Abstract
We evaluated real-time (kinetic) reverse transcription-polymerase chain reaction (RT-PCR) to validate differentially expressed genes identified by DNA arrays. Gene expression of two keratinocyte subclones differing in the physical state of human papillomavirus (episomal or integrated) was used as a model system. High-density filter arrays identified 444 of 588 genes as either negative or expressed with less than twofold difference, and the other 144 genes as expressed uniquely or with more than twofold difference between the two subclones. Real-time RT-PCR used LightCycler-based SYBR Green I dye detection and melting curve analysis to validate the relative change in gene expression. Real-time RT-PCR confirmed the change in expression of 17 of 24 (71%) genes identified by high-density filter arrays. Genes with strong hybridization signals and at least twofold difference were likely to be validated by real-time RT-PCR. This data suggests that (i) both hybridization intensity and the level of differential expression determine the likelihood of validating high-density filter array results and (ii) genes identified by DNA arrays with a two- to fourfold difference in expression cannot be eliminated as false nor be accepted as true without validation. Real-time RT-PCR based on LightCycler technology is well-suited to validate DNA array results because it is quantitative, rapid, and requires 1000-fold less RNA than conventional assays.
High-throughput analysis of gene expression is now feasible with the use of cDNA microarrays and high-density filter arrays (HDFA). However, array results can be influenced by each step of the complex assay, from array manufacturing to sample preparation (extraction, labeling, hybridization) and image analysis. 1, 2, 3 The efficiency of the reverse transcription (RT) reaction is known to be affected by the enzyme, primers, nucleotides, and RNA secondary structure. These factors in turn influence the representation of low-abundance transcripts in the final cDNA probe. 4, 5 Complex cDNA probes can cross-hybridize to related sequences, and low-intensity hybridization signals are difficult to interpret. The field has not reached consensus on the significance of differences in hybridization intensity. Whereas some investigators interpret a twofold difference in hybridization intensity as evidence of differential gene expression, others require fourfold differences. 1, 6, 7
Currently, array technology is most useful in establishing broad patterns of gene expression and in screening for differential gene expression. Validation of expression differences is accomplished with an alternate method such as Northern blot hybridization or RNase protection assay. However, these assays are time-consuming, labor-intensive, and require large amounts of RNA (>5 μg total RNA). Conventional reverse transcription-polymerase chain reaction (RT-PCR) can be done with smaller amounts of RNA (20–40 ng), but quantification is difficult and relies on endpoint analysis of the PCR product. 8, 9, 10 Real-time (kinetic) PCR evaluates product accumulation during the log-linear phase of the reaction and is currently the most accurate and reproducible approach to gene quantification. 9, 10 In this study, we explored the applicability of kinetic RT-PCR as a rapid procedure for the validation of a number of differentially expressed genes identified by HDFA. Because of our interest in the interaction of human papillomavirus (HPV) on cellular gene expression, we used the HDFA expression profiles of two subclones differing in the integration status of HPV (integrated or mixed episomal/integrated) as a model system to test our validation approach. We found that a two-step RT-PCR using SYBR Green I dye detection with product verification by melting curve analysis is rapid, quantitative, and applicable to samples with limited amount of RNA. The method was robust enough to validate relative changes in the expression of a number of genes with varying abundance of transcripts.
Materials and Methods
Cell Culture and RNA Extraction
Two subclones of W12 cervical epithelial cells with HPV16 in differing physical states were a gift from Dr. Paul Lambert (University of Wisconsin, Madison, WI). HPV16 was present in a mixed episomal/integrated state in subclone 20863 and in a multicopy integrated form in subclone 20861. Both subclones were grown as monolayers on γ-irradiated (5000 rads) Swiss Mouse 3T3 fibroblast feeder layers in F-medium (3:1 F12 and Dulbecco’s modified Eagle’s medium) with 5% fetal bovine serum (FBS). 11 CaSki, a human cervical cancer cell line, was obtained from American Type Culture Collection (Manassas, VA). CaSki monolayers were grown in RPMI-1640 medium with 10% FBS and 2.5 mmol/L L-glutamine. Cells were incubated at 37°C in 5% CO2 and harvested at 60 to 70% confluence. Cultures were washed with phosphate-buffered saline, followed by 0.02% EDTA to remove the feeder cells.
All monolayers were lysed with guanidinium thiocyanate for RNA extraction. 12 The total RNA from each sample was divided in half: one half for HDFA after poly(A)+ RNA isolation by using the Oligotex mRNA kit (Qiagen, Santa Clarita, CA) and the other half for HDFA validation by LightCycler (Roche Molecular Biochemicals, Indianapolis, IN). RNA quality and quantity were evaluated by UV spectrophotometry and denaturing formaldehyde agarose gel electrophoresis. 13
Gene Expression Profiling by HDFA
Probe synthesis and hybridization conditions optimized for chemiluminescent detection with HDFA were used as previously described. 14 In brief, cDNA probes were synthesized in a 20 μl RT reaction with 1 μg of poly(A)+ RNA, oligo(dT)12–18, random hexamers, digoxigenin-dUTP (Roche Molecular Biochemicals), and SuperScript II reverse transcriptase enzyme (Life Technologies, Gaithersburg, MD). One half of the labeled cDNA was used to hybridize the Atlas Human Cancer cDNA Expression Array (Clontech, Palo Alto, CA). After an overnight hybridization at 42°C, membranes were washed and hybridization signals were detected with anti-digoxigenin/alkaline phosphatase conjugate and CDP-Star substrate. Membranes were exposed to LumiFilm (Roche Molecular Biochemicals) for 12 minutes after incubating with the substrate for 1 hour.
The films were scanned and the images were analyzed using BioNumerics (Applied Maths, Kortrijk, Belgium). 15 Briefly, images were acquired and converted to Tagged Image File (.tif) format using a flatbed scanner. These array image files were then analyzed in BioNumerics software that subtracted background and normalized intensity on the basis of the lowest negative control as 0 and the highest positive control as 100. The data were copied into Microsoft Excel for further analysis. The lower limit of reliable detection was defined by calculating a threshold value equal to the average intensity of 3 negative controls plus 5 times the SD. Intensities above this threshold were considered positive signals.
Validation of Relative Gene Expression by Kinetic RT-PCR
cDNA Synthesis
Fifty micrograms of total RNA from each sample were treated with DNase I (0.4 units/μg RNA) according to instructions of the MessageClean kit (GenHunter Corp., Nashville, TN). One microgram of DNase-I-treated total RNA was used for cDNA synthesis (20 μl), using conditions described previously except that random hexamers and digoxigenin-dUTP were omitted and all dNTPs were maintained at 0.5 mmol/L. 14
Primers
Gene-specific primers corresponding to the PCR targets on the Atlas Human Cancer cDNA Expression Array were obtained from Clontech. Preliminary experiments were done with each primer pair and CaSki cDNA to determine the annealing temperature that yielded the greatest amount of specific product with melting temperature (Tm) separable from primer-dimer Tm. The acquisition temperature was set 1 to 2°C below the Tm of the specific PCR product. 16 The experimentally determined annealing and fluorescent signal acquisition temperatures for each gene tested in this series of experiments are given in Table 1 .
Table 1.
Validation of Array-Based Gene Expression Profile by LightCycler
| Gene name* (GenBank accession no.) | Annealing; acquisition temp (°C) | Relative expression† | Validation (Yes/No) | |
|---|---|---|---|---|
| HDFA | LightCycler | |||
| Intensity >30‡ | ||||
| Fibronectin (X02761) | 58; 85 | 8.5 | 20.0 | Y |
| Stromelysin-2 (X07820) | 55; 81 | 5.2 | 8.4 | Y |
| Bullous pemphigoid antigen (M63618) | 58; 83 | 4.7 | 4.0 | Y |
| BIGH3 (M77349) | 58; 83 | 4.4 | 6.8 | Y |
| Plasminogen activator inhibitor (X04229) | 60; 86 | 3.9 | 5.1 | Y |
| Collagenase-1 (X05231) | 58; 83 | 3.0 | 4.7 | Y |
| Interleukin (IL)-1 β (K02770) | 55; 86 | 2.8 | 4.6 | Y |
| Integrin α 6 (X59512) | 62; 80 | 2.4 | 3.2 | Y |
| IFN-γ antogonist cytokine (A25270) | 62; 85 | 2.4 | 0.9 | N |
| Zyxin+ Zyxin-2 (X94991) | 58; 90 | 2.3 | 2.0 | Y |
| β-Catenin (X87838) | 60; 87 | 2.1 | 1.0 | N |
| Vimentin (X56134) | 58; 83 | 2.0 | 23.2 | Y |
| Leukocyte interferon inducible peptide§ (X02492) | 58; 81 | 2.2 | 7.3 | Y |
| Cytokeratin 19§ (X00503) | 58; 83 | 2.5 | 4.1 | Y |
| Cyclin (PCNA, J04718) | 60; 88 | 1.1 | 1.0 | Y |
| CDK-interacting protein 1 (U09579) | 58; 86 | 1.0 | 1.0 | Y |
| G3PDH (X01677) | 58; 86 | 1.0 | 1.0 | Y |
| Intensity <30 | ||||
| Desmoplakin I (M77830) | 62; 87 | 4.2 | 2.7 | Y |
| Thrombospondin 1 precursor (X14787) | 60; 88 | 3.5 | 0.8 | N |
| Mitogen-inducible gene 5 (Z30183) | 60; 88 | 2.7 | 1.4 | N |
| Tenacin-C (X78565) | 62; 86 | Unique | Unique | Y |
| Disheveled homolog (U46461) | 65; 86 | Unique | 1.0 | N |
| Bone morphogenetic protein (M22488) | 67; 89 | Unique | 0.8 | N |
| Cell division protein kinase C (X66357) | 62; 88 | Unique | 0.9 | N |
Primer sequences for these genes on HDFA are available from Clontech (Palo Alto, CA).
Relative expression is calculated as ratio (R) of expression levels in subclone 20863/subclone 20861 or the reciprocal of this ratio (1/R) to indicate genes up- or down-regulated, respectively, in subclone 20863. A gene is considered differentially expressed in this report if its relative expression is at least twofold or unique.
Intensity groups are based on the normalized HDFA intensity values of subclone 20863.
Genes down-regulated in subclone 20863. All other genes up-regulated/expressed identically or uniquely in subclone 20863 by HDFA.
Relative Standard Curves
Constructing a standard curve with serial dilutions of known template concentration for each target on the microarray is not feasible. Therefore, dilutions (1:200, 1:2,000, and 1:20,000) of cDNA from the sample with the higher HDFA hybridization intensity for a given target were used to construct a relative standard curve for that target. Template concentrations for reactions in the relative standard were given arbitrary values of 0.5, 0.05, and 0.005. The cDNA (1:200 dilution) from the low-intensity sample was analyzed as unknown.
PCR Assay Conditions
DNA Master SYBR Green I mix (containing Taq DNA polymerase, dNTP, MgCl2, and SYBR Green I dye; Roche Molecular Biochemicals) was incubated with TaqStart Antibody for 5 minutes at room temperature before the addition of primers and cDNA template. Each reaction (20 μl) contained 2 μl of the respective cDNA dilution, primers at 0.4 μmol/L, and MgCl2 at 4 μmol/L. The amplification program consisted of 1 cycle of 95°C with 60-second hold (“hot start”) followed by 50 cycles of 95°C with 0-second hold, specified annealing temperature with 5-second hold, 72°C with 18-second hold, and specified acquisition temperature with 2-second hold (Table 1) . Amplification was followed by melting curve analysis using the program run for one cycle at 95°C with 0-second hold, 65°C with 10-second hold, and 95°C with 0-second hold at the step acquisition mode. A negative control without cDNA template was run with every assay to assess the overall specificity.
LightCycler Data Analysis
Unless otherwise mentioned, each assay included duplicate reactions for each dilution and was repeated once. A relative value for the initial target concentration in each reaction was determined on the basis of the kinetic approach 9, 10 using the LightCycler software, version 3. The mean concentration of glyceraldehyde-3-phosphate dehydrogenase (G3PDH) was used to control for input RNA because it is considered a stable housekeeping gene and was detected at the same level in both subclones by HDFA and LightCycler. The mean G3PDH concentration was determined once for each cDNA sample and used to normalize all other genes tested from the same cDNA sample. The relative change in gene expression was recorded as the ratio of normalized target concentrations in the 1:200 cDNA dilution.
Results
Gene Expression Profile by HDFA
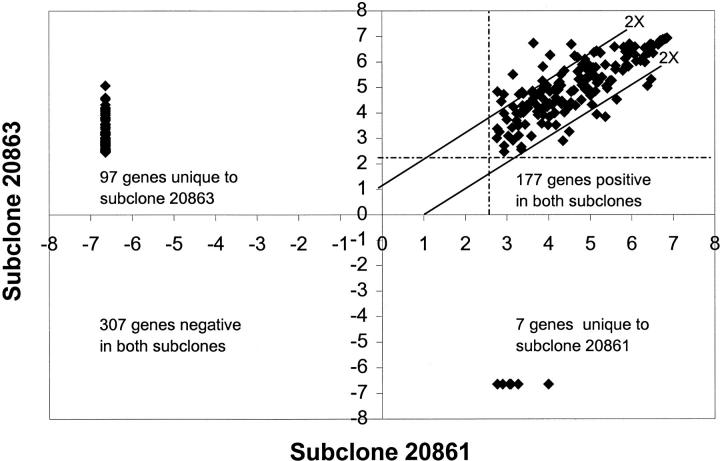
The normalized intensity of the hybridization signals for the 588 genes ranged from 0.03 to 123 for both subclones. The threshold value was slightly higher for subclone 20861 than for subclone 20863 (6.6 vs. 5.4). Based on these thresholds, subclone 20863 expressed 274 genes, whereas subclone 20861 expressed 184 genes. Nearly 60% of the positive genes for both subclones were low intensity (<30). Figure 1is a plot of the log base 2 of the hybridization intensity of each gene in both subclones. The results for subclone 20861 are on the x-axis and those for subclone 20863 are on the y-axis. The two parallel lines indicate the position of twofold difference in intensity. Expression of 444 genes was concordant in both subclones (307 genes with no detectable expression and 137 genes with expression levels differing by less than twofold). Among the 144 discordant genes, 97 genes were detected only in subclone 20863, 7 genes were detected only in subclone 20861, and 40 genes were coexpressed with differential (at least twofold) expression (33 genes up-regulated and 7 genes down-regulated in subclone 20863 compared to 20861). Genes for validation were selected on the basis of their absolute hybridization intensity (subclone 20863 as reference) as well as on their relative intensity (subclone 20863 to subclone 20861). A total of 24 genes with varying expression levels by HDFA, 7 with hybridization intensity <30 and the remainder with intensity >30 (Table 1) , were analyzed by LightCycler.
Figure 1.
Scatterplot of the log base 2 of the hybridization intensities for each gene in subclones 20863 and 20861 as determined by HDFA. Perpendicular lines (dashed) indicate the threshold values for each subclone. The two solid parallel lines indicate the position of twofold differences in intensity. Genes on and outside the parallel lines were identified as differentially expressed by HDFA.
Validation of HDFA results by LightCycler Kinetic RT-PCR
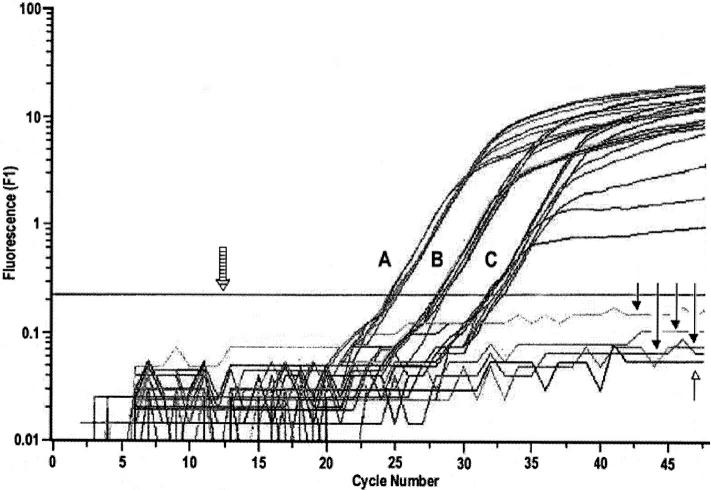
The fidelity of the kinetic RT-PCR assay is demonstrated by the fluorescence versus cycle number plot in Figure 2 , which shows detection of the G3PDH transcript at the same level in both subclones. This type of analysis with a housekeeping gene can be used to control for the amount of cDNA in the RT reaction. The no-RT control is negative, indicating the absence of genomic DNA in the RT products of both subclones. The coefficient of variation (CV) was determined for each gene as an indicator of precision associated with the LightCycler RT-PCR assay. The mean CV for genes with hybridization intensity of >30 was 12% (range, 1–25%), and for those with low intensity (<30) it was 18% (range, 11–28%).
Figure 2.
Fluorescence versus cycle number plot of G3PDH transcript in subclones 20863 and 20861 by LightCycler RT-PCR assay based on SYBR Green I dye detection. Both subclones were evaluated for relative levels of G3PDH transcript using 1:200, 1:2000, and 1:20,000 dilutions of cDNA which are indicated as A, B, and C, respectively, in the amplification plot. There were four reactions per dilution per sample. At each dilution, all reactions overlapped in the log-linear phase, indicating identical levels of G3PDH transcript in both samples. Closed arrowheads indicate no-RT reactions with both subclones. Open arrowhead shows PCR with no template. Striped arrow indicates the position of noise band. All data points below the noise band were excluded from quantification. Annealing and signal acquisition temperatures for G3PDH are given in Table 1 .
Data showing the relative change in expression for several genes as determined by HDFA and LightCycler are presented in Table 1 . The LightCycler results confirmed the majority of HDFA results (88%) when the hybridization intensity was >30 (15 of 17 genes) or when the relative difference in expression between the subclones was greater than fourfold (5 of 5 genes). Uniquely expressed genes detected with HDFA were of low intensity (<30) and only 1 of 4 genes was confirmed. Overall, LightCycler confirmed HDFA-determined relative expression differences for 17 of 24 genes (71%) tested.
Differences were also seen in the magnitude of differential expression detected by the two methods. For example, among the 14 differentially expressed genes confirmed by LightCycler, 10 genes showed expression differences greater than that determined by HDFA. Notable among these are the expression differences between subclones for fibronectin (eightfold by HDFA and 20-fold by LightCycler), vimentin (twofold by HDFA and 23-fold by LightCycler), and leukocyte interferon-inducible peptide (twofold by HDFA and sevenfold by LightCycler).
Discussion
LightCycler-based real-time RT-PCR using SYBR Green I dye detection and product verification by melting curve analysis was examined in this study as a rapid, sensitive, and specific procedure to validate the differential expression of a large number of genes identified by HDFA. This approach determined relative differences in gene expression instead of absolute copy numbers, eliminating the need for many precisely quantified templates as standards. Strategies based on fluorogenic 5′ nuclease chemistry or hybridization probes 9, 10 were not considered because design, synthesis, and optimization of conditions for specific internal fluorescent probes for the large number of genes screened by microarrays are not practical. This kinetic RT-PCR approach is readily applicable only for validation of arrays containing cDNA fragments prepared with gene-specific primers in PCR, although gene-specific primers could be designed or determined for other arrays.
Hot-start PCR mediated by TaqStart antibody and fluorescent signal acquisition at temperatures just below the Tm of specific product allowed SYBR Green I dye-based kinetic-PCR to be sensitive and specific. In this study, the dye-based detection format resulted in an average CV of 12 to 18% for genes with high and low hybridization intensities. This average CV is well below the reported average of 25 to 35% for gene quantification with LightCycler. Most genes could be detected in dilutions of cDNA as low as 1:200 to 1:20,000 from a standard 20-μl RT reaction with 1 μg total RNA. For genes with low expression levels, the amount of cDNA may be increased 10-fold more than suggested in this protocol. Because the assay requires small volumes of cDNA, nearly 100 to 1000 genes can be validated with 1 μg of total RNA. By contrast, validation with Northern blot or RNase protection assay requires at least 5 μg of total RNA per assay, approximately 5000 times more RNA than for the LightCycler assay reported here.
A random selection of genes with varying expression levels detected by HDFA was evaluated by kinetic RT-PCR. Overall, 17 of 24 genes (71%) were confirmed by the LightCycler assay. Both the hybridization intensity and the relative level of gene expression influenced the likelihood that HDFA differences were validated. Genes with weak hybridization signals (<30) and less than fourfold difference in expression were least likely to be validated by the LightCycler. On the other hand, several genes (8 of 10 genes) with high hybridization intensity (>30) and only two- to fourfold difference in expression were validated by the LightCycler. The largest group of differentially expressed genes in many studies using DNA arrays are those with two- to fourfold differences in expression. 1, 6, 7 Our data using kinetic RT-PCR suggests that these genes cannot be eliminated as false nor be accepted as true without validation by a secondary procedure.
For genes confirmed as differentially expressed by LightCycler RT-PCR, the level of gene expression differences could be quite different by the two methods. A notable example of such a discrepancy was vimentin gene expression (23-fold differences by LightCycler as opposed to twofold difference by HDFA). The vimentin PCR product includes nucleotides 1164–1604, a region spanning coding through the 3′ UTR. This sequence has 80 to 90% homology over a stretch of 100 nucleotides with other members of the intermediate filament gene family, such as desmin, plasticin, and internexin. This observation suggests that true expression differences for specific members of gene families may be masked by cross-hybridization in microarrays.
Our findings support the use of microarrays as screening tools and emphasize the need for validation of microarray results. The strength of LightCycler assay as a secondary validation procedure lies in its potential to quantify relative change in expression of large number of genes with limited RNA rapidly and precisely.
Acknowledgments
We wish to acknowledge the expert technical assistance of Daisy Lee and Irina Dimulescu. Use of trade names and commercial sources does not imply endorsement by the Centers for Disease Control or the U.S. Department of Health and Human Services.
Address reprint requests to Mangalathu S. Rajeevan, Division of Viral and Rickettsial Diseases, National Center for Infectious Diseases, Centers for Disease Control and Prevention, U.S. Department of Health and Human Services, Atlanta, GA 30333. E-mail: mor4@cdc.gov.
Footnotes
N. T. was supported by the U.S. Department of Energy and CDC Interagency Agreement administered by the Research Participation Program of the Oak Ridge Institute for Science and Education.
References
- 1.Der SD, Zhou A, Williams BRG, Silverman RH: Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc Natl Acad Sci USA 1998, 95:15623-15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eisen M, Brown PO: DNA arrays for analysis of gene expression. Methods Enzymol 1999, 303:179-205 [DOI] [PubMed] [Google Scholar]
- 3.Winzeler EA, Schena M, Davis RW: Fluorescence based expression monitoring using microarrays. Methods Enzymol 1999, 306:3-19 [DOI] [PubMed] [Google Scholar]
- 4.Brooks EM, Sheflin LG, Spaulding SW: Secondary structure in the 3′UTR of EGF and the choice of reverse transcriptase affect the detection of message diversity by RT-PCR. BioTechniques 1995, 19:806-815 [PubMed] [Google Scholar]
- 5.Gerard GF, Fox DK, Nathan M, D’Alessio JM: Reverse transcriptase, the use of cloned Moloney murine leukemia virus reverse transcriptase enzyme to synthesize DNA from RNA. Mol Biotechnol 1997, 8:61-77 [DOI] [PubMed] [Google Scholar]
- 6.Chang YE, Laimins LA: Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J Virol 2000, 74:4174-4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu H, Cong JP, Mamtora G, Gingeras T, Shenk T: Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci USA 1998, 95:14470-14475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siebert PD: RT-PCR methods and applications. Methods in Molecular Medicine, Vol 13: Molecular Diagnosis of Infectious Diseases. Edited by Reischi U, Totowa, NJ, Humana Press, Inc., 1997, pp 29–53 [DOI] [PubMed]
- 9.Higuchi R, Fockler C, Dollinger G, Watson R: Kinetic PCR analysis-real time monitoring of DNA amplification. Bio/Technology 1993, 11:1026-1030 [DOI] [PubMed] [Google Scholar]
- 10.Wittwer CT, Herrmann MG, Moss AA, Rasmussen RP: Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques 1997, 22:130-138 [DOI] [PubMed] [Google Scholar]
- 11.Jeon S, Allen-Hoffmann BL, Lambert PF: Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J Virol 1995, 69:2989-2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chomczynski P, Sacchi N: Single step-method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987, 162:156-159 [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning, A Laboratory Manual, 2nd ed. 1989, Cold Spring Harbor Laboratory Press Cold Spring Harbor, NY
- 14.Rajeevan MS, Dimulescu IM, Unger ER, Vernon SD: Chemiluminescent analysis of gene expression on high density filter arrays. J Histochem Cytochem 1999, 47:337-342 [DOI] [PubMed] [Google Scholar]
- 15.Vernon SD, Unger ER, Rajeevan MS, Dimulescu IM, Nisenbaum R, Campbell C: Reproducibility of alternate probe synthesis approaches for gene expression profiling with arrays. J Mol Diag 2000, 2:124-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrison TB, Weis JJ, Wittwer CT: Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. BioTechniques 1998, 24:954-962 [PubMed] [Google Scholar]