Abstract
The clinical management of non-small cell lung cancer (NSCLC) would benefit greatly by a test that was able to detect small amounts of NSCLC in the peripheral blood. In this report, we used a novel strategy to enrich tumor cells from the peripheral blood of 24 stage I to IV NSCLC patients and determined expression levels for six cancer-associated genes (lunx, muc1, KS1/4, CEA, CK19, and PSE). Using thresholds established at three standard deviations above the mean observed in 15 normal controls, we observed that lunx (10 of 24, 42%), muc1 (5 of 24, 21%), and CK19 (5 of 24, 21%) were overexpressed in 14 of 24 (58%) peripheral blood samples obtained from NSCLC patients. Patients who overexpressed either KS1/4 (n = 2) or PSE (n = 1) also overexpressed either lunx or muc1. Of patients with presumed curable and resectable stage I to II disease (n = 7), at least one marker was overexpressed in three (43%) patients. In advanced stage III to IV patients (n = 17), at least one marker was overexpressed in 11 patients (65%). These results provide evidence that circulating tumor cells can be detected in NSCLC patients by a high throughput molecular technique. Further studies are needed to determine the clinical relevance of gene overexpression.
Non-small cell lung carcinoma (NSCLC) is the most common cause of cancer-related death among men and women in the United States. Standard therapies for patients with NSCLC include surgery, chemotherapy, and radiation therapy, and the stage of disease dictates the choice of therapy. The current staging system for lung cancer uses the American Joint Committee on Cancer (AJCC) TNM system, and its goal is to classify patients into groups based on the extent of disease. This system relies heavily on the pathological evaluation of the primary tumor (T), regional nodes (N), and distant metastases (M). Patients with early stage disease (no evidence of disease cancer beyond the lung, stage I or II), are considered candidates for surgery for cure with an operation. In contrast, patients with evidence of disease in mediastinal lymph nodes or distant organs (stage III or IV) are not considered candidates for surgery. Thus, surgery is considered primary therapy for patients with early stage disease, while non-surgical therapy is recommended for patients with advanced stage disease. 1 Reliable detection of metastatic disease is therefore critical for appropriate staging and treatment of patients with NSCLC.
Non-small cell lung cancer metastasizes through both lymphatic and hematogenous routes. The current staging evaluation addresses these possibilities by defining stage based on the presence or absence of disease in lymph nodes (hilar and mediastinal) and distant organs (principally bone, brain, adrenal glands, and liver). 2 Following a thorough evaluation, approximately 35% of patients will be considered to have early stage disease and will be candidates for curative lung resection. Unfortunately, 50% of these patients will develop metastases within 5 years of surgery and die from their disease. Further, even in patients with the very earliest stage of disease (pathologically confirmed stage I), the 5-year survival is only 75%. 3 These statistics indicate that conventional staging techniques lack the sensitivity necessary to identify patients who are unlikely to benefit from surgery. 4 Indeed, recent studies of sensitive assays for detection of rare disseminated lung cancer cells have shown that detection of occult metastases by immunocytology or immunohistochemistry in lymph nodes or bone marrow aspirates is a reliable predictor of poor prognosis. 5, 6, 7 However, the detection of circulating tumor cells in peripheral blood has remained a challenge.
Reverse transcriptase polymerase chain reaction (RT-PCR) has been recognized as the method with the highest sensitivity for detection of micrometastatic disease, allowing identification of one cancer cell in 106 to 107 normal cells. 8, 9, 10, 11, 12, 13, 14, 15, 16 Detection of metastatic cancer cells by RT-PCR is based on the fact that cancer cells continue to express genes (or markers) that are specific to the tissue from which they originate, but are not expressed in tissue compartments that frequently harbor metastatic foci, such as lymph nodes and bone marrow. 17, 18 Although RT-PCR-based tumor cell detection assays often yield higher sensitivity than conventional immunohistochemistry, 4, 17, 19, 20 such assays for lung cancer have been limited by the availability of molecular markers. 21 However, two genes have recently shown promise by virtue of the fact that they are overexpressed in metastatic mediastinal lymph nodes of NSCLC patients: 22 lunx, 21 (also known as palate, lung, and nasal epithelium carcinoma-associated (Plunc) gene 23, 24, 25 ) and the epithelial carcinoma-associated gene KS1/4. 26
In addition to aspects of NSCLC marker identification, advances have also been made in tumor cell enrichment. A porous barrier density gradient (PBDG) centrifugation system has recently been developed that results in reliable recovery of tumor cells from peripheral blood with a concomitant >500-fold depletion of white blood cells. 27 This new tumor cell enrichment system can be combined with real-time RT-PCR, a high throughput technology that allows for sensitive detection and quantitation of gene expression. Real-time RT-PCR measures the on-line fluorescence of products as they are amplified from one cycle to the next and is quickly becoming recognized as the technology of choice for the precise measurement of gene expression levels. 8, 28 In this report, we combined real-time RT-PCR with PBDG centrifugation and show that of the five NSCLC-associated markers examined, the one with the highest sensitivity was lunx.
Materials and Methods
Oligonucleotides
All primers were designed using Primer Express software (ABI, Foster City, CA), spanned at least one intron, and failed to amplify negative control cDNA in which reverse transcriptase enzyme was omitted. Sequences of the internal control β2-microglobulin primers were: 5` GCCGTGTGAACCATGTGA (forward) and 5` CCAAATGCGGCATCTTCA (reverse). Other sequences (previously described 22 ) were: lunx, CCCTGGAAGCCTGCAAATT (F) GAACCAACTCAGGCAGGACTTT (R); KS1/4, CGCAGCTCAGGAAGAATGTG (F), TGAAGTACACTGGCATTGACGA(R); CK19, CATGAAAGCTGCCTTGGAAGA (F), TGATTCTGCCGCTCACTATCAG (R); CEA, GGGCCACTGTCGCATCATGATTGG (F), TGTAGCTGTTGCAAATGCTTTAAGAAAGAAGC (R); PSE, AGTGCTCAAGGACATCGAGACG (F), AGCCACTTCTGCACATTGCTG (R). Synthetic lunx fragment for gene copy determination: CCCTGGAAGCCTGCAAATTUCUCUGCUUGAUGGACUUGGCCCCCUCCCCAUUCAAGGUCUUCUGGACAGCCUCACAGGGAUCUUGAAUAAAGTCCTGCCTGAGTTGGTTC.
Peripheral Blood Specimens
This study was approved by the Medical University of South Carolina (MUSC) Institutional Review Board, and informed consent was obtained from all patients enrolled. Peripheral blood specimens (10 to 20 ml) were collected using K3 EDTA tubes (Vacutainer; Becton Dickinson, Franklin Lakes, NJ) and immediately placed in ice. Samples were then processed using a porous barrier density gradient centrifugation media (OncoQuick, Hexal Gentech, Holzkirchen, Germany) per manufacturer’s instructions. Briefly, pre-cooled 50-ml centrifugation tubes containing 15 ml of separation medium below a porous barrier were filled with peripheral blood and centrifuged at 1600 × g for 20 minutes. The entire volume of the upper compartment was then collected and washed for 10 minutes at 200 × g. Cells were pelleted and evaluated as described below.
RNA Isolation and Gene-Specific cDNA Synthesis
Total cellular RNA was isolated from pelleted cells using a guanidinum thiocyanate-phenol-chloroform solution (RNA STAT-60; TEL-TEST, Friendswood, TX). Briefly, pelleted cells recovered from peripheral blood specimens were resuspended in 1 ml of RNA STAT-60. Total RNA was isolated as per manufacturer’s instructions with the exception that 1 μl of a 50 mg/ml solution of glycogen (Sigma, St. Louis, MO) was added to the aqueous phase before addition of isopropanol. Final RNA pellet was dissolved in 50 μl of 1X RNA secure buffer (Ambion, Austin, TX). RNA was quantified by UV absorbance at 260 nm. Complementary DNA (cDNA) was made from 5 μg of total RNA using 200 U of M-MLV reverse transcriptase (Promega, Madison, WI) and the following gene-specific primers (70 ng each): CCAAATGCGGCAT (β2-microglobulin), TGAAGTACACTGG (KS1/4), GAACCAACTCAGGC (lunx), GCCACCATTACCT (muc1), TGATTCTGCCGC (CK19), GTTCCCATCAATCAG (CEA), and AGCCACTTCTGC (PSE). Final reaction volume was 20 μl.
Real-Time RT-PCR
Real-time RT-PCR was performed on a PE Biosystems Gene Amp 5700 Sequence Detection System (Foster City, CA). All reaction components were purchased from PE Biosystems. Standard reaction volume was 10 μl and contained 1X SYBR Green PCR buffer, 3.5 mmol/L MgCl2, 0.2 mmol/L each of dATP, dCTP, dGTP, and 0.4 mmol/L of dUTP, 0.25U AmpliTaq Gold, 0.1U AmpErase UNG enzyme, 0.7 μl cDNA template, and 0.25 mmol/L of forward and reverse primer. Initial step of RT-PCR was 2 minutes at 50°C for AmpErase UNG activation, followed by a 10-minute hold at 95°C. Cycles (n = 40 first round) consisted of a 15-second melt at 95°C, followed by a 1 minute annealing/extension at 60°C. The final step was a 60°C incubation for 1 minute. All reactions were performed in triplicate and a negative control lacking cDNA was included. For a blood sample to be considered evaluable, we set a cutoff value for the β2-microglobulin internal control gene at ≤25 (corresponding to approximately 2 × 104 gene copies).
Results
Reliable Detection of 20 Lunx Gene Copies by Real-Time PCR
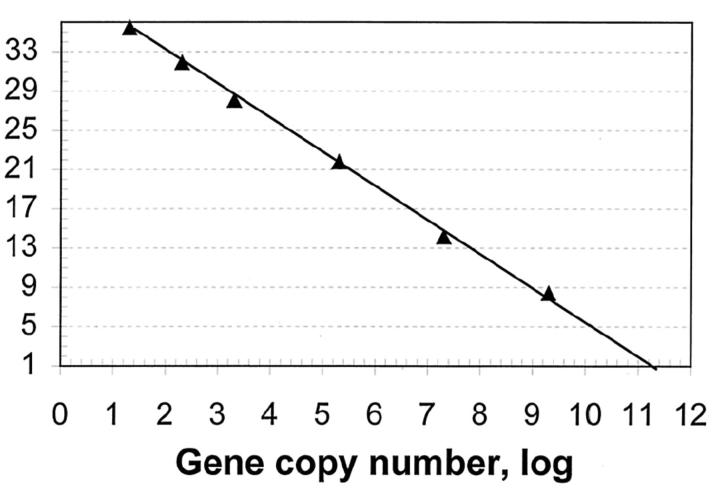
To determine whether a single round of real-time RT-PCR could be used for the sensitive detection of NSCLC, we first performed studies on a synthetic fragment encoding a portion of lunx, a gene previously shown to be expressed in metastatic lymph nodes of NSCLC patients. 21, 22 Ct values for various fragment dilutions were obtained and plotted as a function of initial fragment copy number. Figure 1 demonstrates a strong linear relationship between the Ct value and the log of fragment copy number (R2 = 0.9981). Reliable fluorescent signals were obtained for reactions containing as few as 20 gene copies (Figure 1 , log value = 1.4). In contrast, reliable fluorescent signals were not obtained for samples that contained only two gene copies (data not shown), regardless of the fluorescent threshold setting used for real-time measurements. These data provide evidence that a single round (as opposed to two) of real-time PCR reliably amplifies ≥20 gene copies, a result amenable to detection of circulating tumor cells in peripheral blood.
Figure 1.
Reliable detection of 20 gene copies in a single round of PCR. Real-time RT-PCR reactions were performed in triplicate as described in Materials and Methods using the lunx primer pair and the lunx synthetic sequence listed in Materials and Methods. Gene copy number was determined by UV absorbance measurements at 260 nm. The line through the data points was obtained by linear regression analysis using Microsoft Excel software.
Detection of Lunx Gene Expression in Peripheral Blood of NSCLC Patients
To assess the ability of real-time RT-PCR to detect circulating tumor cells in the peripheral blood of NSCLC patients, samples from 15 healthy volunteers and 24 patients with stage I-IV NSCLC were obtained. Tumor cells were first enriched from peripheral blood by a newly developed porous barrier density gradient (PBDG) centrifugation system. 27 The depletion of mononuclear cells in the enriched cell fraction after PBDG centrifugation is approximately 300- to >500-fold. 27, 29 Mean tumor cell recovery rates for PBDG are comparable to that achieved by Ficoll purification. 27, 29 Previous studies in the breast cancer setting have shown that the upper limit of detection using real-time RT-PCR is one cancer cell among 5 × 108 peripheral blood cells. 29
Using a single round of real-time PCR (40 cycles), we determined expression levels for five genes associated with NSCLC: lunx, KS1/4, muc1, CK19,and CEA, 22 as well as one gene (PSE) associated with prostate 30 and breast cancer. 31, 32 Mean expression levels of the cancer-associated genes were normalized to β2-microglobulin using Q-gene software. 33 We observed that in the normal control peripheral blood samples, expression of the lunx gene was not detectable (Figure 2) . For other genes, expression was detected in a limited number of patients: muc1 and CK19 (four samples), CEA and PSE (three samples), and KS1/4 (two samples) (Figure 2) . Based on data obtained from the normal control population, we set threshold values for marker positivity at three standard deviations beyond the mean normalized expression values of each respective gene (Figure 2 , horizontal lines). Assuming a normal distribution of the control peripheral blood samples, three standard deviations correspond to a test-specificity level of 99.9%. In the control patient group, no gene was overexpressed above threshold levels.
Figure 2.
Multimarker real-time RT-PCR analysis of NSCLC in peripheral blood. Real-time PCR analyses of peripheral blood specimens from 15 healthy volunteers (open triangles), and 24 NSCLC patients (open diamonds) were performed as described in the text using primer pairs for the indicated genes. Threshold levels of marker positivity for each gene were calculated as described in the text and are depicted by the horizontal line on the left side of each data set. Expression levels of each gene were calculated with Q-gene software 33 and are expressed as the ratio of the target gene relative to β2-microglobulin.
In the peripheral blood samples derived from NSCLC patients (n = 24), we observed that 14 of 24 (58%) overexpressed at least one marker gene (Figure 2 , Table 1 ). The gene most highly overexpressed was lunx (10 of 24 samples (42%)). muc1 and CK19 were each overexpressed in 5 of 20 (21%) of patients, three of whom overexpressed both markers. Overexpression of KS1/4 and PSE was observed in two patients and one patient, respectively, all of who overexpressed either lunx or muc1 (Table 1) . Of patients with presumed curable and resectable stage I to II disease (n = 7), lunx was overexpressed in two (29%) blood samples.
Table 1.
Detection of Gene Overexpression in NSCLC Patients
| Patient information | Real-time RT-PCR results* | |||||||
|---|---|---|---|---|---|---|---|---|
| Patient no. | Stage | Age | LUNX | MUC1 | CK19 | KS1/4 | PSE | CEA |
| 1 | IA | 78 | — | — | — | — | — | — |
| 2 | IA | 67 | 1 | — | 1 | — | — | — |
| 3 | IB | 57 | — | — | — | — | — | — |
| 4 | IB | 52 | — | — | — | — | — | — |
| 5 | IB | 75 | 1 | — | — | — | — | — |
| 6 | IIB | 75 | — | — | — | — | — | — |
| 7 | IIB | 41 | — | 1 | 1 | — | — | — |
| 8 | III | 55 | — | — | — | — | — | — |
| 9 | III | 67 | — | — | — | — | — | — |
| 10 | IIIA | 64 | — | — | — | — | — | — |
| 11 | IIIA | 71 | — | — | — | — | — | — |
| 12 | IIIB | 59 | 1 | — | — | — | — | — |
| 13 | IIIB | 63 | 1 | — | — | — | — | — |
| 14 | IIIB | 54 | 1 | 1 | 1 | — | — | — |
| 15 | IIIB | 54 | 1 | — | — | — | — | — |
| 16 | IIIB | 67 | 1 | — | — | — | — | — |
| 17 | IV | 66 | — | — | — | — | — | — |
| 18 | IV | 62 | — | 1 | — | — | — | — |
| 19 | IV | 62 | — | — | — | — | — | — |
| 20 | IV | 74 | 1 | — | — | — | — | — |
| 21 | IV | 46 | 1 | — | — | — | — | — |
| 22 | IV | 77 | — | 1 | — | — | 1 | — |
| 23 | IV | 67 | — | 1 | 1 | 1 | — | — |
| 24 | IV | 52 | 1 | — | 1 | 1 | — | — |
| Total | 10 | 5 | 5 | 2 | 1 | 0 |
No overexpression of the respective gene is indicated by “—”; overexpression is indicated by “1”.
Discussion
The ability to detect nucleic acid fragments by PCR is directly proportional to gene copy number, fragment amplification efficiency, and detection threshold, and inversely proportional to the formation of primer dimers. Due to their extremely low concentration (and hence, gene copy numbers), the molecular detection of cancer cells in peripheral blood has proven challenging compared to other tissues such as lymph node. In an effort to increase the senstivity of NSCLC detection in peripheral blood, reseachers have adopted nested PCR strategies for various genes such as CK19, preproGRP, syndcan 1, collagen 1A2, and CEA. 34, 35, 36, 37, 38, 39 In this study, we provide evidence that the use of real-time PCR and SYBR Green I chemistry allows for reproducible detection of 20 copies of an aritificial lunx sequence by PCR cycle number 36 (Figure 1) . These results agree well with those of Karsai et al, 40 who used SYBR Green I chemistry to detect 10 to 20 copies of UBQ-5 RNA or dsDNA, and Leutenegger et al, 41 who used a real-time TaqMan PCR assay to detect as few as 50 copies of SIV RNA/ml. These results provide evidence that a single round of real-time RT-PCR is sufficient for detection of genes present in low abundance.
Using a single round of real-time PCR, we analyzed the peripheral blood of NSCLC patients for expression of five genes associated with NSCLC:lunx, KS1/4, muc1, CK19,and CEA. 22 With respect to normal control samples, overexpression of at least one gene was observed in 14 of 24 (58%) NSCLC patients. Of stage I to II patients (n = 7), 3 (43%) were positive for at least one marker, while 11 of 17 (65%) stage III to IV patients were marker positive. Ten NSCLC blood samples were positive for lunx, providing evidence that this marker was the most sensitive for detection of circulating NSCLC cells.
In a previous study, we used the same markers described in this paper and observed that KS1/4 had the highest sensitivity for detection of NSCLC in mediastinal lymph nodes. 22 However, in the present study, we observed that KS1/4 was overexpressed in the peripheral blood of only 2 of 24 (8%) patients. The low level of overexpression of this gene in peripheral blood may be due to several factors, including small patient sample size, lack of appropriate growth factors in peripheral blood, and/or increased background gene expression. In support of the later possibility, Zhong and colleagues 42 found by RT-PCR that KS1/4 was expressed in 40% (16 of 40) of normal peripheral blood samples and 100% of normal bone marrow samples (n = 8). DeGraffe et al, 43 using quantitative nested RT-PCR, found a consistently low level of KS1/4 mRNA (4 × 10 −4 copies/cell) in peripheral blood mononuclear cells from normal donors while several breast cancer cell lines expressed 20 to 100 copies/cell. It also should be noted that KS1/4 has been extensively studied as a molecular marker of various cancers. However, this fact is not obvious since the KS1/4 marker and/or the antibody which recognizes its gene product is known by various names (TROP1, 44 AUA1, 45 HeGP314, 46 CO17–1A, 47 EpCAM, 48 MK-1, 49 M4S1, 50 EGP40, 51 EGP2, 52 TACSTD1, 50 KSA, 48 and GA733–2 53 ).
The results described in this paper provide evidence that lunx was the most sensitive marker for detection of circulating NSCLC cells. Examination of the predicted amino acid sequence of plunc, the mouse homologue of lunx, indicates that it is a secreted protein and related to proteins expressed in high abundance in saliva. 23 During mouse embryogenesis, plunc/lunx expression is limited in a temporal manner to the dorso-lateral epithelium of the developing palatal shelves. In the adult (mouse), expression is confined to discrete epithelial bands on the exposed surfaces of the nasal columella, turbinates, and common nasal passage. 23 Recent mass spectroscopy studies indicate that the human lunx gene product is also expressed in normal adult nasal lavage fluid. Further, its expression might be up-regulated in response to certain airway irritants such as cigarette smoke and dimethylbenzylamine. 54, 55 Although lunx mRNA has been detected in metastatic lymph nodes of NSCLC patients, 21, 22 this is the first study to demonstrate expression of lunx in peripheral blood of NSCLC patients. Additional clinical studies with follow-up data are required to determine whether detection of lunx in early stage II NSCLC patients correlates with decreased survival.
Acknowledgments
We thank Angel Anderson and Susan Clarkson Keller for assistance in recruiting patients. We also thank Shelitta Warren and Jennifer Small for drawing patient blood.
Address reprint requests to Michael Mitas, Ph.D., Department of Surgery, Suite 420, 96 Jonathan Lucas Street, Charleston, SC 29425. E-mail: mitasm@musc.edu.
Footnotes
Supported by Department of Defense grant DOD/ONR N6311600MDM0U01-SP0007 (to M.M.)
References
- 1.Miller JD, Gorenstein LA, Patterson GA: Staging: the key to rational management of lung cancer. Ann Thorac Surg 1992, 53:170-178 [PubMed] [Google Scholar]
- 2.Mountain CF, Dresler CM: Regional lymph node classification for lung cancer staging. Chest 1997, 111:1718-1723 [DOI] [PubMed] [Google Scholar]
- 3.Martini N, Bains MS, Burt ME, Zakowski MF, McCormack P, Rusch VW, Ginsberg RJ: Incidence of local recurrence and second primary tumors in resected stage I lung cancer. J Thorac Cardiovasc Surg 1995, 109:120-129 [DOI] [PubMed] [Google Scholar]
- 4.Pantel K, Cote RJ, Fodstad O: Detection and clinical importance of micrometastatic disease. J Natl Cancer Inst 1999, 91:1113-1124 [DOI] [PubMed] [Google Scholar]
- 5.Cote RJ, Beattie EJ, Chaiwun B, Shi SR, Harvey J, Chen SC, Sherrod AE, Groshen S, Taylor CR: Detection of occult bone marrow micrometastases in patients with operable lung carcinoma. Ann Surg 1995, 222:415-423; 423415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kubuschok B, Passlick B, Izbicki JR, Thetter O, Pantel K: Disseminated tumor cells in lymph nodes as a determinant for survival in surgically resected non-small cell lung cancer. J Clin Oncol 1999, 17:19-24 [DOI] [PubMed] [Google Scholar]
- 7.Passlick B, Kubuschok B, Izbicki JR, Thetter O, Pantel K: Isolated tumor cells in bone marrow predict reduced survival in node-negative non-small cell lung cancer. Ann Thorac Surg 1999, 68:2053-2058 [DOI] [PubMed] [Google Scholar]
- 8.Mitas M, Mikhitarian K, Walters C, Baron PL, Elliott BM, Brothers TE, Robison JG, Metcalf JS, Palesch YY, Zhang Z, Gillanders WE, Cole DJ: Quantitative real-time RT-PCR detection of breast cancer micrometastasis using a multi-gene marker panel. Int J Cancer 2001, 93:162-171 [DOI] [PubMed] [Google Scholar]
- 9.Gerhard M, Juhl H, Kalthoff H, Schreiber HW, Wagener C, Neumaier M: Specific detection of carcinoembryonic antigen-expressing tumor cells in bone marrow aspirates by polymerase chain reaction. J Clin Oncol 1994, 12:725-729 [DOI] [PubMed] [Google Scholar]
- 10.Mori M, Mimori K, Ueo H, Karimine N, Barnard GF, Sugimachi K, Akiyoshi T: Molecular detection of circulating solid carcinoma cells in the peripheral blood: the concept of early systemic disease. Int J Cancer 1996, 68:739-743 [DOI] [PubMed] [Google Scholar]
- 11.Battaglia M, Pedrazzoli P, Palermo B, Lanza A, Bertolini F, Gibelli N, Da Prada GA, Zambelli A, Perotti C, Robustelli della Cuna G: Epithelial tumour cell detection and the unsolved problems of nested RT-PCR: a new sensitive one-step method without false positive results. Bone Marrow Transplant 1998, 22:693-698 [DOI] [PubMed] [Google Scholar]
- 12.Zach O, Kasparu H, Krieger O, Hehenwarter W, Girschikofsky M, Lutz D: Detection of circulating mammary carcinoma cells in the peripheral blood of breast cancer patients via a nested reverse transcriptase polymerase chain reaction assay for mammaglobin mRNA. J Clin Oncol 1999, 17:2015-2019 [DOI] [PubMed] [Google Scholar]
- 13.Grunewald K, Haun M, Urbanek M, Fiegl M, Muller-Holzner E, Gunsilius E, Dunser M, Marth C, Gastl G: Mammaglobin gene expression: a superior marker of breast cancer cells in peripheral blood in comparison to epidermal growth factor receptor and cytokeratin-19. Lab Invest 2000, 80:1071-1077 [DOI] [PubMed] [Google Scholar]
- 14.Ballestrero A, Coviello DA, Garuti A, Nencioni A, Fama A, Rocco I, Bertorelli R, Ferrando F, Gonella R, Patrone F: Reverse-transcriptase polymerase chain reaction of the maspin gene in the detection of bone marrow breast carcinoma cell contamination. Cancer 2001, 92:2030-2035 [DOI] [PubMed] [Google Scholar]
- 15.Berois N, Varangot M, Aizen B, Estrugo R, Zarantonelli L, Fernandez P, Krygier G, Simonet F, Barrios E, Muse I, Osinaga E: Molecular detection of cancer cells in bone marrow and peripheral blood of patients with operable breast cancer: comparison of CK19, MUC1, and CEA using RT-PCR. Eur J Cancer 2000, 36:717-723 [DOI] [PubMed] [Google Scholar]
- 16.Slade MJ, Smith BM, Sinnett HD, Cross NC, Coombes RC: Quantitative polymerase chain reaction for the detection of micrometastases in patients with breast cancer. J Clin Oncol 1999, 17:870-879 [DOI] [PubMed] [Google Scholar]
- 17.Ghossein RA, Carusone L, Bhattacharya S: Molecular detection of micrometastases and circulating tumor cells in melanoma prostatic and breast carcinomas. In Vivo 2000, 14:237-250 [PubMed] [Google Scholar]
- 18.Datta YH, Adams PT, Drobyski WR, Ethier SP, Terry VH, Roth MS: Sensitive detection of occult breast cancer by the reverse-transcriptase polymerase chain reaction. J Clin Oncol 1994, 12:475-482 [DOI] [PubMed] [Google Scholar]
- 19.Raj GV, Moreno JG, Gomella LG: Utilization of polymerase chain reaction technology in the detection of solid tumors. Cancer 1998, 82:1419-1442 [DOI] [PubMed] [Google Scholar]
- 20.von Knebel Doeberitz M, Lacroix J: Nucleic acid-based techniques for the detection of rare cancer cells in clinical samples. Cancer Metastasis Rev 1999, 18:43-64 [DOI] [PubMed] [Google Scholar]
- 21.Iwao K, Watanabe T, Fujiwara Y, Takami K, Kodama K, Higashiyama M, Yokouchi H, Ozaki K, Monden M, Tanigami A: Isolation of a novel human lung-specific gene, LUNX, a potential molecular marker for detection of micrometastasis in non-small cell lung cancer. Int J Cancer 2001, 91:433-437 [DOI] [PubMed] [Google Scholar]
- 22.Mitas M, Cole DJ, Hoover L, Fraig MM, Mikhitarian K, Block MI, Hoffman BJ, Hawes RH, Gillanders WE, Wallace MB: Real-time RT-PCR detects KS1/4 mRNA in mediastinal lymph nodes from patients with non-small cell lung cancer. Clin Chem 2003, 49:312-315 [DOI] [PubMed] [Google Scholar]
- 23.Weston W, LeClair E, Trzyna W, McHugh K, Nugent P, Lafferty C, Ma L, Tuan R, RM G: Differential display identification of plunc, a novel gene expressed in embryonic palate, nasal epithelium, and adult lung. J Biol Chem 1999, 274:13698-13703 [DOI] [PubMed] [Google Scholar]
- 24.Bingle CD, Bingle L: Characterisation of the human plunc gene, a gene product with an upper airways and nasopharyngeal restricted expression pattern. Biochim Biophys Acta 2000, 1493:363-367 [DOI] [PubMed] [Google Scholar]
- 25.Sung YK, Moon C, Yoo JY, Pearse D, Pevsner J, Ronnett GV: Plunc, a member of the secretory gland protein family, is up-regulated in nasal respiratory epithelium after olfactory bulbectomy. J Biol Chem 2002, 277:12762-12769 [DOI] [PubMed] [Google Scholar]
- 26.Perez MS, Walker LE: Isolation and characterization of a cDNA encoding the KS1/4 epithelial carcinoma marker. J Immunol 1989, 142:3662-3667 [PubMed] [Google Scholar]
- 27.Rosenberg R, Gertler R, Friederichs J, Fuehrer K, Dahm M, Phelps R, Thorban S, Nekarda H, Siewert JR: Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry 2002, 49:150-158 [DOI] [PubMed] [Google Scholar]
- 28.Bieche I, Olivi M, Champeme MH, Vidaud D, Lidereau R, Vidaud M: Novel approach to quantitative polymerase chain reaction using real-time detection: application to the detection of gene amplification in breast cancer. Int J Cancer 1998, 78:661-666 [DOI] [PubMed] [Google Scholar]
- 29.Baker M, Mikhitarian K, Hoda R, Brescia F, Kneuper-Hall R, Mitas M, Cole D, Gillanders W: Molecular detection of breast cancer cells in the peripheral blood of advanced stage breast cancer patients using multi-marker real-time RT-PCR and a novel porous barrier density gradient centrifugation technology. Clin Cancer Res 2003, in press [PubMed]
- 30.Oettgen P, Finger E, Sun Z, Akbarali Y, Thamrongsak U, Boltax J, Grall F, Dube A, Weiss A, Brown L, Quinn G, Kas K, Endress G, Kunsch C, Libermann TA: PDEF, a novel prostate epithelium-specific ets transcription factor, interacts with the androgen receptor and activates prostate-specific antigen gene expression. J Biol Chem 2000, 275:1216-1225 [DOI] [PubMed] [Google Scholar]
- 31.Ghadersohi A, Sood AK: Prostate epithelium-derived ets transcription factor mRNA is overexpressed in human breast tumors and is a candidate breast tumor marker and a breast tumor antigen. Clin Cancer Res 2001, 7:2731-2738 [PubMed] [Google Scholar]
- 32.Mitas M, Mikhitarian K, Hoover L, Lockett MA, Kelley L, Hill A, Gillanders WE, Cole DJ: Prostate-specific ets (PSE) factor: a novel marker for the detection of metastatic breast cancer in axillary lymph nodes. Br J Cancer 2002, 86:899-905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller PY, Janovjak H, Miserez AR, Dobbie Z: Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 2002, 32:1372-1374, 1376, 13781379 [PubMed] [Google Scholar]
- 34.Peck K, Sher YP, Shih JY, Roffler SR, Wu CW, Yang PC: Detection and quantitation of circulating cancer cells in the peripheral blood of lung cancer patients. Cancer Res 1998, 58:2761-2765 [PubMed] [Google Scholar]
- 35.Lacroix J, Becker HD, Woerner SM, Rittgen W, Drings P, von Knebel Doeberitz M: Sensitive detection of rare cancer cells in sputum and peripheral blood samples of patients with lung cancer by preproGRP-specific RT-PCR. Int J Cancer 2001, 92:1-8 [PubMed] [Google Scholar]
- 36.Kurusu Y, Yamashita J, Ogawa M: Detection of circulating tumor cells by reverse transcriptase-polymerase chain reaction in patients with resectable non-small cell lung cancer. Surgery 1999, 126:820-826 [PubMed] [Google Scholar]
- 37.Yamashita JI, Kurusu Y, Fujino N, Saisyoji T, Ogawa M: Detection of circulating tumor cells in patients with non-small cell lung cancer undergoing lobectomy by video-assisted thoracic surgery: a potential hazard for intraoperative hematogenous tumor cell dissemination. J Thorac Cardiovasc Surg 2000, 119:899-905 [DOI] [PubMed] [Google Scholar]
- 38.Yamashita J, Matsuo A, Kurusu Y, Saishoji T, Hayashi N, Ogawa M: Preoperative evidence of circulating tumor cells by means of reverse transcriptase-polymerase chain reaction for carcinoembryonic antigen messenger RNA is an independent predictor of survival in non-small cell lung cancer: a prospective study. J Thorac Cardiovasc Surg 2002, 124:299-305 [DOI] [PubMed] [Google Scholar]
- 39.Matsunaga H, Hangai N, Aso Y, Okano K, Kawamura M, Kobayashi K, Kambara H, Hoger JH, Mitsuhashi M: Application of differential display to identify genes for lung cancer detection in peripheral blood. Int J Cancer 2002, 100:592-599 [DOI] [PubMed] [Google Scholar]
- 40.Karsai A, Muller S, Platz S, Hauser MT: Evaluation of a homemade SYBR Green I reaction mixture for real-time PCR quantification of gene expression. Biotechniques 2002, 32:790-792, 794796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leutenegger CM, Higgins J, Matthews TB, Tarantal AF, Luciw PA, Pedersen NC, North TW: Real-time TaqMan PCR as a specific and more sensitive alternative to the branched-chain DNA assay for quantitation of simian immunodeficiency virus RNA. AIDS Res Hum Retroviruses 2001, 17:243-251 [DOI] [PubMed] [Google Scholar]
- 42.Zhong X, Kaul S, Eichler A, Bastert G: Evaluating GA733–2 mRNA as a marker for the detection of micrometastatic breast cancer in peripheral blood and bone marrow. Arch Gynecol Obstet 1999, 263:2-6 [DOI] [PubMed] [Google Scholar]
- 43.de Graaf H, Maelandsmo G, Ruud P, Forus A, Oyjord T, Fodstad O, Hovig E: Ectopic expression of target genes may represent an inherent limitation of RT-PCR assays used for micrometastasis detection: studies on the epithelial glycoprotein gene EGP-2. Int J Cancer 1997, 72:191-196 [DOI] [PubMed] [Google Scholar]
- 44.Alberti S, Nutini M, Herzenberg L: DNA methylation prevents the amplification of TROP1, a tumor-associated cell surface antigen gene. Proc Natl Acad Sci USA 1994, 91:5833-5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson R, Royston D: Comparison of monoclonal antibodies AUA1 and BER EP4 with anti-CEA for detecting carcinoma cells in serous effusions and distinguishing them from mesothelial cells. Cytopathology 1993, 4:267-271 [DOI] [PubMed] [Google Scholar]
- 46.Bergsagel P, Victor-Kobrin C, Timblin C, Trepel J, Kuehl W: A murine cDNA encodes a pan-epithelial glycoprotein that is also expressed on plasma cells. J Immunol 1992, 148:590-596 [PubMed] [Google Scholar]
- 47.Ross A, Lubeck M, Steplewski Z, Koprowski H: Identification and characterization of the CO17–1A carcinoma-associated antigen. Hybridoma 1986, 5(Suppl 1):S21-S28 [PubMed] [Google Scholar]
- 48.Strnad J, Hamilton A, Beavers L, Gamboa G, Apelgren L, Taber L, Sportsman J, Bumol T, Sharp J, Gadski R: Molecular cloning and characterization of a human adenocarcinoma/epithelial cell surface antigen complementary DNA. Cancer Res 1989, 49:314-317 [PubMed] [Google Scholar]
- 49.Tomita Y, Arakawa F, Yamamoto T, Kuwahara M, Watanabe R, Iwasaki H, Kikuchi M, Kuroki M: Molecular identification of a human carcinoma-associated glycoprotein antigen recognized by mouse monoclonal antibody FU-MK-1. Jpn J Cancer Res 2000, 91:231-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Calabrese G, Crescenzi C, Morizio E, Palka G, Guerra E, Alberti S: Assignment of TACSTD1 (alias TROP1, M4S1) to human chromosome 2p21 and refinement of mapping of TACSTD2 (alias TROP2, M1S1) to human chromosome 1p32 by in situ hybridization. Cytogenet Cell Genet 2001, 92:164-165 [DOI] [PubMed] [Google Scholar]
- 51.Velders M, Litvinov S, Warnaar S, Gorter A, Fleuren G, Zurawski V, Jr, Coney L: New chimeric anti-pancarcinoma monoclonal antibody with superior cytotoxicity-mediating potency. Cancer Res 1994, 54:1753-1759 [PubMed] [Google Scholar]
- 52.Helfrich W, ten Poele R, Meersma G, Mulder N, de Vries E, de Leij L, Smit E: A quantitative reverse transcriptase polymerase chain reaction-based assay to detect carcinoma cells in peripheral blood. Br J Cancer 1997, 76:29-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szala S, Froehlich M, Scollon M, Kasai Y, Steplewski Z, Koprowski H, Linnenbach A: Molecular cloning of cDNA for the carcinoma-associated antigen GA733–2. Proc Natl Acad Sci USA 1990, 87:3542-3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lindahl M, Stahlbom B, Tagesson C, Weston W, LeClair E, Trzyna W, McHugh K, Nugent P, Lafferty C, Ma L, Tuan R, Greene R: Identification of a new potential airway irritation marker, palate lung nasal epithelial clone protein, in human nasal lavage fluid with two-dimensional electrophoresis and matrix-assisted laser desorption/ionization-time of flight. Electrophoresis 2001, 22:1795-1800 [DOI] [PubMed] [Google Scholar]
- 55.Ghafouri B, Stahlbom B, Tagesson C, Lindahl M, Weston W, LeClair E, Trzyna W, McHugh K, Nugent P, Lafferty C, Ma L, Tuan R, Greene R: Newly identified proteins in human nasal lavage fluid from non-smokers and smokers using two-dimensional gel electrophoresis and peptide mass fingerprinting. Proteomics 2002, 2:112-120 [PubMed] [Google Scholar]