Abstract
Laser capture microdissection (LCM) allows the selective sampling of tissue from histological sections. A prerequisite for this technique is the availability of histological dyes that do not interfere with downstream analysis of the sampled genetic material. We have examined the effect of four histological nuclear dyes (methyl green, hematoxylin, toluidine blue O, azure B) on TaqMan polymerase chain reaction amplification of β-actin genomic DNA prepared from fixed and frozen tissue. Tissue sampled from the histological sections by manual dissection was compared with tissue sampled by LCM. As previously reported, when manually dissected tissue sections were analyzed, polymerase chain reaction amplification of DNA after hematoxylin staining was inferior to that after staining with the other dyes. In contrast, when tissue sampled by LCM was examined, DNA recovery after hematoxylin staining was equivalent to the recovery after methyl green staining. We conclude that DNA recovery from LCM-sampled tissue is independent of the histological stain chosen to highlight nuclear detail.
Laser capture microdissection (LCM) allows the selective sampling of individual cells or groups of cells from histological sections by fusing the targeted cells to a thermoplastic film with a laser pulse, then lifting the thermoplastic film together with the sampled cells from the slide. 1 A prerequisite for this technique is an appropriate histological stain that allows visualization of the target tissue but does not interfere with the downstream molecular analyses of genetic material from the cells. Previous reports have compared the effect of different histological nuclear dyes on recovery of DNA from formalin-fixed, paraffin-embedded tissue samples that were obtained by manual dissection, either by quantitating recovered DNA or examining DNA amplification by polymerase chain reaction (PCR). 2, 3, 4 These studies concluded that hematoxylin staining resulted in unsatisfactory DNA recovery from the tissue samples, suggesting that hematoxylin would not be a suitable dye for microdissection. The results presented here confirm the unsatisfactory performance of hematoxylin on relatively large tissue fragments sampled manually from histological sections. However, when DNA is amplified from the relatively smaller amount of material obtained from LCM, hematoxylin performs as well as other nuclear dyes.
Materials and Methods
Tissue Specimens
Sixteen tissue samples were used for the initial study. Eight cases (two each of breast, lung, colon, and ovarian carcinoma) were archival formalin-fixed, paraffin-embedded surgical tissue obtained from our institution’s Division of Surgical Pathology. Eight additional cases (two each of breast, lung, and colon carcinoma, one case of ovarian carcinoma, and one ovarian cyst) were fresh-frozen tissue specimens obtained from the Alvin J. Siteman Cancer Center Tissue Procurement Core Facility. All human specimens were utilized in accordance with an Institutional Review Board-approved protocol. Representative sections from each case were examined to ensure qualitatively similar degrees of cellularity and absence of necrosis between each specimen.
Processing of Tissue
Five-micron histological sections of formalin-fixed, paraffin-embedded tissue were placed on uncharged, noncoated glass slides. After drying, slides were deparaffinized (4 changes of xylene, 5 minutes each; 3 changes of 100% ethanol, 1 minute each), rinsed in distilled water, and stained as described below. Five-micron histological sections of frozen tissue were also placed on uncharged, noncoated glass slides, immediately fixed in 70% ethanol for 30 seconds, and stained as described below. From each specimen block (frozen and fixed), serial sections were used for each different stain and for the LCM versus manual tissue dissection, so that within-specimen variability due to tissue heterogeneity was minimized. After staining, all sections were rinsed in distilled water, dehydrated in three changes of absolute ethanol (1 minute each) followed by four changes of xylene (5 minutes each). The slides were then air-dried and used for manual dissection or microdissection.
Staining of Tissue
Filtered, aqueous solutions of toluidine blue, azure B, and methyl green (Sigma, St. Louis, MO) were used for staining. Initial experiments varying dye concentrations and staining times established the concentrations and times that gave morphologically satisfactory results. Paraffin-embedded tissue sections were stained in 0.01% toluidine blue for 2 minutes, 0.5% azure B for 30 seconds, 0.5% methyl green for 30 minutes, or Mayer’s hematoxylin solution (Sigma) for 2 minutes. Frozen tissue sections were stained in 0.1% toluidine blue for 30 seconds, 0.5% azure B for 30 seconds, 0.5% methyl green for 30 minutes, or Mayer’s hematoxylin solution for 30 seconds. Staining by Mayer’s hematoxylin was followed by immersion in bluing solution (Richard-Allan Scientific, Kalamazoo, MI) for 30 seconds. For unstained controls, sections were treated identically (deparaffinization, hydration, and dehydration) to the stained slides, except that they were immersed for 30 seconds in deionized water in place of dye solution.
LCM and Manual Dissection of Tissue
LCM was performed with a Pixcell II Laser Capture Microscope (Arcturus Engineering, Mountain View, CA) in the Alvin J. Siteman Cancer Center Tissue Procurement Core Facility. Tissue (5 μm thickness) obtained from an average of 500 firings of a 30-μm laser spot size was captured on the thermoplastic caps and digested in 20 μl proteinase K buffer. Hence, the digestion proceeded at a density of approximately 8.5 × 104 μm3 (= 8.5 × 10−5 μl) of tissue volume per 1 μl of digestion buffer. Based on visual examination of the transfer films and the tissue sections after cell transfer, one laser pulse corresponded to approximately five to six cells. This number varied somewhat, depending on the tissue dissected. In general, however, a total of 2500 to 3000 procured cells were represented in the 20 μl of digestion buffer. For each case, the same area of each differently stained serial section was subjected to LCM, so that the cell population for a single specimen across different staining procedures was equivalent. Tissue obtained by manual dissection was sampled by scraping the entire tissue section from the slide with a razor blade, followed by placing the tissue into proteinase K buffer, at an approximate ratio of 1 square cm tissue area per 100 μl of buffer. This corresponded to a density of approximately 5 × 106 μm3 (= 5 × 10−3 μl) tissue volume per 1 μl of digestion buffer. Thus, the microdissected tissue was digested at a much lower ratio of tissue to digestion buffer than the manually dissected tissue. Nevertheless, for the latter there was still an approximately 200-fold excess of digestion buffer volume over tissue volume.
DNA Preparation
The tissue sampled by LCM or by manual dissection was digested overnight at 60°C in proteinase K buffer. The composition of proteinase K buffer was as follows: 10 mmol/L Tris/HCl, pH 8.0, 1 mmol/L EDTA, 1% Tween 20, and 0.04% (w/v) proteinase K (Boehringer, Indianapolis, IN). Undigested debris was subsequently removed by centrifugation at 10,000 × g for 5 minutes, and proteinase K was inactivated by incubation at 95°C for 10 minutes. For dilution experiments, tissue lysates from manually dissected specimens corresponding to 5 × 106 μm3 tissue volume per 1 μl of digestion buffer (see above) were further diluted in digestion buffer at 1:5, 1:25, and 1:125 after heat inactivation. Lysates were used for DNA analysis by TaqMan PCR or by conventional PCR without further purification and stored at −70°C for up to 2 weeks without loss of amplifiable DNA.
DNA Analysis by TaqMan
Quantitative analysis of genomic β-actin DNA was performed by monitoring in real-time the increase in fluorescence of the dye SYBR green I (Molecular Probes, Inc., Eugene, OR) as described 5 using the TaqMan instrument (ABI Prizm 7700 Sequence Detection System, Perkin Elmer, Norwalk, CT). The sequences of the primers used for amplification of β-actin DNA were 5′-CGAGAAGATGACCCAGATCATGTT-3′ and 5′-CCTCGTAGATGGGCACAGTGT-3′. ROX was used as the reference dye. All reactions were repeated in duplicate. The instrument was calibrated using human genomic DNA standards of known concentration as determined spectrophotometrically at 260 nm wavelength. Selected results obtained by TaqMan amplification were confirmed semiquantitatively by conventional PCR using the same primers, and the amplification product of approximately 150 bp was detected on a 2% agarose gel. All amplification curves generated with TaqMan from stained tissue had the typical sigmoidal appearance of amplification curves generated from unstained tissue, which indicates that traces of these dyes do not interfere with the fluorescence detection system. As noted by others, 4 eosin, a cytoplasmic dye which we had initially included in the study, resulted in highly atypical TaqMan amplification curves due to its innate fluorescence, and we have therefore excluded this dye from this study.
Statistical Analysis
Data sets were analyzed for statistical significance by Student’s t-test in the two-tailed form. Significance was defined as P < 0.05.
Results
All four dyes used resulted in preferential staining of nuclei. Of the four dyes tested, hematoxylin staining resulted in the best discrimination between nuclei and cytoplasm. Toluidine blue O, azure B, and methyl green gave variable cytoplasmic background staining, the intensity of which depended somewhat on the tissue type. In general, cytoplasmic background staining by the three dyes was less pronounced in frozen tissue than in paraffin-embedded tissue. In addition, methyl green strongly stained the cytoplasm of smooth muscle cells, making visualization of nuclei more difficult in this tissue type. None of the stains appeared to adversely affect the cell transfer efficiency from the slide to the LCM transfer film, which was consistently 70 to 90% efficient as judged by visualizing tissue collected on the transfer caps and tissue remaining on the slide in the area of dissection.
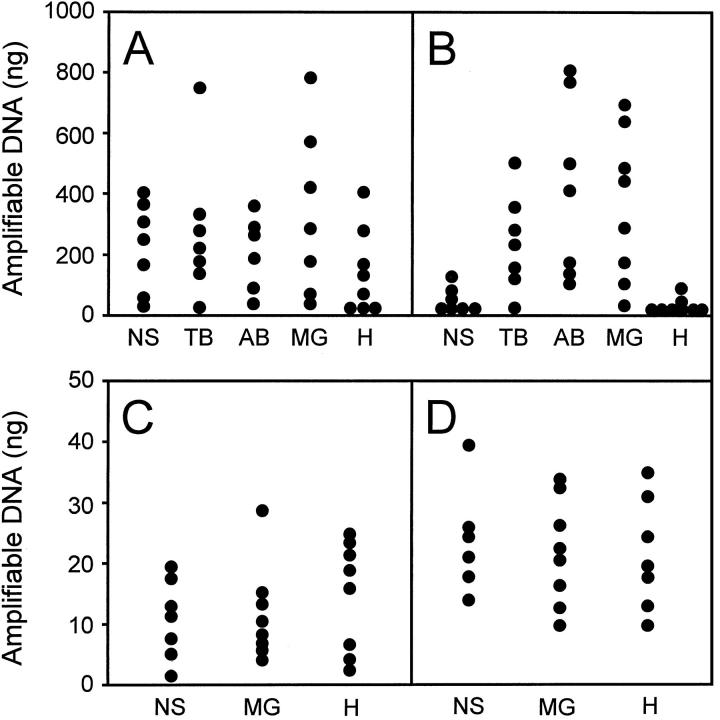
TaqMan PCR was used next to compare the effect of the histological dyes on retrieval of amplifiable DNA. Overall, there was considerable variability in the amount of amplifiable DNA recovered from approximately equal amounts of manually dissected tissue. Specimens from archived tissue blocks could not be controlled for equal fixation conditions. This could be one source of interspecimen variability, although prospectively collected, snap-frozen tissue specimens also demonstrated variability in recovery of amplifiable DNA. Despite attempts to select specimens displaying similar degrees of cellularity and necrosis, another obvious source of this variability is simply the interspecimen cellular heterogeneity. For formalin-fixed, paraffin-embedded tissue harvested by manual dissection, amplification efficiency was approximately equal irrespective of the staining procedure used, although there was a tendency (not statistically significant) for methyl green to yield DNA with increased amplification efficiency (Figure 1A) . The type of stain used had a greater effect in frozen tissue harvested by manual dissection (Figure 1B) . Amplification was consistent and high after staining with azure B, toluidine blue, and methyl green, but was statistically significantly lower after hematoxylin staining or when no stain was used. In our experience, and in accordance with previous reports, 2, 3, 4 the results with hematoxylin-stained tissue obtained by manual dissection were essentially irreproducible. That is, we observed that tissue initially yielding no amplifiable PCR product gave a satisfactory result when repeated on independently sectioned and stained tissue, and vice versa. In this respect, the data with hematoxylin-stained tissue presented in Figure 1 reflect our experience at one given point in time. In contrast, we did not observe this degree of unreliability with the other dyes.
Figure 1.
DNA recoveries evaluated by TaqMan PCR. Staining procedures: NS, no stain; TB, toluidine blue O; AB, azure B; MG, methyl green; H, Mayer’s hematoxylin. Tissue was sampled by manual gross dissection from histological slides (A and B) or by LCM (C and D). The original tissue samples were formalin-fixed, paraffin-embedded archival surgical pathology tissue (A and C) or unfixed, frozen surgical tumor material (B and D). The y-axis gives the amount of template DNA as determined by TaqMan PCR of genomic β-actin DNA. The indicated amount of DNA refers either to a sampled histological tissue section approximately 1 cm2 in area (A and B) or to tissue sampled by 500 LCM firings (C and D). Each data point is the average from two independently processed histological slides. The difference between NS and TB, AB, or MG in B is statistically significant (P < 0.05).
The apparent increase in DNA amplification efficiency after staining with azure B, toluidine blue, and methyl green was observed by two independent detection systems of PCR amplification products: the SYBR green detection method of TaqMan (Figure 1) , and conventional PCR followed by visual inspection of ethidium bromide-stained bands separated electrophoretically on agarose gels (not shown). Therefore, it is unlikely that this effect represents an artifact generated by interference of the histological dyes with the TaqMan detection system. Another possible explanation for the result is that the hematoxylin dye or some other component of the cell lysate is inhibiting PCR amplification. To test this hypothesis, fivefold serial dilutions of cell lysates from hematoxylin-stained, manually dissected tissue were generated. In duplicate reactions of two independently stained sections, cell lysate at a concentration of approximately 5 × 106 μm3 tissue/μl of digestion buffer (comparable to that in the first set of experiments) again yielded no amplifiable DNA. A subsequent fivefold dilution of these lysates resulted in a 200- to 1000-fold increase in the amount of amplifiable DNA, while further fivefold dilutions demonstrated corresponding three- to fourfold decreases in amplifiable material (data not shown). In summary, these data suggest that the effect of hematoxylin-stained tissue on subsequent TaqMan PCR amplification is inhibitory and that this inhibition can be alleviated when the concentration of the lysed tissue falls below 1 × 106 μm/μl of lysis buffer.
For tissue isolated by LCM, DNA amplification was approximately equal, irrespective of the staining procedure, for both paraffin-embedded and frozen tissue (Figure 1 C and 1D) . In particular, hematoxylin was as reliable a stain as methyl green, in contrast to the results obtained with manually dissected tissue.
Discussion
Four different dyes were evaluated in this study: Mayer’s hematoxylin, which is conventionally used as a nuclear stain in pathology laboratories, and three less frequently used nuclear dyes: toluidine blue O, azure B, and methyl green. Hematoxylin is a multicomponent dye with hematein as the active staining ingredient (obtained from hematoxylin by oxidation by sodium iodate) and aluminum ion (Al3+) as the mordant. 6 Other ingredients are citric acid as an acidifier and chloral hydrate, the function of which is unknown. 6 In contrast, toluidine blue O, azure B, and methyl green (active ingredient ethyl green) are essentially one-component nuclear dyes. Because of their cationic nature, these three dyes are believed to bind to the negatively charged DNA in nuclei. 6 The mechanism of interaction of methyl green with DNA has been investigated in detail and is believed to involve not only electrostatic interaction, but also nonionic forces. The nonionic component could be due to intercalation of the dye between the purine and pyrimidine bases. 7, 8, 9
The objective of this study was to investigate the usefulness of different nuclear dyes for microdissection of histological tissue with subsequent amplification of DNA by TaqMan PCR. Both formalin-fixed, paraffin-embedded tissue and frozen tissue were sampled from histological slides either by manual gross dissection or by LCM. The choice of the dye had a marked influence on the outcome of DNA amplification when the entire tissue section was manually sampled from the histological slide. Such sampling, followed by PCR amplification, is commonly done in parallel to microdissection as a quality control to make certain that the content of amplifiable DNA in the tissue has not been unacceptably reduced by autolysis or the staining procedure. Our results show that in this situation, sampling of hematoxylin-stained or unstained tissue would in many instances give false negative, misleading results, whereas staining with methyl green, azure B, or toluidine blue O allows reliable assessment of the DNA content of the tissue (Figure 1 A and B) .
The unsatisfactory amplification of DNA from manually sampled entire tissue sections stained by hematoxylin has been reported previously. 2, 3, 4 In our hands, the average DNA amplification was less after staining with hematoxylin than with other nuclear dyes (Figure 1 A and B) , and TaqMan or conventional PCR failed to yield an amplification product in a substantial number of reactions. Any of the multiple chemical ingredients of the hematoxylin staining solution, which could inhibit the proteinase K digestion or the PCR reaction, may be responsible for this effect. As to the lack of amplifiable DNA from unstained frozen tissue, the mechanism of this effect is unclear. It is not likely due to a washout of tissue inhibitors by the staining solutions, as unstained slides were subjected to similar hydration/dehydration steps. Rather, it may be that the staining solutions, when not inhibitory to the Taq polymerase, render the genomic DNA more accessible, either by releasing it from bound nucleoprotein complexes or chemically cleaving it into smaller fragments that are more readily released from the tissue. This latter hypothesis is partially supported by the finding that unstained tissue from formalin-fixed specimens generally yielded more amplifiable DNA than that from frozen sections.
A previous report has found that after sampling entire tissue sections from the slides by manual dissection, staining with methyl green apparently enhanced DNA recovery relative to unstained tissue. 2 We have made a similar observation (Figure 1A) , although the difference was not statistically significant. We were surprised, however, that the enhancement by methyl green was particularly pronounced and statistically significant when frozen tissue samples were examined (Figure 1B) . The mechanism that leads to this enhancement is not entirely clear and may be multifactorial. Since cationic dyes such as methyl green bind to DNA, it is conceivable that this binding enhances the digestion of DNA-bound proteins by proteinase K, or increases the efficiency of PCR by enhancing the accessibility of the DNA during primer hybridization or extension.
In marked contrast to the situation where entire tissue sections are manually sampled, there was no influence of the staining method on DNA amplification when tissue was sampled by LCM (Figure 1 C and D) . Most likely, this difference is related, at least in part, to the amount of tissue sampled. The tissue pieces obtained by LCM are quite small, resulting in little carryover of dye substances into the proteinase K digestion solution and the PCR reaction. Consequently, inhibitory ingredients of the hematoxylin stain, or enhancing dye substances such as methyl green, will be present in concentrations too low to produce any effect on tissue digestion or DNA amplification.
The main finding of this study is that DNA recovery from manually dissected tissue is not predictive of the recovery after LCM sampling from the same tissue. From our results on tissue sampled by manual dissection, we would have predicted that hematoxylin is an unsuitable stain for microdissection, whereas in fact it gave satisfactory results in the actual LCM experiments. Thus, based on the experience of 16 cases used in this study, hematoxylin can be recommended for LCM-based microdissection.
Address reprint requests to Mark A. Watson, M.D., Ph.D., Assistant Professor of Pathology, Division of Laboratory Medicine, Campus Box 8118, Washington University School of Medicine, 660 S. Euclid Ave., St. Louis, MO 63110. E-mail: watsonm@labmed.wustl.edu.
Footnotes
Supported by Department of Defense grant DAMD 17–99-1–9026 (to J. M.), National Cancer Institute grant K08CA8790101 (to S. A. A.), and the Siteman Cancer Center.
References
- 1.Simone NI, Bonner RF, Gillespie JW, Emmert-Buck MR, Liotta LA: Laser-capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet 1998, 14:272-276 [DOI] [PubMed] [Google Scholar]
- 2.Burton MP, Schneider BG, Brown R, Escamilla-Ponce N, Gulley ML: Comparison of histologic stains for use in PCR analysis of microdissected, paraffin-embedded tissues. BioTechniques 1998, 24:86-92 [DOI] [PubMed] [Google Scholar]
- 3.Murase T, Inagaki H, Eimoto T: Influence of histochemical and immunohistochemical stains on polymerase chain reaction. Mod Pathol 2000, 13:147-151 [DOI] [PubMed] [Google Scholar]
- 4.Serth J, Kuczyk MA, Paeslack U, Lichtinghagen R, Jonas U: Quantitation of DNA extracted after micropreparation of cells from frozen and formalin-fixed tissue sections. Am J Pathol 2000, 156:1189-1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ririe KM, Rasmussen RP, Wittwer CT: Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem 1997, 245:154-160 [DOI] [PubMed] [Google Scholar]
- 6.Kiernan JA: Histological and Histochemical Methods: Theory and Practice. 1990:pp 49-102 Pergamon Press, Oxford
- 7.Lyon H, Jacobson P, Høyer P, Andersen AP: An investigation of new commercial samples of methyl green and pyronin Y. Histochem J 1987, 19:381-384 [DOI] [PubMed] [Google Scholar]
- 8.Scott JE, Willett IH: Binding of cationic dyes to nucleic acids and other biological polyanions. Nature 1966, 209:985-987 [DOI] [PubMed] [Google Scholar]
- 9.Scott JE: On the mechanism of the methyl green-pyronin stain for nucleic acids. Histochemie 1967, 9:30-47 [DOI] [PubMed] [Google Scholar]