Abstract
Epstein-Barr virus (EBV) is the causative agent of infectious mononucleosis, and it may also be found in a wide variety of benign and malignant lesions including oral hairy leukoplakia, inflammatory pseudotumor, Hodgkin’s disease, non-Hodgkin’s lymphoma, nasopharyngeal carcinoma, and gastric carcinoma. Molecular testing is increasingly important in the diagnosis and monitoring of patients affected by these diseases. In biopsy tissues, molecular detection of EBV-encoded RNA transcripts by in situ hybridization remains the gold standard for proving that a histopathological lesion is EBV-related. EBV-encoded RNA hybridization and EBV LMP1 immunostains are used routinely to detect latent EBV in tissues affected by posttransplant lymphoproliferative disorder (PTLD) or in enlarged nodes from patients with infectious mononucleosis. Traditional serology is the best test for evaluating acute versus remote infection in healthy individuals. High serological titers serve as a tumor marker for some EBV-related malignancies, but titers are not a dependable tumor marker in immunocompromised hosts. EBV viral load testing by quantitative DNA amplification of blood samples is a promising new laboratory test that has proven useful for early diagnosis and monitoring patients with PTLD. Recent studies suggest a role for EBV viral load testing in nasopharyngeal carcinoma, Hodgkin’s disease, and AIDS patients with brain lymphoma. Further research is needed to define more fully the clinical utility of viral load tests in the full spectrum of EBV-associated diseases. Gene expression profiling is on the horizon as a means to improve subclassification of EBV-related diseases and to predict response to therapy.
Epstein-Barr virus (EBV) was first identified using electron microscopy of Burkitt’s lymphoma cell cultures in 1964. 1 In subsequent decades, EBV has been linked to a wide variety of benign and neoplastic diseases. Nasopharyngeal carcinomas and posttransplant lymphoproliferative disorders are nearly always EBV-associated, whereas several other tumors, such as Hodgkin’s disease, non-Hodgkin’s lymphoma, lymphoepithelioma-like carcinoma, gastric adenocarcinoma, and several types of sarcoma, are less uniformly EBV-associated. 2, 3, 4, 5, 6, 7 EBV causes benign transient lymphoproliferative lesions at the time of primary infection, and it is found in a benign lesion of the tongue called oral hairy leukoplakia. 8, 9 Patients affected by these benign or malignant diseases may benefit from laboratory detection of EBV to confirm their diagnosis or to monitor disease burden after the initiation of therapy.
Laboratory detection of EBV is accomplished in several ways (Table 1) , and recent progress has focused on the molecular analysis of viral DNA and RNA. In situ hybridization has long been considered the gold standard for detecting tumor-associated viral infection, and EBV viral load assays are now being adopted for clinical evaluation of tumor burden in affected patients. This review article summarizes the pathobiology of EBV infection and describes the clinical laboratory tests that are used to assist in diagnosis and monitoring of patients with EBV-related diseases.
Table 1.
Laboratory Tests for EBV
| Name | Purpose |
|---|---|
| In situ hybridization | Identify EBER transcripts or EBV DNA in specific cell types within histologic lesions |
| EBV clonality assay by Southern blot analysis | Assess clonality of lesions with respect to EBV DNA structure; distinguish latent from replicative infection based on the episomal versus linear structure of the EBV genome |
| EBV DNA amplification | Detect viral DNA in patient tissues; disease specificity is lacking |
| EBV viral load | Quantitate EBV DNA in blood or body fluids to monitor disease status over time |
| Immunohistochemistry (LMP1, EBNA1, EBNA2, LMP2A, BZLF1) | Identify EBV protein expression in specific cell types within histologic lesions; distinguish latent from replicative infection based on expression profiles |
| Culture of EBV or of EBV-infected B lymphocytes | Detect and semiquantitatively measure infectious virions or latently-infected B lymphocytes; impractical for routine clinical use |
| Electron microscopy | Identify whole virions representing replicative viral infection; impractical for routine clinical use |
| Serology (VCA, EBNA, EA, heterophile antibodies) | Measure antibody response to viral proteins in serum samples; distinguish acute from remote infection; monitor disease status over time |
The Pathobiology of EBV Infection
EBV has a 173-kb DNA genome for which the nucleotide sequence and predominant transcripts are well characterized. EBV is capable of infecting B and T lymphocytes, squamous epithelial cells of the oropharynx and nasopharynx, glandular epithelium of the thyroid, stomach, and salivary gland, smooth muscle cells, and follicular dendritic cells. Healthy virus carriers harbor 1 to 50 EBV genomes per million blood mononuclear cells, with B lymphocytes representing the major cellular reservoir. 10 Beyond B cells, it is nearly impossible to find infected cells of other lineages in healthy carriers, but we presume that the other cell types listed above are capable of being infected based on the identification of EBV DNA in lesions arising from them. Investigation of patients with EBV-infected tumors provides reasonable evidence that EBV was present before neoplastic transformation, raising the still unresolved question of the extent to which EBV contributes to tumorigenesis.
EBV infects nearly all humans by the time they reach adulthood. Primary infection results in transient viremia followed by rapid immune response. The virus persists for life in its human host by cleverly balancing its ability to hide from the immune system via latent infection of B lymphocytes with its ability to replicate and shed from oral mucosa. At any given time, about 20% of carriers are shedding salivary virions, leading to nearly universal propagation of the virus in human populations.
EBV infection of B lymphocytes leads to two alternate outcomes mimicking the physiological effects of antigen stimulation. One outcome culminates in the production of memory B cells that persist long-term; the other outcome results in differentiation toward plasma cells that are destined to die. These two outcomes support latent viral persistence and lytic viral replication, respectively. Lifelong infection of the human host relies on these dual phases of infection whereby the virus hides from the immune system in memory B cells, and a subset of these cells are diverted to produce thousands of virions that not only infect more of the host’s own lymphocytes but also are shed in saliva to infect other individuals. Viral replication is naturally enriched in the oral mucosa where memory B cells are routinely stimulated to differentiate after exposure to foreign antigens.
Lytic viral replication is accompanied by expression of about 90 viral proteins, including BZLF1 (also known as ZEBRA), and complexes of viral proteins collectively referred to as early antigen and viral capsid antigen. These lytic antigens elicit a humoral immune response, resulting in elevated antibody titers that quell rampant lytic virus production in the healthy carrier.
Latent infection is characterized by abundant production of EBV-encoded RNA (EBER), but it is important to mention that EBER transcripts remain untranslated. EBER transcripts are thought to function in controlling translation. Also expressed in latently infected cells are EBV nuclear antigen (EBNA) 1 and latent membrane protein (LMP) 2A , neither of which elicits an effective immune response. EBNA1 functions to ensure that the viral genome is propagated to daughter cells upon cell division, whereas LMP2A keeps other viral proteins from being expressed. Limited protein expression helps avert immune destruction in vivo.
In vitro where immune surveillance is absent, infected cell cultures tend to express a broader spectrum of EBV proteins, such as LMP1, -2A, and -2B, and EBNA2, -3A, -3B, -3C, and -LP. LMP1 and EBNA2 are critical for the unique ability of EBV to immortalize B cells in vitro. In this immortalization process, EBV can be cultured by cocultivating virions with B cells from uninfected persons (usually neonatal umbilical cord lymphocytes). The resulting lymphoblastoid cell lines are capable of being propagated indefinitely in culture media. Naturally infected B lymphocytes can likewise be cultured from the blood of viral carriers. Viral culture represents an accurate and semiquantitative measure of EBV in clinical samples, but it is rarely used in clinical laboratories due to high costs and slow turnaround time.
More practical laboratory tests for EBV rely on detection of viral DNA and its gene products. In EBV-infected tissues, three different patterns of latent viral gene expression are seen. Type I latency refers to a very limited spectrum of latent viral gene expression, namely EBER transcripts along with EBNA1 and LMP2A proteins. This pattern is found in circulating lymphocytes of healthy viral carriers, and it is also characteristic of Burkitt’s lymphoma and gastric carcinoma. Type II latency, characterized additionally by LMP1 and LMP2B coexpression, is seen in Hodgkin’s disease, T cell lymphoma, and nasopharyngeal carcinoma, all of which tend to occur in immunocompetent hosts. Type III latency refers to the full spectrum of latent viral gene expression, as found transiently in acute infectious mononucleosis, and as seen in EBV-driven lymphoproliferations arising in immunocompromised hosts. Viral genes expressed in Type III latency include all of the EBNAs (1, 2, 3A, 3B, 3C, LP), the LMPs (1, 2A, 2B), and EBER.
Although the patterns of gene expression described above are useful for characterizing various histopathological entities, in practice there is heterogeneity of expression among different tumors of the same histological type, and even among cells within a given tumor. For example, lymphoid tumors arising in AIDS or transplant patients tend to express more viral products than do their histological look-alikes arising in immunocompetent hosts. Therefore, the typical expression patterns described here provide only rough guidelines to assist in the clinicopathological diagnosis of each entity.
EBER in Situ Hybridization
EBER in situ hybridization is considered the gold standard for detecting and localizing latent EBV in tissue samples. 11 After all, EBER transcripts are consistently expressed in virtually every EBV-infected tumor, and they are likewise expressed in lymphoid tissues taken from patients with infectious mononucleosis, and in the rare infected cell representing normal flora in healthy virus carriers. The only EBV-related lesion that lacks EBER is oral hairy leukoplakia, a purely lytic infection of oral epithelial cells. 12
EBER actually represents two RNA species, EBER1 and EBER2, encoded from two separate but homologous viral genes. EBER transcripts are expressed in latently infected cells at levels approaching a million copies per cell. 13 Because EBER transcripts are naturally amplified, they represent a reliable target for detecting and localizing EBV in tissue sections by in situ hybridization. The literature is replete with EBER hybridization protocols that rely on either oligonucleotide DNA probes, RNA probes (riboprobes), or peptide nucleic acid (PNA) probes. 14, 15, 16, 17, 18, 19, 20 Commercially available EBER probes are labeled with biotin, digoxigenin, or fluorescein (Dako, Glostrup, Denmark; Enzo Diagnostics, Farmingdale, NY; Kreatech Diagnostics, Amsterdam, The Netherlands; Novocastra Laboratories Ltd., Newcastle, UK; Shandon Lipshaw, Pittsburgh, PA; Innogenex, San Ramon, CA; Ventana Medical Systems, Tucson, AZ).
EBER in situ hybridization can be accomplished on paraffin sections or on cytology preparations. A typical 1-day procedure begins with removal of any paraffin followed by treatment with proteinase K and detergent to enhance probe entry into the nucleus where EBER transcripts are located. Any unbound probe is washed away, and then colorization and counterstaining are performed. Interpretation of EBER stains relies on microscopic visualization of the nuclear EBER signal in latently infected cells. Evaluation of cell type and distribution is helpful in evaluating the clinical significance of the result (Figure 1) .
Figure 1.
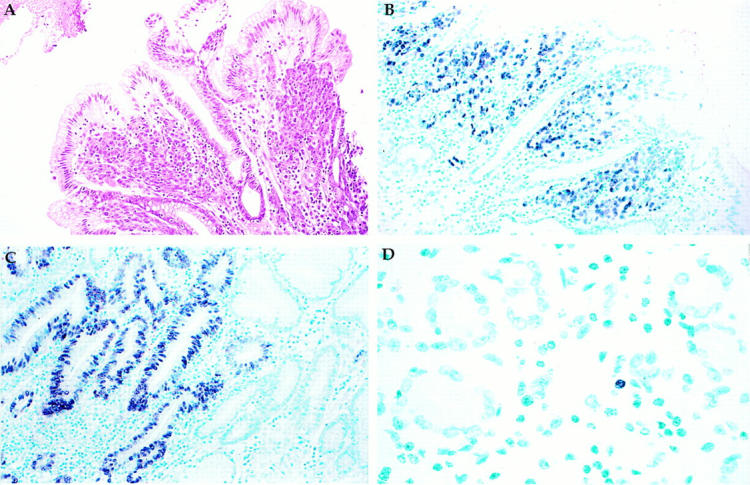
A: H&E stain of invasive gastric adenocarcinoma surrounded by normal surface epithelium. B: EBER in situ hybridization reveals EBER transcripts in the nucleus of the carcinoma cells, but not in the overlying normal surface epithelium, nor in the surrounding benign stromal cells. C: EBER is localized to dysplastic gastric epithelium but not to adjacent normal-appearing glands, implying that EBV infection is an early event in gastric carcinogenesis. D: EBER is localized to the nucleus of a single small lymphoid cell, representing the rare infected lymphocyte that might be found in any previously infected individual. Original magnifications, ×50 (A and B), ×80 (C), and ×150 (D).
Even though EBER transcripts are usually abundantly produced in latently infected cells, users are cautioned about the possibility of false negative EBER hybridization results as a consequence of RNA degradation. A control hybridization must be run in parallel to ensure that RNA is preserved and available for probe binding. This control might target the polyA mRNA tail using a polyT probe, or it might target a ubiquitous cell-derived transcript such as U6 RNA. U6 RNA is a particularly appropriate control because it is similar to EBER in size, abundance, and intranuclear localization, but it is encoded by a cellular gene that is constitutively transcribed. With such a control, the likelihood of false negative EBER interpretation is markedly diminished. Accurate interpretation of results relies on the ability of the morphologist to distinguish tumor cells from background lymphocytes or artifact. When proper attention is paid to these quality control issues, EBER in situ hybridization is the most reliable method for determining if a lesion is EBV-associated.
The primary advantage of EBER in situ hybridization is its ability to localize EBV in the context of cytological and histopathological features of the tissue. Enlarged lymph nodes from infectious mononucleosis patients typically contain EBER in a high fraction of lymphoid cells, including small and large lymphocytes and immunoblasts. 21, 22 In contrast, lymphoid tissues from remotely infected virus carriers harbor EBV in only rare (<0.1%) scattered small to medium lymphoid cells. 21 In EBV-related Hodgkin’s disease, EBER is localized to the malignant Reed-Sternberg/Hodgkin’s cells, whereas the background small lymphocytes are almost completely negative (<0.1%). Likewise, the remaining EBV-associated malignancies, including carcinomas, sarcomas, and lymphomas, exhibit EBER signal in virtually all of the tumor cells, whereas EBER is absent from the adjacent normal tissue, except perhaps for rare scattered lymphoid cells. Premalignant lesions of the gastric epithelium and nasopharyngeal epithelium have also been shown to harbor EBV, suggesting the EBV infection occurs early during carcinogenesis.
EBER hybridizations are used diagnostically in several specific clinical situations. They are used routinely for confirming a diagnosis of EBV-driven posttransplant lymphoproliferative disorder (PTLD). 3, 23 PTLD is a potentially fatal complication of allogeneic transplantation that requires prompt diagnosis and therapy. About 95% of all PTLDs are EBV-associated, as shown by EBER expression by tissue-infiltrating lymphocytes and/or immunoblasts. Treatment involves cutting back or withdrawing immunosuppressives so that natural immunity is allowed to destroy virally infected tumor cells. In recent years, therapeutic success has been reported following infusion of EBV-specific T cells. The occasional EBV-negative PTLD occurs later (usually >2 years) after transplant and does not respond as well to withdrawal of immunosuppression. 24, 25
PTLD-like tumors occasionally occur in patients who have not undergone transplant but who are immunosuppressed for other reasons, such as rheumatoid arthritis patients on methotrexate therapy. 26 As with PTLD, these tumors are often EBER-positive and respond favorably to immune reconstitution.
In biopsies where the differential diagnosis includes infectious mononucleosis, Hodgkin’s disease, and/or non-Hodgkin’s lymphoma, EBER hybridization is often helpful in making the correct diagnosis. In EBV-related Hodgkin’s disease, EBER is largely restricted to Reed-Sternberg cells and mononuclear variants, whereas infectious mononucleosis is characterized by a mixture of small and large EBER-positive cells including immunoblasts rimming necrotic zones. 22 EBER is not expressed in Kikuchi’s lymphadenitis, a lesion that shares some clinical and histological features with infectious mononucleosis. 27
Nearly half of all classical Hodgkin’s disease and T cell lymphomas have EBER-positive tumor cells, whereas only 5% of diffuse large B cell or anaplastic large cell lymphomas express EBER. 4, 5, 28 Certain subsets of these lymphomas are more likely than others to harbor EBV, such as nasal T/NK lymphoma (Table 2) . In classes of tumors that are only fractionally associated with EBV, further investigation of the prognostic value of EBV testing is warranted.
Table 2.
EBV-Associated Diseases
| Disease | Proportion of cases EBV-related | Reference |
|---|---|---|
| Benign, reactive infections | ||
| Infectious mononucleosis | >99 | 22 |
| Oral hairy leukoplakia | >95 | 8 |
| Inflammatory pseudotumor | 40 | 60 |
| Non-Hodgkin’s lymphomas and immunodeficiency-related neoplasms | ||
| Non-Hodgkin’s lymphoma, all subtypes | 5 | 5 |
| Non-Hodgkin’s lymphoma, AIDS-related | 40 | 61 |
| Brain lymphoma, AIDS-related | 95 | 62 |
| Brain lymphoma, immunocompetent hosts | 5 | 63 |
| Post transplant lymphoproliferative disorder (PTLD) | 95 | 3 |
| Burkitt’s lymphoma, African | >95 | 64 |
| Burkitt’s lymphoma, North American | 20 | 64 |
| Burkitt’s lymphoma, AIDS-related | 30 | 65 |
| Lymphoma, primary immunodeficiency | most | 66 |
| Lymphomatoid granulomatosis (B cell) | most | 67 |
| Peripheral T cell lymphoma | 40 | 28 |
| Nasal T/NK cell lymphoma | >95 | 68 |
| Smooth muscle tumors in AIDS or transplant patients | >95 | 69 |
| Hodgkin’s disease | ||
| Hodgkin’s disease, all subtypes | 40 | 4 |
| Hodgkin’s disease, mixed cellularity | 70 | 4 |
| Hodgkin’s disease, nodular sclerosis | 20 | 4 |
| Hodgkin’s disease, lymphocyte predominant | <5% | 70 |
| Hodgkin’s disease, lymphocyte depleted | 50 | 4 |
| Hodgkin’s disease, AIDS-related | >95 | 71 |
| Carcinomas | ||
| Nasopharyngeal carcinoma, Asian | >95 | 2 |
| Nasopharyngeal carcinoma, North American | 75 | 29 |
| Lymphoepithelioma-like carcinoma, foregut derived | most | 6 |
| Gastric adenocarcinoma | 7 | 7 |
Selected subtypes of carcinoma express EBER, notably nasopharyngeal carcinomas and lymphoepithelioma-like carcinomas of the thymus, thyroid, salivary gland, lung, or stomach. 2, 6, 29 The majority of nasopharyngeal carcinoma patients initially present with enlarged lymph nodes containing metastatic undifferentiated carcinoma of unknown primary, and EBER expression is touted as an indicator of nasopharyngeal origin.
EBER is usually expressed uniformly in all of the tumor cells comprising an EBV-associated malignancy, although occasional tumors have only focal EBER expression. Lack of uniform expression could be a technical artifact related to focal preservation of RNA, or it could represent true biological variability in EBER levels. John Sixbey and colleagues have proposed a “hit-and-run” hypothesis whereby the virus is lost from some or all cells within a tumor. 30 Further research on this topic is warranted. In the meantime, it is prudent to interpret focal EBER hybridization results in conjunction with control assays for RNA preservation and in conjunction with other tests for EBV.
There is intriguing geographic variability in the incidence of EBV-related tumors. For example, Burkitt’s lymphoma is the most common pediatric cancer in tropical Africa, where it is almost always EBV-related, whereas it is 50-fold less common in the United States and only 20% EBV-related. As another example, EBV-related nasopharyngeal carcinoma is the most common cancer in parts of Southern Asia, where it is nearly always EBV-related, whereas the tumor is 50-fold less common in the United States and only 75% EBV-related. There appears to be an inverse correlation between the incidence of gastric cancer and its EBV relatedness, unlike what is observed with nasopharyngeal carcinoma. These striking geographic variations have yet to be fully explained, but preliminary studies implicate environmental cofactors over genetic predisposition or oncogenic viral strains.
Once identified in a patient’s tumor, EBER can be used as a marker of recurrent disease. For example, when looking for recurrence in patients treated for nasopharyngeal carcinoma, EBER hybridizations can be used to complement microscopic examination of nasopharyngeal biopsies. As described below, blood tests for EBV viral load are also useful markers of tumor burden after therapy.
In Situ Hybridization to EBV DNA
Probes targeting the BamHIW internal repeat sequence, which is reiterated up to 11 times in each EBV genome, can be used to detect and localize EBV DNA in tissue sections. 16 Single-copy viral sequences could also be targeted, but assay sensitivity is relatively diminished. In a practical sense, there is little reason to target EBV DNA rather than EBER RNA, except perhaps in samples where the RNA has been selectively destroyed. In clinical situations, EBER transcripts remain the more common target for in situ detection of EBV.
LMP1 Immunohistochemistry
The relative merits of immunohistochemistry versus EBER in situ hybridization deserve attention. In fact, LMP1 immunostains are nearly as effective as EBER in situ hybridization for identifying EBV in PTLD cases, in Hodgkin’s disease, and in infectious mononucleosis. 31 Such is not the case for non-Hodgkin’s lymphomas or carcinomas, however, in which LMP1 is often undetectable even when EBER is clearly positive.
Some important differences are seen in the distribution of EBER versus LMP1 expression in tumor samples. In PTLD samples, LMP1 is typically expressed in about 5% of lesional immunoblasts (range, 0–100%). When the same PTLD samples are stained for EBER, it becomes apparent that many more lymphoid cells are EBV-infected, but only a fraction of those cells coexpress LMP1. Immunoblasts are often the subtype of lymphocyte that coexpress LMP1, whereas small lymphocytes are more likely to express EBER alone. Occasional PTLDs lack LMP1 entirely, even though EBER is clearly positive, implying that EBER is a more reliable target than is LMP1. Nevertheless, LMP1 immunostains are economical and rapid; therefore, they retain a role in clinical evaluation of suspected PTLD cases.
LMP1 stains reliably identify EBV in Reed-Sternberg/ Hodgkin’s cells, although sometimes only a fraction of the EBER-positive tumor cells coexpress LMP1. LMP1 is reliably expressed in lymph nodes from infectious mononucleosis patients, with some EBER-positive small lymphocytes failing to coexpress LMP1, but immunoblasts coexpressing both markers. Therefore, EBER and LMP1 stains appear to be equally informative in confirming a diagnosis of infectious mononucleosis. 22, 32
LMP1 immunostains can be performed on paraffin sections using commercially available antibodies (CS1–4 monoclonal cocktail, Dako, or S12 monoclonal, Organon-Teknika, Boxtel, The Netherlands). 33 True LMP1 signal is granular in character and is localized to the cytoplasm and surface membrane. Results should be interpreted by a morphologist who is confident in discerning tumor cells from other cells in which a false positive signal has been described, namely eosinophils, plasma cells, cells of the nervous system, and poorly fixed cells.
Measuring EBV Gene Expression by Immunohistochemistry, Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR), and Nucleic Acid Sequence-Based Amplification (NASBA)
Detection of viral proteins can be achieved by immunohistochemical stains of paraffin sections. Common targets include EBNA1, EBNA2, LMP2A, and BZLF1. 34, 35, 36 Of these, BZLF1, also called ZEBRA, is the only factor that is characteristic of lytic viral replication. In fact, BZLF1 immunostains are quite useful in confirming a diagnosis of oral hairy leukoplakia in tongue biopsies from AIDS patients using commercially available antibody (clone BZ.1, Dako, Carpinteria, CA). Interpreting pathologists are cautioned that BZLF1 staining is localized to the nucleus of ballooned epithelial cells of oral hairy leukoplakia, whereas cytoplasmic cross-reactivity of the antibody should be disregarded.
Alternative approaches to detecting these viral gene products are RT-PCR and NASBA. 37 Though not yet used routinely in clinical laboratories, there is much to recommend them as disease-specific markers, especially if they can be applied in multiplex or array format. In theory, this should facilitate diagnosis of each class of EBV-associated disease based on the unique expression profile of viral and cellular genes that characterizes each disease.
Progress continues to be made in profiling the expression pattern of each EBV-associated disease. For example, a recent study used NASBA to identify EBV BARF1 transcripts in gastric carcinomas. 38 BARF1 is likewise expressed in nasopharyngeal carcinomas but apparently not in lymphocytes, implying that BARF1 might serve as a marker of these epithelial malignancies without concern for interference from the occasional bystander lymphocyte that might be infected. 39 If validation studies pan out, it is feasible that quantitative measurement of this and other viral transcripts will prove useful for diagnosis and monitoring of affected patients.
Southern Blot Analysis of EBV DNA
Southern blot analysis can be used to determine the clonality of EBV-infected tissues with respect to the structure of EBV DNA. This assay, first described by Raab-Traub and Flynn in 1986, 40 is based on the presence of variable numbers of terminal repeat sequences at the ends of each EBV DNA molecule. A given cell is apparently infected only once, and each infecting genome contains up to 20 terminal repeat sequences. The relatively unique terminal repeat structure that is present in a given cell is passed along to cellular progeny upon cell division. Analysis of clinical samples has provided interesting results. Oral hairy leukoplakia, representing an infectious process, produces polyclonal viral genomes indicative of lytic viral replication. On the other hand, EBV-associated tumors harbor monoclonal EBV DNA.
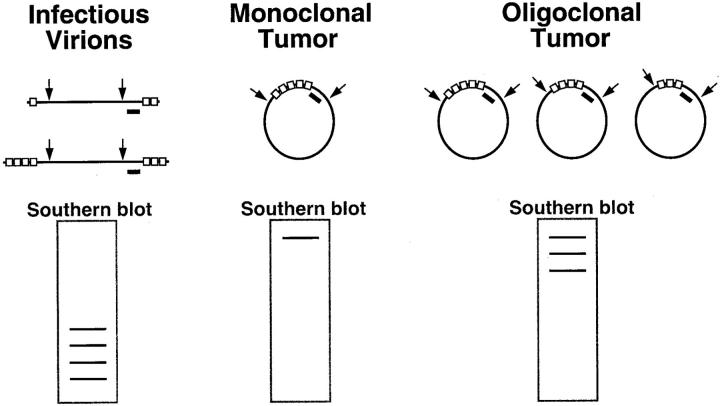
To perform the EBV clonality assay, lesional DNA is first subjected to digestion by BamHI restriction enzyme, which cuts at sequences flanking the region where the terminal repeats are located. After electrophoresis and transfer, a labeled internal probe is applied to detect the fragment(s) containing the terminal repeats. 14 Analysis of the band pattern distinguishes monoclonal from oligoclonal, polyclonal, and uninfected tumors, and also reveals whether the sample contains substantial amounts of linear EBV genomes as a consequence of active viral replication (Figure 2) .
Figure 2.
The EBV clonality assay evaluates clonality with respect to the structure of the EBV genome. The assay is based on the presence of variable numbers of tandem repeat sequences (shown as open boxes) at the ends of the linear viral genome. On infection of a cell, these ends join to form an episome by fusing up to 20 terminal repeat sequences. When an infected cell undergoes malignant transformation, the same fused terminal repeat structure is inherited by all progeny of the malignant clone. The clonality assay is accomplished by Southern blot analysis of DNA extracted from patient tissue and digested with BamHI restriction endonuclease (shown by arrows) to cut the EBV genome at sites flanking the terminal repeats. This results in restriction fragments that are recognized by a DNA probe (black bar). Examination of the band pattern on Southern blots reveals that infectious virions produce a ladder array of small bands. In contrast, a monoclonal tumor exhibits a single band of high molecular weight, and an oligoclonal tumor has several such bands.
Application of this clonality assay reveals monoclonal EBV DNA in nearly all infected carcinomas, sarcomas, and Hodgkin’s and non-Hodgkin lymphomas. 40, 41, 42, 43 A subset of immunocompromised patients have either oligoclonal or polyclonal lymphoid proliferations, and these patients apparently have a better prognosis. 3, 44 Even so, monoclonal tumors may respond to immune reconstitution, leading many clinicians to treat their patients the same regardless of clonality status.
Amplification of EBV DNA
Amplification methods have been used by many clinical laboratories for detecting EBV in blood, body fluid, or tissue samples. For example, detection of EBV in biopsies of metastatic undifferentiated carcinoma of unknown primary narrows the differential diagnosis and focuses attention on the nasopharynx. As another example, a study of HIV-infected patients with persistent generalized lymphadenopathy showed that amplifiable EBV DNA was associated with a heightened risk of developing lymphoma. 45 Most remarkably, amplification of EBV DNA from the cerebrospinal fluid of AIDS patients is nearly always indicative of a brain lymphoma, leading oncologists to proceed with lymphoma treatment without the need for brain biopsy (assuming an appropriate clinical setting and radiographic support for the diagnosis). 46 After treatment, disappearance of EBV DNA from the cerebrospinal fluid is associated with better outcomes. 47
From a technical standpoint, PCR amplification of EBV DNA is accomplished using primers spanning conserved EBV sequences, whereas strain typing relies on amplification of polymorphic regions of the viral genome. Strain typing will not be discussed in any detail, since there are no solid clinical indications for such testing. Even qualitative amplification assays are difficult to justify because of their inability to distinguish lesion-specific EBV from that representing normal flora. After all, EBV DNA is present in a small fraction of lymphoid cells from every healthy virus carriers, which means that nearly every adult and a substantial fraction of all children harbor amplifiable EBV DNA. The inability to distinguish EBV disease from background infection led many laboratory scientists to abandon PCR in favor of EBER in situ hybridization for the reliable detection of lesion-associated EBV in biopsy specimens. Indeed, EBER studies remain a mainstay of diagnostic surgical pathology. But improvements in quantitative amplification technology are stimulating a resurgence of interest in amplification strategies for detecting EBV in patient samples.
EBV Viral Load Measurement by Quantitative DNA Amplification
EBV viral load testing involves quantitative measurement of EBV DNA in patient samples. A typical viral load assay employs PCR to coamplify EBV DNA and a spiked control sequence in nucleic acid extracted from blood samples. 14 The amount of amplification product, measured either at the end point of the assay or in real time, can be used to calculate the EBV viral load in copies per milliliter of blood.
The EBV viral load assay has several technical and clinical advantages over other methods of viral detection. First, the test is rapid, with with a turnaround time of only 1 to 2 days. Second, it appears that patients with several subsets of EBV-related diseases are massively and systemically infected by EBV, allowing us to screen for these diseases by viral load assays of blood or body fluid, potentially alleviating the need for invasive tissue biopsy. And finally, recent clinical studies reveal that several EBV-related diseases can be monitored by sequential measurement of EBV viral load.
EBV viral load testing appears to be more reliable than serology for evaluating the EBV status of immunocompromised hosts. In fact, recent studies of transplant patients showed that those affected by EBV-driven PTLD have extremely high EBV viral loads, sometimes exceeding 1 million copies per milliliter of blood. 48, 49 Furthermore, viral load rises as early as several months before the clinical onset of PTLD, suggesting that the assay might be used to screen high risk populations for purposes of early intervention. 50, 51, 52, 53 And finally, EBV viral load decreases on successful therapy, suggesting that the assay should be used to monitor therapeutic efficacy. 52, 54
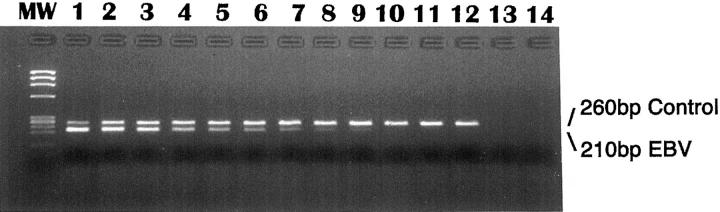
From a technical standpoint, EBV viral load assays have been shown to be sensitive, specific, and quantitative across a wide dynamic range. A commercial kit (BioSource International, Camarillo, CA) is available to facilitate PCR amplification of EBV EBER genomic sequences. After coamplification of EBV and a spiked competitor using biotinylated primers, products are detected in an automated enzyme-linked immunosorbent assay plate system. Comparison between the amount of EBV product and the amount of control product permits calculation of EBV viral load in the patient specimen (Figure 3) . 14
Figure 3.
The EBV viral load assay is accomplished by coamplification of EBV DNA and a control sequence that is spiked into the sample before DNA extraction. In the experiment shown here, assay linearity was tested on serial twofold dilutions of EBV DNA. PCR products at the endpoint of amplification were evaluated by agarose gel electrophoresis. In lanes 1–8, the EBV product is seen as a 210-bp band at template levels as low as 5 copies. The control product, visible at 260 bp, ensures that no inhibitors are present, and it also serves as a gauge by which to extrapolate the amount of EBV template in each sample. A molecular weight (MW) marker is shown on the left, and lanes 13 and 14 represent control reactions to which no template was added.
An alternative procedure for EBV viral load measurement involves real-time measurement of PCR products, a procedure that has the potential to reduce labor costs, diminish the risk of amplicon contamination, and reduce turnaround time. 49, 51, 55 Additional laboratory strategies will undoubtedly be developed as molecular technology continues to advance.
In nasopharyngeal carcinoma patients, EBV viral load shows promise as a marker of tumor burden that will facilitate monitoring of patients after therapy. 56 Because about half of all affected patients are destined to relapse, further investigation of the impact of EBV viral load assays is important to distinguish those patients in long-term remission from those destined to relapse.
In patients with EBV-related Hodgkin’s disease, a recent study suggests that EBV viral load might likewise serve as a marker of tumor burden. 57 More research is needed on this and other EBV-related diseases to define more fully the clinical utility of EBV viral load assays.
EBV Serology
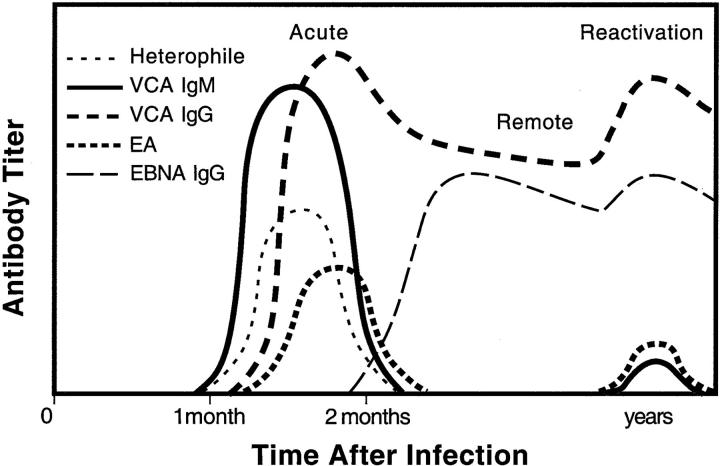
No article about laboratory testing for EBV would be complete without a discussion of serological testing, which is the gold standard for confirming acute versus remote EBV infection in immunocompetent hosts. The heterophile test (also known by a commercial trade name, the Monospot test) was introduced in 1932 as a marker for infectious mononucleosis, even though it would be several decades before EBV was discovered as the causative agent. Heterophile tests are still used today, often in the form of a 2-minute horse red cell agglutination test (Seradyn Color Slide II, Seradyn, Indianapolis, IN). EBV-specific serological assays by enzyme-linked immunosorbent assay or by immunofluorescent assay are used for more accurate confirmation of acute or convalescent EBV infection. 9 Figure 4 displays a typical serological response to EBV infection.
Figure 4.
Primary EBV infection in healthy hosts is accompanied by an orchestrated serological response. IgM antibody against viral capsid antigen (VCA) rises first. Antibodies against EBNA appear at least 1 month after primary infection and are measured, along with IgG anti-VCA, as markers of prior infection and as indicators of EBV reactivation. Titers against early antigen (EA) rise on primary infection and again in pathological states of EBV reactivation.
EBV-associated tumors are often characterized by abnormally high titers against early antigen and IgG viral capsid antigen with diminished EBNA titers. However, this pattern is not specific for malignancy and can be seen in patients with autoimmune diseases or other immune dysfunction, implying that serology alone is inadequate for diagnosis of EBV-related malignancy.
Nasopharyngeal carcinoma patients usually have elevated titers against multiple viral antigens, particularly IgA antibodies against lytic antigens, reflecting the tumor’s origin in the mucosa of the nasopharynx. 58 In fact, a panel of serological tests is used fairly successfully to screen for nasopharyngeal carcinoma in high risk populations, to assign prognosis in those patients who are affected, and to detect early relapse after therapy. 59 Analogous studies are underway in gastric carcinoma patients who likewise harbor high serological titers against EBV. 7
Immunosuppressed patients have inconsistent humoral responses against EBV; therefore, serology is not as reliable a marker of clinical status. In these patients, direct detection of viral nucleic acid or protein is more reliable for identifying clinically relevant EBV infection.
Summary
Molecular diagnostics is increasingly important for diagnosis and monitoring of patients affected by EBV-related diseases. These diseases represent a wide spectrum of clinical manifestations, from transient benign infection to aggressive malignancies. As virus-specific treatments continue to be investigated, it becomes even more important to recognize these EBV-associated diseases so that proper clinical management decisions can be made.
New molecular tests combined with traditional serological or histochemical assays are helpful for diagnosis and monitoring of EBV-related diseases, depending on the clinical setting and the types of samples available for testing. EBER in situ hybridization on biopsy samples and, more recently, EBV viral load testing of blood samples provide an accurate measure of clinical status in PTLD patients. Investigations are underway to better define the utility of these assays across the full spectrum of EBV-associated diseases. On the horizon are gene expression profiling and array technology, which likely will improve our ability to subclassify these diseases and predict responses to therapy.
Address reprint requests to Margaret L. Gulley, M.D., Department of Pathology, University of Texas Health Science Center at San Antonio, 7703 Floyd Curl Drive, San Antonio, TX 78284-7750. E-mail: gulleym@uthscsa.edu.
References
- 1.Barr YM, Epstein MA: Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet 1964, I:702-703 [DOI] [PubMed] [Google Scholar]
- 2.Chen CL, Wen WN, Chen JY, Hsu MM, Hsu HC: Detection of Epstein-Barr virus genome in nasopharyngeal carcinoma by in situ DNA hybridization. Intervirology 1993, 36:91-98 [DOI] [PubMed] [Google Scholar]
- 3.Chadburn A, Cesarman E, Knowles DM: Molecular pathology of posttransplantation lymphoproliferative disorders. Semin Diagn Pathol 1997, 14:15-26 [PubMed] [Google Scholar]
- 4.Glaser SL, Lin RJ, Stewart SL, Ambinder RF, Jarrett RF, Brousset P, Pallesen G, Gulley ML, Khan G, O’Grady J, Hummel M, Preciado MV, Knecht H, Chan JK, Claviez A: Epstein-Barr virus-associated Hodgkin’s disease: epidemiologic characteristics in international data. Int J Cancer 1997, 70:375-382 [DOI] [PubMed] [Google Scholar]
- 5.Hummel M, Anagnostopoulos I, Korbjuhn P, Stein H: Epstein-Barr virus in B-cell non-Hodgkin’s lymphomas: unexpected infection patterns and different infection incidence in low- and high-grade types. J Pathology 1995, 175:263-271 [DOI] [PubMed] [Google Scholar]
- 6.Iezzoni JC, Gaffey MJ, Weiss LM: The role of Epstein-Barr virus in lymphoepithelioma-like carcinomas. Am J Clin Pathol 1995, 103:308-315 [DOI] [PubMed] [Google Scholar]
- 7.Imai S, Koizumi S, Sugiura M, Tokunaga M, Uemura Y, Yamamoto N, Tanaka S, Sato E, Osato T: Gastric carcinoma: monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proc Natl Acad Sci USA 1994, 91:9131-9135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenspan JS, Greenspan D: Oral hairy leukoplakia: diagnosis and management. Oral Surg Oral Med Oral Pathol 1989, 67:396-403 [DOI] [PubMed] [Google Scholar]
- 9.Svahn A, Magnusson M, Jagdahl L, Schloss L, Kahlmeter G, Linde A: Evaluation of three commercial enzyme-linked immunosorbent assays and two latex agglutination assays for diagnosis of primary Epstein-Barr virus infection. J Clin Microbiol 1997, 35:2728-2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan G, Miyashita EM, Yang B, Babcock GJ, Thorley-Lawson DA: Is EBV persistence in vivo a model for B cell homeostasis? Immunity 1996, 5:173-179 [DOI] [PubMed] [Google Scholar]
- 11.Ambinder RF, Mann RB: Epstein-Barr-encoded RNA in situ hybridization: diagnostic applications. Hum Pathol 1994, 25:602-605 [DOI] [PubMed] [Google Scholar]
- 12.Gilligan K, Rajadurai P, Resnick L, Raab-Traub N: Epstein-Barr virus small nuclear RNAs are not expressed in permissively infected cells in AIDS-associated leukoplakia. Proc Natl Acad Sci USA 1990, 87:8790-8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clemens MJ: The small RNAs of Epstein-Barr virus. Mol Biol Rep 1993, 17:81-92 [DOI] [PubMed] [Google Scholar]
- 14.Fan H, Gulley ML: Molecular methods for detecting Epstein-Barr virus. Molecular Pathology Protocols. Edited by Killeen AA. Totowa, NJ, Humana Press, 2001
- 15.Barletta JM, Kingma DW, Ling Y, Charache P, Mann RB, Ambinder RF: Rapid in situ hybridization for the diagnosis of latent Epstein-Barr virus infection. Mol Cell Probes 1993, 7:105-109 [DOI] [PubMed] [Google Scholar]
- 16.Brousset P, Butet V, Chittal S, Selves J, Delsol G: Comparison of in situ hybridization using different nonisotopic probes for detection of Epstein-Barr virus in nasopharyngeal carcinoma and immunohistochemical correlation with anti-latent membrane protein antibody. Lab Invest 1992, 67:457-464 [PubMed] [Google Scholar]
- 17.Hamilton-Dutoit SJ, Pallesen G: Detection of Epstein-Barr virus small RNAs in routine paraffin sections using non-isotopic RNA/RNA in situ hybridization. Histopathology 1994, 25:101-111 [DOI] [PubMed] [Google Scholar]
- 18.Permeen AM, Sam CK, Pathmanathan R, Prasad U, Wolf H: Detection of Epstein-Barr virus DNA in nasopharyngeal carcinoma using a non-radioactive digoxigenin-labelled probe. J Virol Methods 1990, 27:261-267 [DOI] [PubMed] [Google Scholar]
- 19.Khan G, Coates PJ, Kangro HO, Slavin G: Epstein Barr virus (EBV) encoded small RNAs: targets for detection by in situ hybridisation with oligonucleotide probes. J Clin Pathol 1992, 45:616-620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang KL, Chen YY, Shibata D, Weiss LM: Description of an in situ hybridization methodology for detection of Epstein-Barr virus RNA in paraffin-embedded tissues, with a survey of normal and neoplastic tissues. Diagn Mol Pathol 1992, 1:246-255 [PubMed] [Google Scholar]
- 21.Niedobitek G, Herbst H, Young LS, Brooks L, Masucci MG, Crocker J, Rickinson AB, Stein H: Patterns of Epstein-Barr virus infection in non-neoplastic lymphoid tissue. Blood 1992, 79:2520-2526 [PubMed] [Google Scholar]
- 22.Reynolds DJ, Banks PM, Gulley ML: New characterization of infectious mononucleosis and a phenotypic comparison with Hodgkin’s disease. Am J Pathol 1995, 146:379-388 [PMC free article] [PubMed] [Google Scholar]
- 23.Harris NL, Ferry JA, Swerdlow SH: Posttransplant lymphoproliferative disorders: summary of Society for Hematopathology workshop. Semin Diagn Pathol 1997, 14:8-14 [PubMed] [Google Scholar]
- 24.Leblond V, Davi F, Charlotte F, Dorent R, Bitker MO, Sutton L, Gandjbakhch I, Binet JL, Raphael M: Posttransplant lymphoproliferative disorders not associated with Epstein-Barr virus: a distinct entity? J Clin Oncol 1998, 16:2052-2059 [DOI] [PubMed] [Google Scholar]
- 25.Nelson BP, Nalesnik MA, Bahler DW, Locker J, Fung JJ, Swerdlow SH: Epstein-Barr virus-negative post-transplant lymphoproliferative disorders: a distinct entity? Am J Surg Pathol 2000, 24:375-385 [DOI] [PubMed] [Google Scholar]
- 26.Kamel OW: Iatrogenic lymphoproliferative disorders in nontransplantation settings. Semin Diagn Pathol 1997, 14:27-34 [PubMed] [Google Scholar]
- 27.Hollingsworth HC, Peiper SC, Weiss LM, Raffeld M, Jaffe ES: An investigation of the viral pathogenesis of Kikuchi-Fujimoto disease: lack of evidence for Epstein-Barr virus or human herpesvirus type 6 as the causative agents. Arch Pathol Lab Med 1994, 118:134-140 [PubMed] [Google Scholar]
- 28.d’Amore F, Johansen P, Houmand A, Weisenburger DD, Mortensen LS: Epstein-Barr virus genome in non-Hodgkin’s lymphomas occurring in immunocompetent patients: highest prevalence in nonlymphoblastic T-cell lymphoma and correlation with a poor prognosis. Danish Lymphoma Study Group, LYFO. Blood 1996, 87:1045-1055 [PubMed] [Google Scholar]
- 29.Gulley ML, Amin MB, Nicholls JM, Banks PM, Ayala AG, Srigley JR, Eagan PA, Ro JY: Epstein-Barr virus is detected in undifferentiated nasopharyngeal carcinoma but not in lymphoepithelioma-like carcinoma of the urinary bladder. Hum Pathol 1995, 26:1207-1214 [DOI] [PubMed] [Google Scholar]
- 30.Sixbey JW: Epstein-Barr virus DNA loss from tumor cells and the geography of Burkitt’s lymphoma. Epstein-Barr Virus Rep 2000, 7:37-40 [Google Scholar]
- 31.Lones MA, Shintaku IP, Weiss LM, Thung SN, Nichols WS, Geller SA: Posttransplant lymphoproliferative disorder in liver allograft biopsies: a comparison of three methods for the demonstration of Epstein-Barr virus. Hum Pathol 1997, 28:533-539 [DOI] [PubMed] [Google Scholar]
- 32.Hummel M, Anagnostopoulos I, Dallenbach F, Korbjuhn P, Dimmler C, Stein H: EBV infection patterns in Hodgkin’s disease and normal lymphoid tissue: expression and cellular localization of EBV gene products. Br J Haematol 1992, 82:689-694 [DOI] [PubMed] [Google Scholar]
- 33.Brink AA, Dukers DF, van den Brule AJ, Oudejans JJ, Middeldorp JM, Meijer CJ, Jiwa M: Presence of Epstein-Barr virus latency type III at the single cell level in post-transplantation lymphoproliferative disorders and AIDS related lymphomas. J Clin Pathol 1997, 50:911-918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grasser FA, Murray PG, Kremmer E, Klein K, Remberger K, Feiden W, Reynolds G, Niedobitek G, Young LS, Mueller-Lantzsch N: Monoclonal antibodies directed against the Epstein-Barr virus-encoded nuclear antigen 1 (EBNA1): immunohistologic detection of EBNA1 in the malignant cells of Hodgkin’s disease. Blood 1994, 84:3792-3798 [PubMed] [Google Scholar]
- 35.Niedobitek G, Kremmer E, Herbst H, Whitehead L, Dawson CW, Niedobitek E, von Ostau C, Rooney N, Grasser FA, Young LS: Immunohistochemical detection of the Epstein-Barr virus-encoded latent membrane protein 2A in Hodgkin’s disease and infectious mononucleosis. Blood 1997, 90:1664-1672 [PubMed] [Google Scholar]
- 36.Cruchley AT, Murray PG, Niedobitek G, Reynolds GM, Williams DM, Young LS: The expression of the Epstein-Barr virus nuclear antigen (EBNA-I) in oral hairy leukoplakia. Oral Diseases 1997, 3:S177-S179 [DOI] [PubMed] [Google Scholar]
- 37.Brink AA, Oudejans JJ, Jiwa M, Walboomers JM, Meijer CJ, van den Brule AJ: Multiprimed cDNA synthesis followed by PCR is the most suitable method for Epstein-Barr virus transcript analysis in small lymphoma biopsies. Mol Cell Probes 1997, 11:39-47 [DOI] [PubMed] [Google Scholar]
- 38.zur Hausen A, Brink AA, Craanen ME, Middeldorp JM, Meijer CJ, van den Brule AJ: Unique transcription pattern of Epstein-Barr virus (EBV) in EBV-carrying gastric adenocarcinomas: expression of the transforming BARF1 gene. Cancer Res 2000, 60:2745-2748 [PubMed] [Google Scholar]
- 39.Hayes DP, Brink AA, Vervoort MB, Middeldorp JM, Meijer CJ, van den Brule AJ: Expression of Epstein-Barr virus (EBV) transcripts encoding homologues to important human proteins in diverse EBV associated diseases [published erratum appears in Mol Pathol 1999, 52: 305]. Mol Pathol 1999, 52:97-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raab-Traub N, Flynn K: The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell 1986, 47:883-889 [DOI] [PubMed] [Google Scholar]
- 41.Gulley ML, Eagan PA, Quintanilla-Martinez L, Picado AL, Smir BN, Childs C, Dunn CD, Craig FE, Williams J, Jr, Banks PM: Epstein-Barr virus DNA is abundant and monoclonal in the Reed-Sternberg cells of Hodgkin’s disease: association with mixed cellularity subtype and Hispanic American ethnicity. Blood 1994, 83:1595-1602 [PubMed] [Google Scholar]
- 42.Gulley ML, Raphael M, Lutz CT, Ross DW, Raab-Traub N: Epstein-Barr virus integration in human lymphomas and lymphoid cell lines. Cancer 1992, 70:185-191 [DOI] [PubMed] [Google Scholar]
- 43.Gulley ML, Pulitzer DR, Eagan PA, Schneider BG: Epstein-Barr virus infection is an early event in gastric carcinogenesis and is independent of bcl-2 expression and p53 accumulation. Hum Pathol 1996, 27:20-27 [DOI] [PubMed] [Google Scholar]
- 44.Locker J, Nalesnik M: Molecular genetic analysis of lymphoid tumors arising after organ transplantation. Am J Pathol 1989, 135:977-987 [PMC free article] [PubMed] [Google Scholar]
- 45.Shibata D, Weiss LM, Nathwani BN, Brynes RK, Levine AM: Epstein-Barr virus in benign lymph node biopsies from individuals infected with the human immunodeficiency virus is associated with concurrent or subsequent development of non-Hodgkin’s lymphoma. Blood 1991, 77:1527-1533 [PubMed] [Google Scholar]
- 46.Antinori A, De Rossi G, Ammassari A, Cingolani A, Murri R, Di Giuda D, De Luca A, Pierconti F, Tartaglione T, Scerrati M, Larocca LM, Ortona L: Value of combined approach with thallium-201 single-photon emission computed tomography and Epstein-Barr virus DNA polymerase chain reaction in CSF for the diagnosis of AIDS-related primary CNS lymphoma. J Clin Oncol 1999, 17:554-560 [DOI] [PubMed] [Google Scholar]
- 47.Antinori A, Cingolani A, De Luca A, Gaidano G, Ammassari A, Larocca LM, Ortona L: Epstein-Barr virus in monitoring the response to therapy of acquired immunodeficiency syndrome-related primary central nervous system lymphoma. Ann Neurol 1999, 45:259-261 [PubMed] [Google Scholar]
- 48.Bai X, Hosler G, Rogers BB, Dawson DB, Scheuermann RH: Quantitative polymerase chain reaction for human herpesvirus diagnosis and measurement of Epstein-Barr virus burden in posttransplant lymphoproliferative disorder. Clin Chem 1997, 43:1843-1849 [PubMed] [Google Scholar]
- 49.Niesters HG, van Esser J, Fries E, Wolthers KC, Cornelissen J, Osterhaus AD: Development of a real-time quantitative assay for detection of Epstein-Barr virus. J Clin Microbiol 2000, 38:712-715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riddler SA, Breinig MC, McKnight JL: Increased levels of circulating Epstein-Barr virus (EBV)-infected lymphocytes and decreased EBV nuclear antigen antibody responses are associated with the development of posttransplant lymphoproliferative disease in solid-organ transplant recipients. Blood 1994, 84:972-984 [PubMed] [Google Scholar]
- 51.Kimura H, Morita M, Yabuta Y, Kuzushima K, Kato K, Kojima S, Matsuyama T, Morishima T: Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J Clin Microbiol 1999, 37:132-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rooney CM, Smith CA, Ng CY, Loftin SK, Sixbey JW, Gan Y, Srivastava DK, Bowman LC, Krance RA, Brenner MK, Heslop HE: Infusion of cytotoxic T cells for the prevention and treatment of Epstein-Barr virus-induced lymphoma in allogeneic transplant recipients. Blood 1998, 92:1549-1555 [PubMed] [Google Scholar]
- 53.Lucas KG, Filo F, Heilman DK, Lee CH, Emanuel DJ: Semiquantitative Epstein-Barr virus polymerase chain reaction analysis of peripheral blood from organ transplant patients and risk for the development of lymphoproliferative disease. Blood 1998, 92:3977-3978 [PubMed] [Google Scholar]
- 54.Gustafsson A, Levitsky V, Zou JZ, Frisan T, Dalianis T, Ljungman P, Ringden O, Winiarski J, Ernberg I, Masucci MG: Epstein-Barr virus (EBV) load in bone marrow transplant recipients at risk to develop posttransplant lymphoproliferative disease: prophylactic infusion of EBV-specific cytotoxic T cells. Blood 2000, 95:807-814 [PubMed] [Google Scholar]
- 55.Lo YMD, Chan LYS, Lo KW, Leung SF, Zhang J, Chan ATC, Lee JCK, Hjelm NM, Johnson PJ, Huang DP: Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res 1999, 59:1188-1191 [PubMed] [Google Scholar]
- 56.Lo YMD, Chan LYS, Chan ATC, Leung SF, Lo KW, Zhang J, Lee JCK, Hjelm NM, Johnson PJ, Huang DP: Quantitative and temporal correlation between circulating cell-free Epstein-Barr virus DNA and tumor recurrence in nasopharyngeal carcinoma. Cancer Res 1999, 59:5452-5455 [PubMed] [Google Scholar]
- 57.Gallagher A, Armstrong AA, MacKenzie J, Shield L, Khan G, Lake A, Proctor S, Taylor P, Clements GB, Jarrett RF: Detection of Epstein-Barr virus (EBV) genomes in the serum of patients with EBV-associated Hodgkin’s disease. Int J Cancer 1999, 84:442-448 [DOI] [PubMed] [Google Scholar]
- 58.Deng H, Zeng Y, Lei Y, Zhao Z, Wang P, Li B, Pi Z, Tan B, Zheng Y, Pan W: Serological survey of nasopharyngeal carcinoma in 21 cities of south China. Chinese Med J 1995, 108:300-303 [PubMed] [Google Scholar]
- 59.Liu MY, Chang YL, Ma J, Yang HL, Hsu MM, Chen CJ, Chen JY, Yang CS: Evaluation of multiple antibodies to Epstein-Barr virus as markers for detecting patients with nasopharyngeal carcinoma. J Med Virol 1997, 52:262-269 [PubMed] [Google Scholar]
- 60.Arber DA, Weiss LM, Chang KL: Detection of Epstein-Barr virus in inflammatory pseudotumor. Semin Diagn Pathol 1998, 15:155-160 [PubMed] [Google Scholar]
- 61.Knowles DM: Molecular pathology of acquired immunodeficiency syndrome-related non-Hodgkin’s lymphoma. Semin Diagn Pathol 1997, 14:67-82 [PubMed] [Google Scholar]
- 62.Camilleri-Broet S, Davi F, Feuillard J, Seilhean D, Michiels JF, Brousset P, Epardeau B, Navratil E, Mokhtari K, Bourgeois C, Marelle L, Raphael M, Hauw JJ: LAIDS-related primary brain lymphomas: histopathologic and immunohistochemical study of 51 cases. The French Study Group for HIV-Associated Tumors. Hum Pathol 1997, 28:367-374 [DOI] [PubMed] [Google Scholar]
- 63.Camilleri-Broet S, Martin A, Moreau A, Angonin R, Henin D, Gontier MF, Rousselet MC, Caulet-Maugendre S, Cuilliere P, Lefrancq T, Mokhtari K, Morcos M, Broet P, Kujas M, Hauw JJ: Primary central nervous system lymphomas in 72 immunocompetent patients: pathologic findings and clinical correlations. Groupe Ouest Est d’etude des Leucenies et Autres Maladies du Sang (GOELAMS). Am J Clin Pathol 1998, 110:607-612 [DOI] [PubMed] [Google Scholar]
- 64.Magrath I, Jain V, Bhatia K: Epstein-Barr virus and Burkitt’s lymphoma. Semin Cancer Biol 1992, 3:285-295 [PubMed] [Google Scholar]
- 65.Hamilton-Dutoit SJ, Rea D, Raphael M, Sandvej K, Delecluse HJ, Gisselbrecht C, Marelle L, van Krieken HJ, Pallesen G: Epstein-Barr virus-latent gene expression and tumor cell phenotype in acquired immunodeficiency syndrome-related non-Hodgkin’s lymphoma: correlation of lymphoma phenotype with three distinct patterns of viral latency. Am J Pathol 1993, 143:1072-1085 [PMC free article] [PubMed] [Google Scholar]
- 66.Elenitoba-Johnson KS, Jaffe ES: Lymphoproliferative disorders associated with congenital immunodeficiencies. Semin Diagn Pathol 1997, 14:35-47 [PubMed] [Google Scholar]
- 67.Jaffe ES, Wilson WH: Lymphomatoid granulomatosis: pathogenesis, pathology and clinical implications. Cancer Surv 1997, 30:233-248 [PubMed] [Google Scholar]
- 68.Chiang AK, Tao Q, Srivastava G, Ho FC: Nasal NK- and T-cell lymphomas share the same type of Epstein-Barr virus latency as nasopharyngeal carcinoma and Hodgkin’s disease. Int J Cancer 1996, 68:285-290 [DOI] [PubMed] [Google Scholar]
- 69.Jenson HB, Leach CT, McClain KL, Joshi VV, Pollock BH, Parmley RT, Chadwick EG, Murphy SB: Benign and malignant smooth muscle tumors containing Epstein-Barr virus in children with AIDS. Leuk Lymphoma 1997, 27:303-314 [DOI] [PubMed] [Google Scholar]
- 70.von Wasielewski R, Werner M, Fischer R, Hansmann ML, Hubner K, Hasenclever D, Franklin J, Sextro M, Diehl V, Georgii A: Lymphocyte-predominant Hodgkin’s disease: an immunohistochemical analysis of 208 reviewed Hodgkin’s disease cases from the German Hodgkin Study Group. Am J Pathol 1997, 150:793-803 [PMC free article] [PubMed] [Google Scholar]
- 71.Siebert JD, Ambinder RF, Napoli VM, Quintanilla-Martinez L, Banks PM, Gulley ML: Human immunodeficiency virus-associated Hodgkin’s disease contains latent, not replicative, Epstein-Barr virus. Hum Pathol 1995, 26:1191-1195 [DOI] [PubMed] [Google Scholar]