Abstract
The finding of possibly contaminant tissues or cells in surgical or cytology case material can be a challenging problem in diagnostic anatomical pathology samples. The reported rates of occurrence have ranged from 0 to 8.8% (including prospective and retrospective cases). A diagnostically dissimilar tissue fragment, whether contiguous with other tissue or among other fragments within a paraffin section, and which is not incompatible with the case tissue, often requires a rigorous investigation to confirm or deny its relevance to the case. Fluorescence in situ hybridization using dual red and green DNA probes to regions of the X and Y chromosomes, respectively, were used in one case where the potential contaminant was suspected to have originated from a male patient. The putative contaminant tissue fragment was confirmed as male, with cells having one X and one Y chromosome, unlike the other tissue fragments on the slide with two X chromosomes. In a second case, DNA polymorphisms were used to compare allelic patterns that were informative not only in proving the extraneous tissue as a contaminant, but in addition, could be used to trace the latter to its original tissue source. The molecular tools of fluorescence in situ hybridization in sex-mismatched cases and of DNA microsatellite probes that are applicable to paraffin sections can provide definitive identifiers of tissues and individual cells. They are important adjuncts to histology for the anatomical pathologist when faced with the diagnostic problems of tissue contamination encountered in routine practice.
The finding of potentially contaminating tissue in a surgical or cytology case can be a vexing diagnostic problem in anatomical pathology. The ambient rate of this occurrence demonstrates considerable variation, dependent on the method of detection. In a 1994 College of American Pathologists Q-Probes study of data from 275 laboratories, an overall extraneous tissue frequency of 0.6% (range, 0–1.8%) was detected based on prospective review at the time of case sign-out. However, the frequency rose to 2.9% (range, 0–8.8%) when slides were reviewed retrospectively with the specific intent to find contaminants. 1
On occasion, however, an apparently inconsistent tissue fragment that is not incompatible with the case tissue may be encountered within the section. Frequently, the source of contamination can be traced within the laboratory itself or, less commonly, to the physician’s office or operating room. But in 4 to 7% of cases the origin of the extraneous tissue is uncertain. 1 In the Q-Probes study, roughly 30% of the extraneous tissues encountered prospectively were abnormal or neoplastic, 10% presented some degree of diagnostic difficulty, and in 0.6% it could not be determined whether the tissue was truly alien. Such findings raise the possibility of disease with serious medical consequences, requiring the clinician to subject the patient to additional diagnostic studies, possibly necessitating an additional tissue biopsy, or to initiate close clinical surveillance. The pathologist must use every available means to pursue the origin of such tissue fragments in hopes of determining their contaminant status.
Although visual assessment may yield an educated guess, molecular approaches to selected cases can result in definitive exclusion of tissue identity. In this paper we present two cases where molecular approaches were used to decipher the origins of diagnostically problematic tissue contaminants encountered in routine surgical pathology practice.
Materials and Methods
Patient 1
Molecular Approach: Fluorescence in Situ Hybridization (FISH)
A routine hematoxylin and eosin section reviewed in Surgical Pathology included several fragments of gastric mucosa and submucosa with mild chronic inflammatory infiltrate and a smaller fragment showing poorly differentiated adenocarcinoma. The specimen (Figure 1A , Case A) was appropriately labeled as an endoscopic gastric biopsy from an elderly male with an endoscopic appearance of diffuse gastric mucosal erythema and pebbling, but no mass or ulcer. However, several years earlier, this individual had had a gastric adenocarcinoma in situ in a fundic polyp that was treated by local excision/polypectomy. In the same tray of slides for a pathologist’s review was a gastric biopsy from a female whose endoscopic examination was highly suggestive of malignant ulcer. This specimen (Figure 1B) was also appropriately labeled (Case B) and showed a poorly differentiated adenocarcinoma that was histologically similar to that seen in the smaller fragment of Case A.
Figure 1.
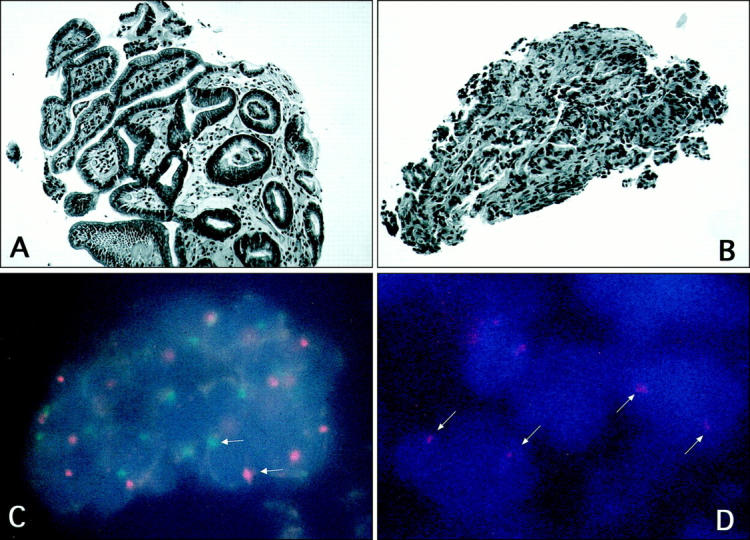
A: A routine H&E section of an endoscopic gastric biopsy from an elderly male with diffuse gastric mucosal erythema and pebbling, labeled Case A. B: A routine H&E section of a gastric biopsy from a female whose endoscopic examination was highly suggestive of malignant ulcer (contaminant), labeled Case B. C: FISH on tissue section derived from the gastric biopsy illustrated in A. Note the presence of one red signal (X chromosome) and one green signal (Y chromosome) in gastric mucosal cells. D: FISH on the tissue section derived from the gastric biopsy shown in B (contaminant). Note the presence of two red signals (representing X chromosomes) and absence of green signals.
The original slides and tissue blocks from both cases were available for study. Additional sections were cut for examination by fluorescence in situ hybridization (FISH). Hybridization was performed on 5-μm sections from both cases according to previously published procedures. 2, 3 Before hybridization, the region corresponding to the smaller questionable fragment in the Case A section was circled with a diamond pencil. Dual probes for the X and Y chromosomes were used (DXZ1 for the centromeric region of the X and DYZ3 for the distal heterochromatic region of the Y (Oncor, Inc., Gaithersburg, MD). The X probe was labeled with digoxigenin and a rhodamine (red) fluorophore and the Y probe with avidin and a fluorescein (green) fluorophore.
Patient 2
Molecular Approach: DNA Polymorphisms-Microsatellite Markers
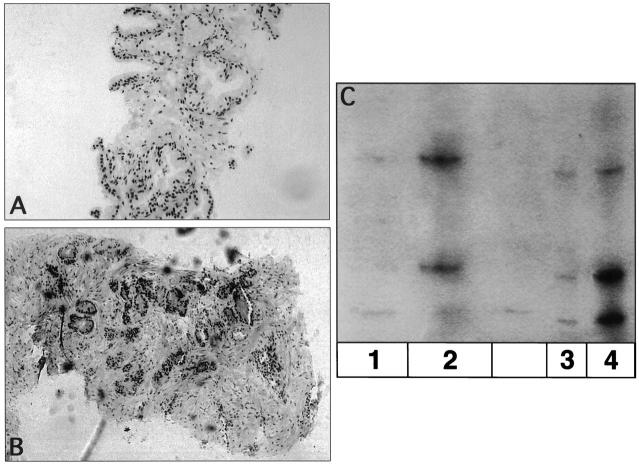
Three needle core biopsy samples of the prostate examined at several levels of a hematoxylin and eosin (H&E)-stained section were composed of benign prostatic tissue (Case C, Figure 2A ). Included on only one of the three levels was a much smaller fragment infiltrated with prostatic adenocarcinoma (Figure 2B) . No residual tissue from this small piece was evident in the tissue block Case C for retrieval or recuts. However, the malignant area was noted to be morphologically similar to that observed in another prostate needle biopsy case processed simultaneously (Case D). The Case D sections (Figure 2B) showed near total replacement by prostatic adenocarcinoma, with Gleason score 8 (patterns 3 + 5) showing glandular perineural invasion. Both surgical samples were needle biopsies of the prostate from elderly males, and neither clinical nor serological prostate-specific antigen findings were helpful.
Figure 2.
A: H&E section from Case C composed of benign prostatic epithelium. B: H&E section from Case D showing moderately to poorly differentiated prostatic adenocarcinoma, with Gleason pattern 3 + 5 (Gleason score = 8), and with perineural invasion. C: Autoradiograph of microsatellite analysis of DNA from Case C and Case D (putative source of contaminant). Lane 1: DNA-1 microdissected from the H&E-stained tissue section area of alleged contaminant labeled Case D present on the slide from Case C. Lane 2: DNA-1′ microdissected from tissue sections of a paraffin block of prostate adenocarcinoma from which Case D was suspected to originate. Lane 3: DNA-2 microdissected from the H&E-stained tissue section area of normal prostatic epithelium from Case C. Lane 4: DNA-2′ microdissected from tissue sections originating from a paraffin block of Case C.
The fragments from the original H&E-stained slide (Case C) were microdissected individually and placed in separate sterile tubes. The smaller fragment (prostatic adenocarcinoma, probable contaminant) was labeled DNA-1, and a larger fragment (benign prostatic epithelium) labeled DNA-2. DNA was extracted using a lysis buffer with Tween and 5 μl of proteinase K, with a 2-hour incubation at 55°C followed by boiling for 15 minutes. DNA (3–6 μml) was amplified by polymerase chain reaction (PCR) using a polymorphic VNTR marker D1S80, and incorporating radioactive 32dCTP. The reaction products were run on a 6% denaturing polyacrylamide gel and subsequently exposed to X-ray film.
Results
In Patient 1, cells of the gastric mucosa in larger fragments of Case A (Figure 1A) contained one red signal (one copy of the X chromosome) and one green signal (Figure 1C) . The section from Case B (Figure 1B) demonstrated 2 red signals by FISH in glandular epithelial nuclei. The fragment within the circled area showed only red signals (Figure 1D) , indicating two copies of the X chromosome per cell and absence of the Y chromosome. The chromosomal discrepancy confirmed the fragment as a contaminant. On the basis of both the morphological similarities and sex chromosome correspondence, the fragment was considered to be a contaminant of Case A, most likely originating from Case B.
In Patient 2, the bands of DNA-1 and DNA-2 migrated differently, indicating that the two samples were genetically distinct (Figure 2C) . Additional DNA samples were extracted from separate sections recut from different paraffin blocks of the original benign prostatic epithelium Case C, and labeled DNA-2′ and from the original potentially contaminating tissue (Case D), and labeled DNA-1′. The DNA alleles extracted from Case C (DNA-2′) showed a pattern identical to that of the larger, benign fragment (DNA-2), and those of Case D, DNA-1′ were identical to the bands seen in the DNA-1 sample from the stained original fragment that contained the focus of adenocarcinoma. Thus, it was concluded that the small fragment seen in one section from Case C represented a contaminant from Case D.
Discussion
Surgical specimen mix-ups, even in the most stringently controlled clinical laboratory setting, are often inevitable. Potential tissue mismatches can be resolved in several ways. The immunostaining method of blood group antigens A, B, and O determinants in tissue sections can identify patients’ tissues correctly. 4 Recently, the use of DNA-based PCR techniques performed on DNA isolated from paraffin-embedded tissues can correctly determine whether tissue samples have been interchanged, and can correctly assign specimens to a patient. Kit-based PCR assays, which amplify and distinguish different genotypes at the highly polymorphic human leukocyte antigen locus 4, 5 can be applied routinely to fixed-tissue specimens to confirm the identity of cases where there is potential tissue contamination. Short tandem repeat sequences or microsatellites that vary in their repeat number between individuals are well suited to PCR of minute tissue sections and are an effective method to confirm surgical specimen mix-ups. 6
In this study we have used relatively simple molecular approaches employing FISH and microsatellite marker analyses to confirm the contaminant status of suspect tissue fragments in surgical pathology slides. These approaches are useful both for gender-mismatched tissues, where simple evaluation of X and Y chromosomes is sufficient for confirmation of contamination, and for same-gender contaminants which require the additional steps of microdissection and DNA extraction, followed by microsatellite marker analysis. The latter, which reveals distinct DNA fingerprints, can lead to positive identification of the source of contamination (depending on availability of source tissue and DNA and on the extent of fingerprinting), whereas the former simply excludes portions of tissue or certain cells from the diagnostic evaluation of an individual case without definitive identification of the source of contamination.
The assessment of DNA fingerprint patterns is limited in certain respects. The suspected contaminant usually presents as a very small area, which may be lost in subsequent cuts from the block and, therefore, lost to additional analyses. The potential number of blocks from which the contaminant may have originated may be numerous, necessitating laborious, costly, and extensive microsatellite analyses and DNA extractions. In Patient 2, although the allelic pattern of marker DS180 was informative, permitting the recognition of the extraneous tissue as different, the finding of identity with a single PCR marker is not unequivocal. Positive identification may require the use of several markers to exclude a tissue fragment as a possible contaminant, adding to the labor and expense.
The issue of patient sample misidentification and tissue contamination is an important one. The Q-Probes quality improvement databases, derived from many institutions, provide a glimpse of the magnitude of this problem encountered in surgical pathology laboratories in the 1990s. The overall extent of specimen identification deficiencies approaches 50% in the poorest performing (10th percentile) laboratories. 7 Types of deficiencies that could lead to the incorrect assignment of patient tissues include processing specimens with (i) no label on container, (ii) no requisition slip, (iii) no patient identification on either container or requisition slip, (iv) patient name on container or requisition slip does not match that on master patient index, (v) wrong patient name on both container and requisition slip, and (vi) incorrect patient name keyed by remote order entry. Many of these identification deficiencies in the pre-analytic aspect of surgical pathology testing would be unknown to the pathologist examining the tissues, assuming the error had not been detected in the process of accessioning. The laboratory should have in place specific quality control criteria for specimen rejection from the accessioning process to prevent such errors from entering the system. Laboratory users guides should specify that either the clinician or his/her designee must rectify these types of deficiencies when detected before the specimen is processed. Such errors may also be caught after the fact by the clinicians who recognize an inconsistent or unexpected result or a diagnosis returned on a patient who had had no prior biopsy. Often, most of the latter error types originate in the clinical setting, where the tissue sample is placed in an unlabeled container and subsequently mislabeled by ancillary personnel.
In prospective slide evaluation, the situation that approximates actual case sign-out by the pathologist, the frequency of contaminant tissue is quite high, approaching 2.9%. 1 Although most contaminant tissues are loosely referred to as “floaters,” in truth, contaminants derived from the water bath during slide preparation pose less of a problem than tissue contaminants that are present within the paraffin block. 1 The origin of a floater is more readily detected as it usually is present only once, implying that it floated onto the slide, usually from a contaminated water bath from sections previously cut by the same microtome. The tissue contaminant present within the block is more difficult to assess, but when present on subsequent tissue sections, tissues are available for molecular probing. Furthermore, preservation of the morphological context allows the geographic localization of even minute contaminants during microscopic fluorescence examination and subsequent verification with conventional stains by comparing pre- and post-sectioned slide levels.
From the Q-Probes data, it appears that the two most common conditions after normal extraneous tissue are tissue that is neoplastic or tissue that is non-neoplastic but otherwise abnormal. Although in over 70% of cases, the source of contamination is a different case, even the 15% of same-case between- or within-specimen contaminants may pose serious problems. 1 A common example of this is the alleged knife blade-induced focus of vascular invasion, which is a subject of lively debate. In this study, we have shown how FISH in one case, and molecular genetic techniques in another, can be used to clarify potential cases of tissue contamination arising in routine surgical pathology practice.
Address reprint requests to Maria J. Worsham, Ph.D., Diplomate ABMG, Department of Pathology, Henry Ford Hospital, 2799 W. Grand Blvd., Detroit, MI 48202. E-mail: mworsha1@hfhs.org.
Footnotes
Supported by National Institutes of Health grant RO1 CA 70923 and American Cancer Society grant RPG-96–093.
References
- 1.Gephardt GN, Zarbo RJ: Extraneous tissue in surgical pathology: a College of American Pathologists Q-Probes study of 275 laboratories. Arch Pathol Lab Med 1996, 120:1009-1014 [PubMed] [Google Scholar]
- 2.Worsham MJ, Wolman SR, Carey TE, Zarbo RJ, Benninger MS, Van Dyke DL: Common clonal origin of synchronous primary head and neck squamous cell carcinomas: analysis by tumor karyotypes and fluorescence in situ hybridization. Hum Pathol 1995, 26:251-261 [DOI] [PubMed] [Google Scholar]
- 3.Worsham MJ, Wolman SR, Carey TE, Zarbo RJ, Benninger MS, Van Dyke DL: Chromosomal aberrations identified in culture of squamous carcinomas are confirmed by fluorescence in situ hybridization. Mol Pathol 1999, 52:42-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ota M, Fukushima H, Akamatsu T, Nakayama J, Katusyam T, Hasekura H: Availability of immunostaining methods for identification of mixed-up tissue specimens. Am J Clin Pathol 1989, 92:665-669 [DOI] [PubMed] [Google Scholar]
- 5.Shibata D: Identification of mismatched fixed specimens with a commercially available kit based on the polymerase chain reaction. Am J Clin Pathol 1993, 100:666-670 [DOI] [PubMed] [Google Scholar]
- 6.Tsongalis GJ, Berman MM: Application of forensic identity testing in a clinical setting: specimen identification. Pathology 1997, 18:385-389 [DOI] [PubMed] [Google Scholar]
- 7.Nahkleh RE, Baker PB, Zarbo RJ: Autopsy result utilization: a College of American Pathologists Q-probes study of 256 laboratories. Arch Pathol Lab Med 1999, 123:290-295 [DOI] [PubMed] [Google Scholar]