biosystems), 900 nmol/L each primer, 250 nmol/L each probe in a 50 μl final reaction volume. Reactions were cycled at 50°C for 2 minutes, 95°C for 10 minutes followed by 35 cycles of 95°C for 15 seconds and annealing temperature for 1 minute. Annealing temperatures tested were 54°C, 56°C, 58°C, 60°C, and 62°C. Data were collected in real time followed by a plate read on an ABI Prism 7900HT sequence detection system (Applied Biosystems).
Results and Discussion
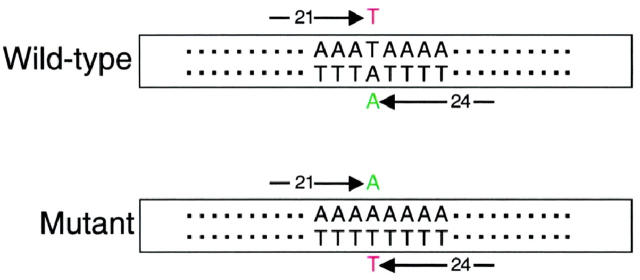
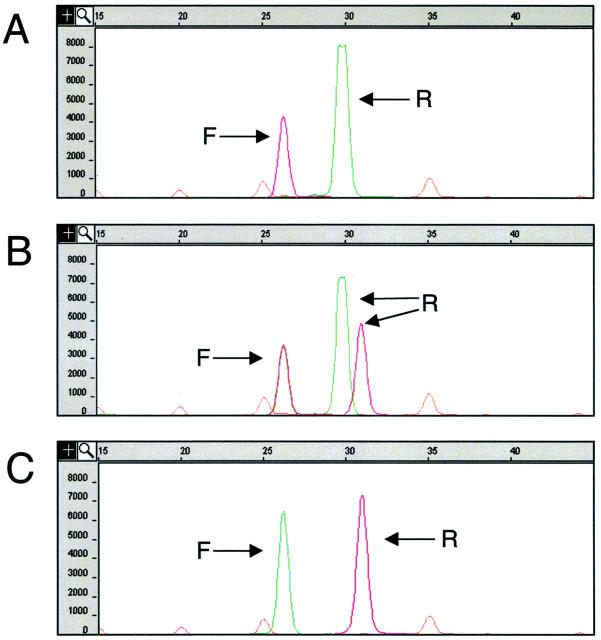
We have designed a single nucleotide primer extension (SNaPshot) assay to detect the APC I1307K gene variant. Genomic samples first were subjected to PCR amplification of a portion of exon 15 of the APC gene. Following PCR, the amplicons underwent thermal cycling with two oligonucleotide primers, DNA polymerase, and fluorescently labeled dideoxynucleotide (ddNTP) terminators using a SNaPshot kit (Applied Biosystems). The SNaPshot oligonucleotide primers were designed such that one annealed immediately 5′ to the variant position in the forward direction and the second annealed immediately 5′ to the variant position in the reverse direction (Figure 1) . The primers were designed to be of different lengths so that they could be distinguished by size during CE separation following the SNaPshot reaction. Using this approach, the APC I1307K locus was interrogated in the forward and reverse direction in a single, multiplex reaction, providing two independent assessments of the base position in question. Because the addition of a non-polar fluor to a short oligonucleotide can significantly alter its electrophoretic mobility it was necessary to determine the electrophoretic mobility of each extended primer empirically. Monoplex reactions using each individual SNaPshot primer were initially performed to determine the mobility of each extended primer before multiplexing the forward and reverse primers into a single reaction (data not shown). The forward SNaPshot primer is a 21-base oligomer. With the addition of either the wild-type (T) or mutant (A) base, the forward primer migrates at ∼26 bases during CE. The reverse SNaPshot primer is a 24-base oligomer that migrates during CE at ∼30 bases with addition of the wild-type (A) base or at ∼31 bases with addition of the mutant (T) base. Figure 2 demonstrates typical examples of wild-type (Figure 2A) , heterozygous mutant (Figure 2B) , and homozygous mutant (Figure 2C) results.
Figure 1.
Schematic of the single nucleotide primer extension assay. The boxes represent the PCR product, the sequence surrounding the I1307K variant is demonstrated. The forward (21-base oligomer) and reverse (24-base oligomer) SNaPshot primers are indicated by arrows. A wild-type and mutant allele are depicted demonstrating the fluorescent ddNTP which extends each primer.
Figure 2.
Examples of SNaPshot results. Capillary electrophoresis pherograms, x axis is size in bases, y axis is fluorescence intensity. The orange peaks represent the internal size standard. The arrows labeled “F” indicate SNaPshot products resulting from extension of the forward primer, sizing at 26 bases. The arrows labeled “R” indicate SNaPshot products resulting from extension of the reverse primer, sizing at either 30 or 31 bases. The addition of a thymidine (T) ddNTP to a primer results in a red-colored product/peak, the addition of an adenine (A) ddNTP results in a green-colored peak. A: Wild-type result demonstrating a red (T) peak at 26 bases resulting from extension of the forward primer and a green (A) peak at 30 bases resulting from extension of the reverse primer. B: Heterozygous mutant result demonstrating superimposed green (A) and red (T) peaks at 26 bases resulting from extension of the forward primer. The green (A) peak at 30 bases and the red (T) peak at 31 bases result from extension of the reverse primer. The green and red reverse primer peaks travel at different sizes due to the effect of the fluorophore on migration. C: Homozygous mutant result demonstrating a green (A) peak at 26 bases resulting from extension of the forward primer and a red peak at 31 bases resulting from extension of the reverse primer.
Sixty samples that had previously been characterized for the APC I1307K genetic variant by an allele-specific oligonucleotide (ASO) assay were analyzed using the described SNaPshot assay. The ASO assay used was originally described by Laken et al 6 and has been used by others to detect the APC I1307K gene variant. 6, 8, 9, 16 The assay involves PCR amplification of the region of the gene of interest followed by slot-blotting of the PCR product onto two nylon membranes and hybridization of each with a radioactively labeled probe specific for either the wild-type or variant APC gene sequence. Examples of results from the ASO assay are shown in Figure 3 . To validate the SNaPshot assay, 40 wild-type, 19 heterozygous mutants, and one homozygous mutant samples were tested by both the ASO and SNaPshot assays. Results demonstrated 100% concordance between the two assays (Table 1) .
Figure 3.
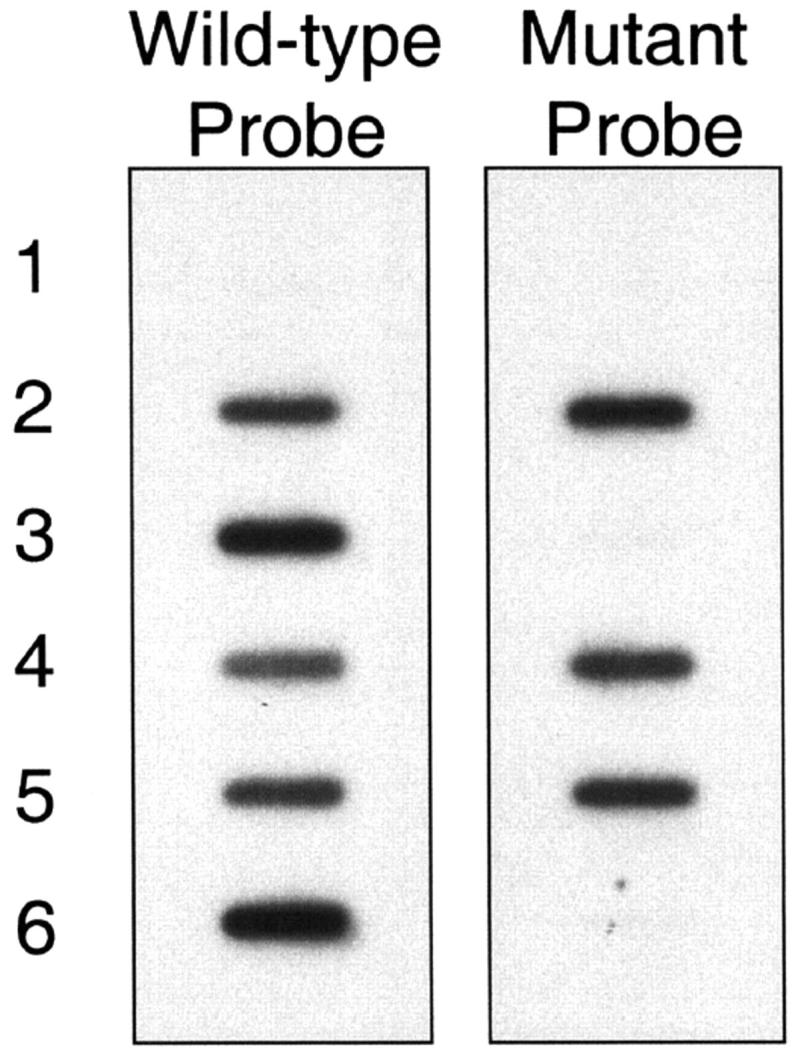
Results from the ASO assay. PCR products were slot-blotted onto two nylon membranes and probed with either a wild-type or mutant radioactive probe. After incubation and washing, the membranes were exposed to film and the results were compared. Lane 1 is a no template control. Lanes 2, 4,and 5 hybridized with both the wild-type and mutant probes and are thus interpreted as heterozygous positive for the APC I1307K variant. Lanes 3 and 6 hybridized with the wild-type probe only and are thus interpreted as wild-type (negative for the I1307K variant).
Table 1.
Comparison of ASO and SNaPshot Results
| SNaPshot results | ||||
|---|---|---|---|---|
| Wild-type (WT/WT) | Heterozygous mutant (WT/MT) | Homozygous mutant (MT/MT) | ||
| ASO results | Wild-type (WT/WT) | 40 | — | — |
| Heterozygous mutant (WT/MT) | — | 19 | — | |
| Homozygous mutant (MT/MT) | — | — | 1 |
The SNaPshot assay has several advantages over the ASO assay, including an improved primer design for the PCR reaction. The reverse primer described by Laken et al 6 is positioned to hybridize within another polymorphic site of the APC gene, namely the subsequently described E1317Q variant. Because the APC E1317Q variant position is four bases from the 3′ end of the Laken et al 6 reverse primer, it is theoretically possible that the presence of the E1317Q variant could affect PCR primer binding and extension and lead to erroneous I1307K genotyping results. For the SNaPshot assay we have chosen an alternate position for the reverse PCR primer that does not encompass the E1317Q position, but instead includes this polymorphic site within the amplified product.
The SNaPshot assay is a very specific assay which has been favorably compared to a 5′ nuclease allelic discrimination assay (TaqMan) and oligonucleotide ligation assay (OLA) for the detection of five different single nucleotide polymorphisms (SNPs). 17 In the assay described here, nucleotide 3920 of the APC gene is interrogated using all four available nucleotide terminators such that all sequence variants are specifically identified with direct sequence data. In addition, each sample is independently interrogated in both the forward and reverse directions, essentially testing each sample twice. The use of two different primers to interrogate the I1307K base has the additional advantage that “private mutations”/SNPs that adversely affect binding/extension of one primer, should not lead to erroneous results.
Another advantage of the SNaPshot assay is the increased throughput capacity and decreased technical time compared to the ASO assay. SNaPshot reactions can be performed in 96- or 384-well plates, greatly increasing the throughput capacity compared to that achievable by hybridization techniques. Indeed, others have used SNaPshot reactions for high-throughput genotyping to characterize polymorphisms in the macrophage migration inhibitory factor (MIF) and tumor necrosis factor (TNF) genes. 18, 19 In addition, SNaPshot reactions can be multiplexed to interrogate up to 10 SNPs in a single reaction, further increasing the potential throughput capacity. 20 Finally, the elimination of radioactivity as part of the detection methodology is highly convenient.
Other assays have been described for research-based APC I1307K genotyping including amplification refractory mutation system (ARMS), 21, 22 heteroduplex analysis, 23 and single-strand conformational polymorphism (SSCP). 21, 24 We are not aware of another report validating a method for testing for this genetic variant in a clinical laboratory setting. The disadvantage of heteroduplex and SSCP analyses is that confirmatory testing (usually by sequencing) is required for all positive results to definitively identify the sequence variant. In comparison, the SNaPshot assay is a highly specific assay, producing sequence data such that confirmatory testing is not required. Before developing the SNaPshot assay, we attempted to develop a TaqMan allelic discrimination assay to detect the APC I1307K gene variant (described in Materials and Methods). Although the assay was designed according to the manufacturer’s specifications and aggressive attempts at optimization were made, the assay showed poor discrimination between wild-type and mutant samples (data not shown). Difficulty in achieving high specificity of the TaqMan probes was likely due to the repetitive nature of the variant locus (homopolymer A8), and may suggest an inherent problem in assaying for variants within repetitive sequences and/or within A/T rich regions, using the TaqMan allelic discrimination approach.
An appealing feature of SNaPshot reactions is that they can be multiplexed to interrogate multiple SNPs simultaneously. As other polymorphisms that increase risk for CRC are identified it may be possible to multiplex a panel of multiple SNPs for CRC risk in a single SNaPshot reaction. Indeed, using the PCR product described here, the APC I1307K and E1317Q genetic variants could be genotyped simultaneously in a single PCR and SNaPshot reaction. Currently there are conflicting data in the literature regarding the risk of CRC for APC E1317Q carriers. These data will need to be clarified before clinical APC E1317Q testing is warranted. 21, 25, 26, 27, 28
Testing for the APC I1307K may facilitate improved clinical management of patients with a positive test result. More frequent screening for the detection of colorectal tumors is indicated for individuals at increased risk because such screening can decrease mortality from CRC. 29 An individual’s compliance with screening recommendations is influenced by many factors, including their proclivity to follow other preventative health behaviors, perception of benefit from screening, physician recommendation for screening, and knowledge of others with colorectal cancer. Patients with higher concern about CRC are more likely to undergo screening, and for some individuals, genetic testing can help clarify that risk. Following the initial report of discovery of the I1307K mutation, we found that acceptance of testing for the APC I1307K mutation was high. 16 Follow-up data from patients seen in our colon cancer risk assessment clinic has shown that patients who were mutation-positive were more likely to adhere to screening guidelines than those with negative gene tests (100% versus 40.5%). 30 The SNaPshot assay described here provides a highly sensitive and specific assay to identify individuals with the APC I1307K gene variant who may benefit from increased screening. Multiplexed SNaPshot assays may become increasingly useful as other genetic variant risk factors for CRC are identified.
Acknowledgments
We thank Ms. Lisa Cooper and Mr. Patrick Pearson for technical assistance.
Address reprint requests to Kathleen M. Murphy, Ph.D., Johns Hopkins Medical Institutions, Carnegie Bldg, Room 367, 600 North Wolfe Street, Baltimore, MD 21287. E-mail: kmurphy4@jhmi.edu.
References
- 1.Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ: Cancer statistics, 2003. CA Cancer J Clin 2003, 53:5-26 [DOI] [PubMed] [Google Scholar]
- 2.Cannon-Albright LA, Skolnick MH, Bishop DT, Lee RG, Burt RW: Common inheritance of susceptibility to colonic adenomatous polyps and associated colorectal cancers. N Engl J Med 1988, 319:533-537 [DOI] [PubMed] [Google Scholar]
- 3.Fuchs CS, Giovannucci EL, Colditz GA, Hunter DJ, Speizer FE, Willett WC: A prospective study of family history and the risk of colorectal cancer. N Engl J Med 1994, 331:1669-1674 [DOI] [PubMed] [Google Scholar]
- 4.Kinzler KW, Vogelstein B: Lessons from hereditary colorectal cancer. Cell 1996, 87:159-170 [DOI] [PubMed] [Google Scholar]
- 5.Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, Vogelstein B, Kinzler KW: APC mutations occur early during colorectal tumorigenesis. Nature 1992, 359:235-237 [DOI] [PubMed] [Google Scholar]
- 6.Laken SJ, Petersen GM, Gruber SB, Oddoux C, Ostrer H, Giardiello FM, Hamilton SR, Hampel H, Markowitz A, Klimstra D, Jhanwar S, Winawer S, Offit K, Luce MC, Kinzler KW, Vogelstein B: Familial colorectal cancer in Ashkenazim due to a hypermutable tract in APC. Nat Genet 1997, 17:79-83 [DOI] [PubMed] [Google Scholar]
- 7.Gryfe R, Di Nicola N, Gallinger S, Redston M: Somatic instability of the APC I1307K allele in colorectal neoplasia. Cancer Res 1998, 58:4040-4043 [PubMed] [Google Scholar]
- 8.Woodage T, King SM, Wacholder S, Hartge P, Struewing JP, McAdams M, Laken SJ, Tucker MA, Brody LC: The APCI1307K allele and cancer risk in a community-based study of Ashkenazi Jews. Nat Genet 1998, 20:62-65 [DOI] [PubMed] [Google Scholar]
- 9.Rozen P, Shomrat R, Strul H, Naiman T, Karminsky N, Legum C, Orr-Urtreger A: Prevalence of the I1307K APC gene variant in Israeli Jews of differing ethnic origin and risk for colorectal cancer. Gastroenterology 1999, 116:54-57 [DOI] [PubMed] [Google Scholar]
- 10.Gryfe R, Di Nicola N, Lal G, Gallinger S, Redston M: Inherited colorectal polyposis and cancer risk of the APC I1307K polymorphism. Am J Hum Genet 1999, 64:378-384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stern HS, Viertelhausen S, Hunter AG, O’Rourke K, Cappelli M, Perras H, Serfas K, Blumenthall A, Dewar D, Baumann E, Lagarde AE: APC I1307K increases risk of transition from polyp to colorectal carcinoma in Ashkenazi Jews. Gastroenterology 2001, 120:392-400 [DOI] [PubMed] [Google Scholar]
- 12.Redston M, Nathanson KL, Yuan ZQ, Neuhausen SL, Satagopan J, Wong N, Yang D, Nafa D, Abrahamson J, Ozcelik H, Antin-Ozerkis D, Andrulis I, Daly M, Pinsky L, Schrag D, Gallinger S, Kaback M, King MC, Woodage T, Brody LC, Godwin A, Warner E, Weber B, Foulkes W, Offit K: The APCI1307K allele and breast cancer risk. Nat Genet 1998, 20:13-14 [DOI] [PubMed] [Google Scholar]
- 13.Shtoyerman-Chen R, Friedman E, Figer A, Carmel M, Patael Y, Rath P, Fidder HH, Bar-Meir S, Theodor L: The I1307K APC polymorphism: prevalence in non-Ashkenazi Jews and evidence for a founder effect. Genet Test 2001, 5:141-146 [DOI] [PubMed] [Google Scholar]
- 14.Yuan ZQ, Kasprzak L, Gordon PH, Pinsky L, Foulkes WD: I1307K APC and hMLH1 mutations in a non-Jewish family with hereditary non-polyposis colorectal cancer. Clin Genet 1998, 54:368-370 [DOI] [PubMed] [Google Scholar]
- 15.Nathanson KL, Antin-Ozerkis D, Couch FJ, Weber BL: I1307K APC variant in non-Ashkenazi Jewish women affected with breast cancer. Am J Med Genet 1999, 85:189-190 [PubMed] [Google Scholar]
- 16.Johnson KA, Rosenblum-Vos L, Petersen GM, Brensinger JD, Giardiello FM, Griffin CA: Response to genetic counseling and testing for the APC I1307K mutation. Am J Med Genet 2000, 91:207-211 [DOI] [PubMed] [Google Scholar]
- 17.Aydin A, Baron H, Bahring S, Schuster H, Luft FC: Efficient and cost-effective single nucleotide polymorphism detection with different fluorescent applications. Biotechniques 2001, 31:920-922, 924, 926–928 [DOI] [PubMed] [Google Scholar]
- 18.Donn R, Alourfi Z, De Benedetti F, Meazza C, Zeggini E, Lunt M, Stevens A, Shelley E, Lamb R, Ollier WE, Thomson W, Ray D: Mutation screening of the macrophage migration inhibitory factor gene: positive association of a functional polymorphism of macrophage migration inhibitory factor with juvenile idiopathic arthritis. Arthritis Rheum 2002, 46:2402-2409 [DOI] [PubMed] [Google Scholar]
- 19.Zeggini E, Thomson W, Kwiatkowski D, Richardson A, Ollier W, Donn R: Linkage and association studies of single-nucleotide polymorphism-tagged tumor necrosis factor haplotypes in juvenile oligoarthritis. Arthritis Rheum 2002, 46:3304-3311 [DOI] [PubMed] [Google Scholar]
- 20.Makridakis NM, Reichardt JK: Multiplex automated primer extension analysis: simultaneous genotyping of several polymorphisms. Biotechniques 2001, 31:1374-1380 [DOI] [PubMed] [Google Scholar]
- 21.Frayling IM, Beck NE, Ilyas M, Dove-Edwin I, Goodman P, Pack K, Bell JA, Williams CB, Hodgson SV, Thomas HJ, Talbot IC, Bodmer WF, Tomlinson IP: The APC variants I1307K and E1317Q are associated with colorectal tumors, but not always with a family history. Proc Natl Acad Sci USA 1998, 95:10722-10727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drucker L, Shpilberg O, Neumann A, Shapira J, Stackievicz R, Beyth Y, Yarkoni S: Adenomatous polyposis coli I1307K mutation in Jewish patients with different ethnicity: prevalence and phenotype. Cancer 2000, 88:755-760 [PubMed] [Google Scholar]
- 23.Petrukhin L, Dangel J, Vanderveer L, Costalas J, Bellacosa A, Grana G, Daly M, Godwin AK: The I1307K APC mutation does not predispose to colorectal cancer in Jewish Ashkenazi breast and breast-ovarian cancer kindreds. Cancer Res 1997, 57:5480-5484 [PubMed] [Google Scholar]
- 24.Abrahamson J, Moslehi R, Vesprini D, Karlan B, Fishman D, Smotkin D, Ben David Y, Biran H, Fields A, Brunet JS, Narod SA: No association of the I1307K APC allele with ovarian cancer risk in Ashkenazi Jews. Cancer Res 1998, 58:2919-2922 [PubMed] [Google Scholar]
- 25.Popat S, Stone J, Coleman G, Marshall G, Peto J, Frayling I, Houlston R: Prevalence of the APC E1317Q variant in colorectal cancer patients. Cancer Lett 2000, 149:203-206 [DOI] [PubMed] [Google Scholar]
- 26.Lamlum H, Al Tassan N, Jaeger E, Frayling I, Sieber O, Reza FB, Eckert M, Rowan A, Barclay E, Atkin W, Williams C, Gilbert J, Cheadle J, Bell J, Houlston R, Bodmer W, Sampson J, Tomlinson I: Germ-line APC variants in patients with multiple colorectal adenomas, with evidence for the particular importance of E1317Q. Hum Mol Genet 2000, 9:2215-2221 [DOI] [PubMed] [Google Scholar]
- 27.Michils G, Tejpar S, Fryns JP, Legius E, Van Cutsem E, Cassiman JJ, Matthijs G: Pathogenic mutations and rare variants of the APC gene identified in 75 Belgian patients with familial adenomatous polyposis by fluorescent enzymatic mutation detection (EMD). Eur J Hum Genet 2002, 10:505-510 [DOI] [PubMed] [Google Scholar]
- 28.Heinimann K, Thompson A, Locher A, Furlanetto T, Bader E, Wolf A, Meier R, Walter K, Bauerfeind P, Marra G, Muller H, Foernzler D, Dobbie Z: Non-truncating APC germ-line mutations and mismatch repair deficiency play a minor role in APC mutation-negative polyposis. Cancer Res 2001, 61:7616-7622 [PubMed] [Google Scholar]
- 29.Winawer SJ, Fletcher RH, Miller L, Godlee F, Stolar MH, Mulrow CD, Woolf SH, Glick SN, Ganiats TG, Bond JH, Rosen L, Zapka JG, Olsen SJ, Giardiello FM, Sisk JE, Van Antwerp R, Brown-Davis C, Marciniak DA, Mayer RJ: Colorectal cancer screening: clinical guidelines and rationale. Gastroenterology 1997, 112:594-642 [DOI] [PubMed] [Google Scholar]
- 30.Johnson KA, Trimbath JD, Petersen GM, Griffin CA, Giardiello FM: Impact of genetic counseling and testing on colorectal cancer screening behavior. Genet Test 2002, 6:303-306 [DOI] [PubMed] [Google Scholar]