Abstract
Although the Vienna classification has been introduced to resolve discrepancies in histological diagnoses of colorectal tumors between Western and Japanese pathologists, practical applications of this classification scheme have been problematic because invasion of the lamina propria of tumor cells is often difficult to recognize. Therefore, the following refinements of the classification criteria are needed: category 3, low-grade adenoma/dysplasia; category 4, intramucosal borderline neoplasia; 4-a, high-grade adenoma/dysplasia; 4-b, well-differentiated adenocarcinoma; category 5, definite carcinoma; 5-a, intramucosal moderately-differentiated adenocarcinoma; and 5-b, submucosal carcinoma. We attempted to test whether molecular genetic alterations are related to the modified classification scheme and whether they may help to further categorize the various intramucosal neoplasia grades of colorectal tumors. Two-hundred-thirty-two colorectal tumors were examined using flow cytometric analysis of DNA content, polymerase chain reaction microsatellite assays, and single-strand conformational polymorphism assays to detect abnormalities of DNA content, chromosomal allelic loss, and Ki-ras and p53 gene mutations. Microsatellite instability (MSI) was also examined. Frequencies of genetic alterations and DNA aneuploid states increased with an increase in the grade assigned according to the modified Vienna classification. MSI was a rare event in colorectal adenomas and their frequency of MSI did not correlate with tumor grade. The combined genetic and DNA ploidy data support the conclusion that analysis of genetic alterations and DNA aneuploid states may help in appropriate categorization of colorectal tumors according to the modified Vienna scheme. In addition, MSI-positive tumors may represent a specific subtype of colorectal adenomas.
It is well known that there have been large discrepancies in the diagnosis of colorectal neoplasia by Western and Japanese pathologists. 1 These discrepancies are caused by differences in the histological criteria applied to colorectal intramucosal tumors according to accepted Western or Japanese protocols. 1, 2 The presence of submucosal invasion is the most important diagnostic criterion for most Western pathologists, whereas in Japan, nuclear features and glandular structures are more important to the diagnosis. 2 In fact, the actual degree of histological concordance between Western and Japanese pathologists is reported to be poor. 1, 2 Schlemper et al 2 indicated that the actual rate of concordance between Western and Japanese pathologists is less than 20% (3 of 20 cases). Recently, to resolve these discrepancies, an international panel of experts in gastrointestinal pathology proposed a new classification of these lesions on the basis of common histological criteria. The lesions were divided into five categories of colorectal neoplasia (Vienna classification of gastrointestinal neoplasia). 1 In brief, category 1 and 2 lesions are negative for neoplasia/dysplasia or indefinite for neoplasia/dysplasia, respectively. Category 3 means low-grade adenoma/dysplasia, in which the risk of developing invasive carcinoma is low. Clinicians may consider local treatment of the lesion or may choose to follow up. Category 4 includes lesions of high-grade adenoma/dysplasia that are either non-invasive or that carry a suspicion of invasive carcinoma; the risk of invasion is increased in this category. Local treatment such as endoscopic mucosal resection would be indicated here. Finally, category 5 includes invasive lesions carrying the risk of subsequent deeper invasion; the rate of metastases is so high that treatment is urgently needed. Indeed, it appears to be easy for the experienced general pathologist to categorize colorectal tumors into the appropriate group. However, there are some diagnostic problems associated with this classification. In particular, judgment of invasion beyond the basement membrane is very difficult to define. 1 Therefore, in many cases, neither Western nor Japanese pathologists were able to distinguish between high-grade adenoma/dysplasias versus most intramucosal carcinomas in a reproducible manner. 1, 2 These results suggested that further refinement of the Vienna classification scheme was needed, and particular emphasis was placed on a better definition of the somewhat problematic category 4 and 5 lesions. In addition, Western and Japanese pathologists may not be in agreement even with the diagnosis of low-grade adenoma versus high-grade adenoma. When a pathologist differentiates a tumor from a low- or a high- grade adenoma, the criteria used to differentiate these adenomas should be derived from objective data, such as genetic and DNA ploidy data.
Colorectal tumors are thought to follow a model of multi-step tumorigenesis, in which abnormalities in dominant oncogenes and tumor suppressor genes have been identified as frequent events. 3, 4 Although many genetic alterations in colorectal tumors have been studied, 3, 4, 5 mutations of APC (adenomatous polyposis coli), p53 and Ki-ras genes, as well as allelic losses of chromosome 5q, 17p, 18q, 1p, 8p, or 22q, are most frequently associated with the colorectal tumorigenesis. 3, 4, 5, 6 On the other hand, recent molecular analysis has shown that there are two distinct genetic pathways in colorectal tumorigenesis that are characterized by genetic instability: chromosomal instability and microsatellite instability (MSI), the latter of which represents mutations in the short tandem repeat sequences distributed within the genome. 7 Chromosomal instability is frequently found in association with a loss of heterozygosity (LOH) of cancer-related chromosomal loci, or with p53 and Ki-ras gene mutations. These changes characterize the chromosomal instability (CIN) or LOH genotype, which is often associated with DNA aneuploidy. 7, 8, 9 Aneuploidy is well known as an indicator used to predict the malignant potential of colorectal adenomas. Most sporadic colorectal carcinomas arising in Western countries are believed to arise from adenoma 10 and belong genetically to the LOH genotype. 9, 11 On the other hand, a few sporadic colorectal adenomas that occur with MSI do occur. Although MSI characterizes one of the two major genetic pathways, its role during development of colorectal tumors is not fully understood. Genetic data on lesions classified according to the various modified Vienna categories could conceivably provide the necessary means to further refine the histopathological Vienna classification criteria, and may also be useful for early detection of high-risk lesions. We attempted to identify such criteria through an extensive analysis of allelic loss of cancer-related chromosomal loci, mutations of p53 and Ki-ras genes, DNA ploidy, and MSI. An effective crypt isolation method was used to obtain pure tumor crypts. 9, 12, 13 The aim of the present study is to clarify the usefulness of this modified classification scheme that is based on an objective molecular analysis.
Materials and Methods
In the present study, category 1 and 2 cases were excluded, given the facts that category 1 cases are thought to involve no genetic alterations, and category 2 lesions are rarely encountered during routine pathological examination. The objective of this study is to improve classification of lesions in categories 3 to 5. We excluded colorectal tumors that occurred in patients with a family history of colorectal tumors.
Patients
All colorectal tumors are composed of a mixture of neoplastic and non-neoplastic cells. To analyze such tumors for specific genetic alterations accurately, the neoplastic glands must be separated from most of the non-neoplastic cells. 12, 13 Genetic alterations of tumor cells were analyzed after using a crypt isolation technique. This technique allowed for an accurate genetic analysis because pure tumor crypts were thus obtained.
A total of 155 colorectal adenomas (from 102 men and 43 women ranging in age from 39 to 85 years; mean, 62.7) were obtained from Iwate Medical University and related hospitals (low-grade adenoma, n = 118; high-grade adenoma, n = 37). Thirty-two early carcinomas (characterized by invasion of malignant cells that were limited to the mucosa or submucosa) were also examined to investigate relationships between genetic alterations and high- grade dysplastic adenomas and early carcinomas. Furthermore, two groups of early carcinomas were sub-classified according to their depth of invasion, as follows: intramucosal (n = 22), and submucosal carcinoma (n = 10). The term “intramucosal” carcinoma was defined as a cancer limited to the lamina propria. The patients consisted of 22 men and 10 women (age 41 to 88 years; mean, 62.1). Histological diagnoses of the adenomas and intramucosal carcinomas were performed according to our hospital criteria, which were modified from those of the Japanese Research Society for cancer of the colon and rectum. 14 In brief, low-grade dysplasia was diagnosed by the presence of cells with enlarged, hyperchromatic nuclei that were largely confined to the basal portions of the cells, but that had some loss of polarity. Lesions containing cells with round-to-ovoid crypts and an obvious loss of polarity, but no architectural or glandular distortion, were classified as high-grade dysplastic adenomas. The intramucosal carcinomas were subdivided into two groups: well-differentiated and moderately-differentiated. A well-differentiated intramucosal carcinoma was defined by an obvious derangement of glandular architecture, with irregular branches of glands or “back to back” formations. Moderately-differentiated adenocarcinomas were defined by “gland in gland” or cribriform patterns of glands. Differentiation of the former from the latter is histologically easy for general pathologists. In the present study, moderately-differentiated adenocarcinomas often showed a higher cellular atypia than was seen in the intramucosal well-differentiated intramucosal carcinomas. However, it was often difficult to accurately distinguish the high dysplastic adenomas from the well-differentiated intramucosal carcinomas. A submucosal carcinoma was defined as a cancer characterized by submucosal invasion. In addition, an advanced carcinoma was defined as a cancer that has invaded beyond submucosa. Forty-five advanced colorectal carcinomas were analyzed as a positive control. The tumors were classified according to the Japanese Research Society for cancer of the colon and rectum 14 and modified Dukes’ stage. 15 Histological features of adenomas and carcinomas are illustrated in Figure 1 . Clinicopathological features of colorectal adenomas and carcinomas are listed in Table 1 . Each tumor was classified into either category of modified Vienna classification by two experienced pathologists (S. T. and N. S.) according to our histological criteria. The Vienna and modified Vienna classification schemes are shown in Table 2 .
Figure 1.
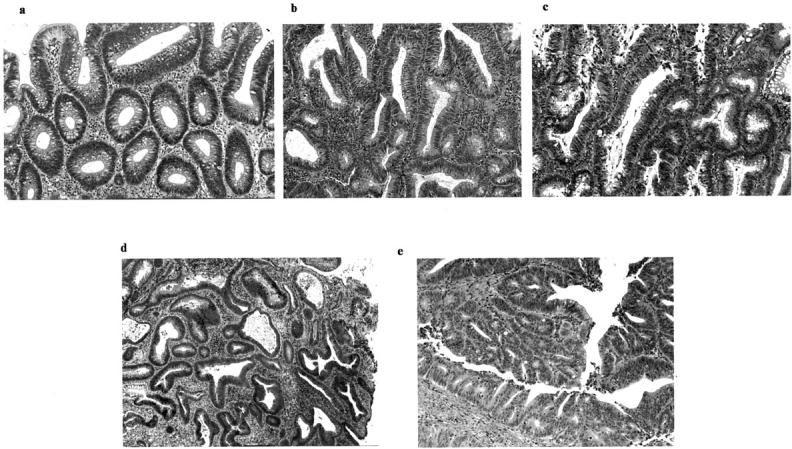
a and b: Low-grade dysplastic adenoma (category 3). 1-b was treated as moderate dysplasia according to the previous classification scheme. c: High-grade dysplastic adenoma showing irregular glands and hyperchromatic nuclei (category 4-a). d: Well-differentiated adenocarcinoma showing severe irregular branching of glands and hyperchromatic nuclei (category 4-b). e: Moderately-differentiated adenocarcinoma (category 5-a) showing higher cellular and structural atypia (cribriform pattern) than the well-differentiated adenocarcinoma.
Table 1.
Clinicopathological Features of Colorectal Neoplasms
| Adenoma | Early cancer | Advanced cancer | |
|---|---|---|---|
| Total | 155 | 32 | 45 |
| Age (mean) | 62.7 | 62.1 | 65.3 |
| Gender (man/woman) | 102/43 | 22/10 | 33/12 |
| Location of tumor (right/left) | 51/104 | 7/25 | 13/32 |
| Size of tumor (mean, mm) | 5–28 (13.2) | 9–37 (18.4) | 20–87 (48.5) |
| Grade of dysplasia | |||
| Low dysplastic | 118 | ||
| High dysplastic | 54 | ||
| Differentiation | |||
| Well | 20 | 15 | |
| Moderately | 12 | 30 |
Table 2.
Vienna or Modified Vienna Classification of Gastrointestinal Neoplasia
| Vienna classification | Modified Vienna classification | |
|---|---|---|
| Category 1 | Negative for neoplasia/dysplasia | Negative for neoplasia/dysplasia |
| Category 2 | Indefinite for neoplasia/dysplasia | Indefinite for neoplasia/dysplasia |
| Category 3 | Non-invasive low grade neoplasia (low grade adenoma/dysplasia) | Low grade neoplasia (low grade adenoma/dysplasia) |
| Category 4 | Non-invasive high grade neoplasia | Intramucosal borderline neoplasia |
| 4.1 High grade adenoma/dysplasia | 4.1 High grade adenoma/dysplasia | |
| 4.2 Non-invasive carcinoma (carcinoma in situ) | 4.2 Intramucosal carcinoma, well differentiated | |
| 4.3 Suspicion of invasive carcinoma | ||
| Category 5 | Invasive neoplasia | Definite carcinoma |
| 5.1 Intramucosal carcinoma | 5.1 Intramucosal carcinoma, moderately or poorly differentiated | |
| 5.2 Submucosal carcinoma or beyond | 5.2 Submucosal carcinoma or beyond |
Crypt Isolation Method and DNA Extraction
Tumor DNAs from adenomatous and cancer tissues were extracted from isolated adenomatous and cancer crypts. The tumor crypts were isolated from mucosa propria using a crypt isolation technique, as described elsewhere. 16, 17, 18 Briefly, fresh tumor samples and normal colonic mucosa were minced into small pieces using a razor, then incubated at 37°C for 50 minutes in Ca2+/Mg2+-free Hanks’ balanced salt solution containing 30 mmol/L EDTA. The tissue was stirred in the salt solution for 20 minutes × 2. Crypts were separated from the lamina propria mucosa or fibrous stroma. These isolated tumor crypts were identified using a dissecting microscope (SZ60, Olympus, Tokyo, Japan). Normal crypts were clearly distinguishable from tumor crypts by characteristic features as described elsewhere. 13 A proportion of the isolated crypts were fixed in 70% ethanol and stored at 4°C until they were used for DNA extraction. The remaining isolated crypts were fixed with paraffin and examined by light microscopy to confirm the histopathological findings. There was no evidence of contamination by interstitial cells using this technique. However, it should be appreciated that this method cannot reliably differentiate between the crypts of adenomas with high-grade dysplasia and those with low-grade dysplasia. Accordingly, we confirmed the histological diagnosis by light microscopy of paraffin-embedded sections of these crypts. In general, colorectal adenomas tend to have a heterogeneous appearance, and in such cases the tumor grade was determined by the ratio of high-grade to low-grade dysplasia.
DNAs from tumors and from some corresponding normal crypts were extracted by standard SDS proteinase K treatment. Samples were resuspended in TE buffer [10 mmol/L Tris-HCl, 1 mmol/L EDTA (pH 8.0)] to the equivalent of 1000 cells/μl. In adenomatous lesions and most early carcinomas, normal genomic DNA, obtained from blood samples from the corresponding patients, was isolated using DrGTL (TAKARA, Tokyo, Japan).
Analysis of DNA Ploidy Pattern
A portion of each sample was prepared for flow cytometric analysis (flow cytometer; EPICS XL, Coulter, CA) as previously described. 9, 19 A total of 50 to 300 tumor crypts were isolated and examined in the present study. Briefly, normal and tumor crypts were incubated with 0.0125% pepsin (pH 2.0, Sigma, St. Louis, MO) for 5 minutes at 37°C, then washed twice with 0.2 mol/L Tris chloride-buffered saline and passed through a 27-gauge needle. The nuclei (from normal and tumor cells) were stained with propidium iodide (50 μg/ml, Sigma) containing ribonuclease (0.25 mg/ml, Sigma) in 0.2 mol/L Tris chloride-buffered saline at room temperature in the dark for 30 minutes. After filtration through a 37-μm nylon mesh (Tokyo Screen, Tokyo, Japan), samples were analyzed using a flow cytometer. At least 10,000 nuclei from each sample were examined. These samples were classified as diploid (D), multiploid (M), or aneuploid (A) carcinomas as determined by previous criteria. 9, 13, 19 Diploid and aneuploid carcinomas were defined as either a diploid tumor population alone or an aneuploid tumor population alone within a tumor. Multiploid carcinomas consisted of both diploid and aneuploid tumor populations. The DNA index (DI) was also calculated as defined previously. 9, 19 The DNA ploidy pattern of each of these samples was reconfirmed using cells remaining after extraction of the DNA used for genetic analysis.
Analysis of Allelic Imbalances at Chromosomal Loci
Normal and tumor DNA were analyzed for allelic loss by polymerase chain reaction (PCR) amplification of polymorphic dinucleotide repeat sequences. Seventeen markers on chromosomes 17p (TP53), 5q (D5S107; D5S346; D5S82; D5S299), 18q (D18S487; D18S34; DCC), 1p (D1S228; D1S548; D1S507), 8q (D8S201; D8S513; D8S532) and 22q (D22S274; D22S1140; D22S1168) were used. The sequences of these primers were obtained from “The Genome Database” (http://gdbwww.gdb.org/gdb/). PCR was carried out using a DNA autosequencer (Applied Biosystems 373A sequencer; Applied Biosystems, CA), as previously described. 9, 16, 17, 18, 19 The data from the PCR analysis were collected automatically and analyzed by GeneScan software (Applied Biosystems) for allele scoring and assessment of allelic loss as described previously. 16, 17, 18, 19 In this study, we define allelic imbalance as more than a 50% change in the ratio (q value). Samples were regarded as uninformative if they showed constitutional homozygosity or microsatellite instability.
Analysis of Microsatellite Instability
The three dinucleotide markers (D2S123, D3S1611, and D17S250), and three adenine mononucleotide repeats (BAT 25, BAT 26, and TGFβRII), were used for determination of MSI. MSI-positive status of a tumor was defined as previously described. 20 MSI-positive colorectal carcinomas in the present study were divided into two groups, those with high-level instability (ie, MSI at ≧ 33% of loci) and those with low-level instability (ie, MSI at <17% of loci). However, tumors that showed only one alteration of the three markers and were categorized as MSI-low were considered MSI-negative tumors in this study.
Polymerase Chain Reaction-Single-Strand Conformation Polymorphism Analysis and Sequencing
Single-strand conformational polymorphism (SSCP) analysis was used to screen PCR products derived from exons 5 to 8 of the p53 gene and from exon 1 of the Ki-ras gene in patient tumor and normal mucosal DNA samples. PCR conditions, PCR-SSCP, and sequencing of the two gene mutations were performed according to previously described methods. 16, 18 Direct sequencing was performed using fluorescent-labeled dideoxynucleotide triphosphates for automated DNA sequence analysis (Applied Biosystems 373A sequencer).
Statistical Analysis
The data were analyzed using the χ2 test with the aid of StatView-IV software (Abacus Concepts, Berkeley, CA). Samples were determined to be significantly different at P < 0.05.
Results
The adenomas were subdivided into two groups according to the modified Vienna classification criteria. Each of the 232 tumor samples was precisely diagnosed histopathologically, and was classified into one of four groups: (1) category 3 tumors (low-grade dysplastic adenoma, 118 tumors); (2) category 4 tumors, 4-a (high-grade dysplastic adenoma, 37 tumors), 4-b (well-differentiated intramucosal adenocarcinoma, 17 tumors); (3) category 5 tumors, 5-a (moderately-differentiated intramucosal adenocarcinoma, 5 tumors), 5-b (submucosal carcinoma, 10 tumors); and (4) advanced carcinomas, 45 tumors.
Analysis of DNA Ploidy Pattern
The frequency of DNA multiploidy was 6.8% (8 of 118) in category 3 tumors but was significantly elevated in both category 4 and category 5 tumors, at 40.7% (22 of 54) and 73.3% (11 of 15), respectively (P < 0.001, Table 3 * and †). The frequency of DNA multiploidy in colorectal adenomas was significantly correlated with the tumor grade (category 3 and category 4-a, P < 0.01, Table 3 ¶). In addition, a significant difference in the frequency of DNA multiploidy between category 4 (40.7%) and category 5 tumors (73.3%) was found (P < 0.05) (Table 3 ∥). However, there was no significant difference in the DNA multiploidy frequency between category 4-a (37.8%, high-grade adenomas) and category 4-b (47.1%, intramucosal carcinoma, well-differentiated). There was a significant difference in the frequency of DNA multiploidy between advanced carcinomas (82.2%) and category 3 tumors (6.8%, P < 0.001) or category 4 tumors (40.7%, P < 0.001) (Table 3 ‡ and §). Thirty-seven of 45 advanced carcinomas exhibited a multiploid or aneuploid state (30 and 7 cases, respectively).
Table 3.
Frequency of DNA Aneuploidy in Colorectal Adenomas and Carcinomas Based on a Modified Vienna Classification
| Diploidy (%) | Multiploidy/ Aneuploidy (%) | Total | |
|---|---|---|---|
| Category 3 | 110 (93.2) | 8 (6.8)*†‡§ | 118 |
| Category 4 | 32 (59.3) | 22 (40.7)*§∥ | 54 |
| Category 4-a | 23 (63.2) | 14 (37.8)‡ | 37 |
| Category 4-b | 9 (52.9) | 8 (47.1) | 17 |
| Category 5 | 4 (26.7) | 11 (73.3)†¶ | 15 |
| Category 5-a | 1 (20) | 4 (80) | 5 |
| Category 5-b | 3 (30) | 7 (70) | 10 |
| Advanced cancer | 8 (21.8) | 37 (82.2)§∥ | 45 |
Lower case super-scripted symbols indicate a statistical difference between the two values. For example, * represents a difference in the frequency of DNA multiploidy/aneuploidy between category 3 and category 4 tumors. * †,‡,§ and ∥, p < 0.01; ¶, p < 0.05; ∥.
Analysis of Genetic Alterations in Colorectal Adenomas and Carcinomas
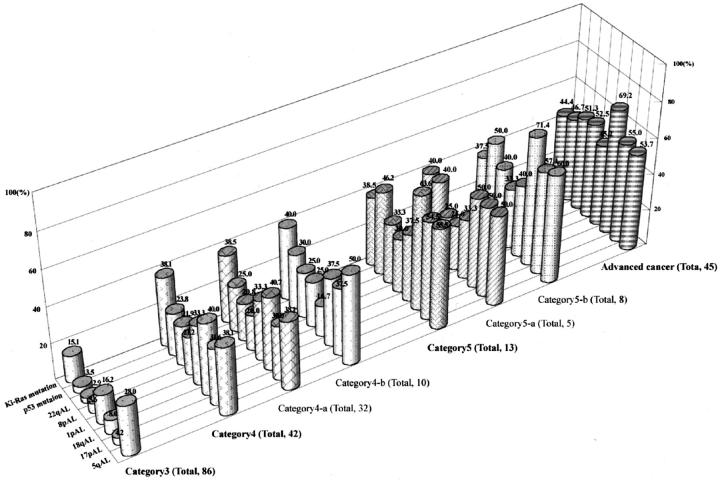
Of 187 colorectal adenomas and early carcinomas, 118 adenomas and 23 early carcinomas were available for genetic analysis. The detailed data on genetic alterations is shown in Figure 2 . The frequency of allelic loss in low-grade dysplastic adenomas (category 3) was less than 10% [17p, 3/71/86 (allelic loss/informative cases/total cases); 18q, 6/75/86; 8p, 4/71/86; 22q, 2/69/86], with the exception of 5q and 1p allelic loss (21/75/86; 11/68/86). In high-grade dysplastic adenomas (category 4-a), the frequency of allelic loss on chromosomal loci lay between 20% and 40% (5q, 8/21/32; 17p, 9/30/32; 18q, 11/27/32; 1p, 7/21/32; 8p, 5/25/32; 22q, 5/24/32). Although differences in frequencies of 5q and 1p allelic loss between low-grade adenomas and high-grade adenomas did not reach a statistically significant level, significant differences in 17p, 18q, 8p, and 22q allelic loss as well as in p53 and Ki-ras genes mutation rates were found between low- and high-grade dysplastic adenomas (8p and 22q allelic loss, P < 0.05; 17p, 18q allelic loss, and p53 and Ki-ras gene mutation, P < 0.01). In category 4 (high-grade adenoma and intramucosal carcinoma, well-differentiated) and category 5 (intramucosal carcinoma, moderately-differentiated and submucosal carcinoma), the frequencies of genetic alterations were higher in category 5 tumors than in category 4 tumors, although these differences were not statistically significant.
Figure 2.
Frequencies of genetic alterations in colorectal adenomas and carcinomas. Frequencies of 17p, 18q, 8p, and 22q allelic losses and mutations of p53 and Ki-ras genes in category 4 tumors are significantly higher than are those of category 3 tumors. The frequencies denoted at the top of the column indicate those frequencies (%) showing allelic loss or mutations.
Genetic alterations in advanced carcinomas were examined as a positive control. Numerous genetic alterations were found in the carcinomas, as expected. However, there were no differences in the genetic frequency between category 5 tumors and advanced carcinomas. In particular, no differences in genetic frequency were detected in category 5-b (submucosal carcinoma) and advanced carcinomas. In more advanced carcinomas, allelic losses on chromosomes 8q and 22q tended to be associated with a late stage of the progression of colon carcinomas. A representative example is shown in Figure 3 .
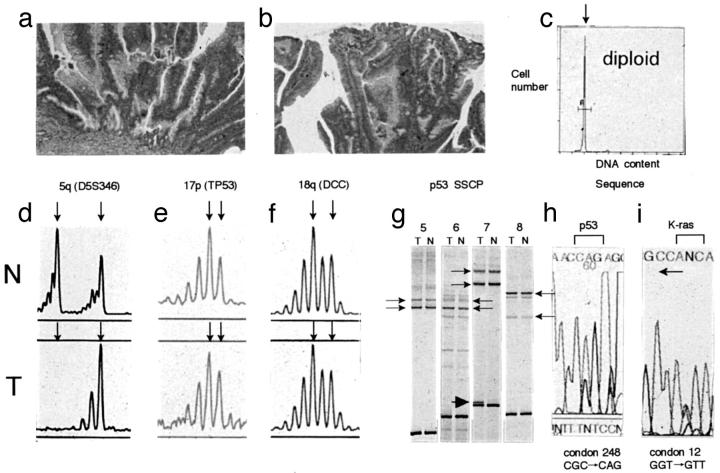
Figure 3.
Analysis of allelic losses of chromosomal loci on chromosomes 17p, 5q, and 18q and p53 and Ki-ras gene mutation in high-grade dysplastic adenomas (category 4-a). a and b: Histological features of high-grade dysplastic adenoma. c: DNA histogram indicates a diploid cell. d–f: Electrophoregram of allelic losses at 5q, 17p, and 18q chromosomal loci. Arrows show the two alleles for each chromosomal locus. 5q allelic losses were found in this adenoma. In addition, 1p, 8p, and 22q allelic losses were not observed (electrophoregram not shown). g: A SSCP analysis of the p53 gene (exons 5–8). Arrows show the two normal alleles for each exon. An anomalously migrating band is seen corresponding to exon 7 (arrowheads). h and i: DNA sequence showing p53 and Ki-ras gene mutations. A CGC to CAG transition in codon 248 was found in h. In i, a GGT to GTT transversion occurred in codon 12 (sequenced by reverse primer). N, normal; T, tumor.
Accumulation of Genetic Alterations in Colorectal Tumors
The cumulative number of genetic changes, including allelic losses on chromosomes 5, 8, 17, 18, and 22, and Ki-ras and p53 mutations, increased with an increase in the histological grade of sporadic colorectal tumors (Table 4) . Whereas either none or only a single genetic alteration was found in 76 of 82 category 3 tumors (88.4%), genetic alterations were detected in 24 of 42 category 4 tumors (57.1%), 3 of 13 category 5 tumors (23.1%) and 5 of 45 advanced carcinomas (11.1%), respectively. The majority of submucosal and advanced carcinomas had multiple genetic alterations (data not shown). In addition, there were no differences of the cumulative number of genetic changes between categories 4-a and 4-b, and 5-a and 5-b, respectively (data not shown).
Table 4.
Accumulations of Genetic Alterations in Colorectal Tumors Based on a Modified Vienna Classification
| Accumulation | Category 3 (%) | Category 4 (%) | Category 5 (%) | Advanced cancer (%) |
|---|---|---|---|---|
| Total | 86 | 42 | 15 | 45 |
| 0 | 44 (51.2) | 9 (21.4) | 1 (6.7) | 2 (4.4) |
| 1 | 32 (37.2) | 15 (35.7) | 2 (13.3) | 3 (6.7) |
| 2 | 7 (8.1) | 10 (23.8) | 5 (33.3) | 4 (8.9) |
| 3 | 3 (3.5) | 5 (11.9) | 4 (26.7) | 6 (13.3) |
| 4 | 0 | 2 (4.8) | 2 (13.3) | 14 (31.1) |
| 5 | 0 | 0 | 1 (6.7) | 8 (17.7) |
| 6 | 0 | 0 | 0 | 5 (11.1) |
| 7 | 0 | 0 | 0 | 3 (6.7) |
| 8 | 0 | 0 | 0 | 0 |
Genetic alterations examined: allelic loss, 17p; 5q; 18q; 1p; 8p; 22p; mutation, p53; Ki-ras.
Analysis of Microsatellite Instability in Colorectal Adenomas and Carcinomas
The summary of MSI analysis is shown in Table 5 . High levels of MSI were observed in 3 of 118 adenomas (2.5%, category 3 and 4-a tumors), whereas MSI was not detected in early colorectal carcinomas. Of the three high MSI adenomas, all were low-grade. In addition, whereas the two adenomas with high MSI were found in a right-sided colon (ascending), the other adenoma was observed in a left-sided colon (sigmoid). On the other hand, low levels of MSI were detected in two low-grade adenomas and in one high-grade adenoma. In advanced carcinomas, high levels of MSI were observed in four of 45 tumors. There were seven MSI-H tumors (3 adenomas and 4 carcinomas) and all demonstrated instability at the mononucleotide markers (BAT 25 and BAT 26). In addition, the seven MSI-H tumors were diploid, whereas none of the MSI was detected in aneuploid or multiploid tumors. Five of the seven MSI-positive tumors had alterations within the RII gene. Of these five tumors, four contained a one-base deletion and one contained a two-base deletion. Allelic losses and mutations of Ki-ras and p53 genes were inversely related to MSI. None of the colorectal tumors that demonstrated MSI had allelic losses, with the exception of one low-grade adenoma, which showed multiple genetic alterations (17p, 18q, and 8p allelic losses, and p53 gene mutation).
Table 5.
Frequency of Microsatellite Instability in Colorectal Tumors Based on a Modified Vienna Classification
| MSI-positive | MSI-negative | Total | |
|---|---|---|---|
| Category 3 | 3 (3.5) | 83 (96.5) | 86 |
| Category 4 | 0 | 42 (100) | 42 |
| Category 5 | 0 | 15 (100) | 15 |
| Advanced cancer | 4 (8.9) | 37 (91.1) | 45 |
MSI, microsatellite instability.
Discussion
During the World Congresses of Gastroeneterology held in 1998, a new standardized nomenclature and classification for gastrointestinal neoplasia was introduced. 1 In this classification scheme, intramucosal neoplasia is categorized according to defined histopathological criteria. When tested among expert gastrointestinal pathologists, however, the reproducibility of the intramucosal classifications was found to be poor for category 4 and 5 lesions (and even among category 3 tumors), indicating that additional criteria were needed. This is due to the fact that invasion of the lamina propria is not always easy to recognize, for either Western or Japanese pathologists, resulting in poor agreement among the histological diagnoses. 1 The term “intramucosal carcinoma” is not synonymous with “carcinoma in situ,” but rather reflects an invasive growth that penetrates the lamina propria and does not break through the muscularis mucosae. 21 If Vienna classification were applied to colorectal tumors without refinement, determination of invasion into the lamina propria would inevitably be necessary to classify them. However, as mentioned above, invasion into the lamina propria is very difficult to define. The modified Vienna classification of colorectal neoplasia makes it easy to classify a given tumor, given that for both Western and Japanese pathologists, to recognize a morphological difference between well- and moderately-differentiated carcinomas (mainly according to cribriform pattern) is easy. Therefore, both Western and Japanese pathologists could come to a common consensus of colorectal tumors classification by use of the modified classification scheme. Hypothesizing that genetic alterations in various colorectal intramucosal neoplasia could provide this criteria, we analyzed allelic losses at chromosomes 5q, 17p, 18q, 1p, 8p, and 22q, and mutations of Ki-ras and p53 genes in a series of colorectal intramucosal neoplasias. In addition, the DNA ploidy patterns were examined to assess biological behavior associated with intramucosal neoplasia.
In the present study, we found progressive increases in allelic losses at all chromosomal loci as well as mutations of Ki-ras and p53 genes in colorectal adenomas from low- (category 3) to high-grade (category 4), with the exception of 1p and 5q allelic losses. Also, the DNA multiploidy frequency was higher in category 4 than in category 3 tumors. Furthermore, the accumulation of genetic alterations was more pronounced in category 4 tumors than in category 3 tumors. These data are consistent with earlier molecular studies showing that the successive accumulation of genetic changes paralleled the severity of dysplasia of adenoma cells. 3, 4, 5 These findings suggest that our histological judgment as to the various dysplastic grades of colorectal adenomas is accurate for differentiating low-grade (category 3) from high-grade adenomas (category 4). Our data also support the previously suggested precursor nature of the adenomatous lesions, and the proposed tumor progression model for colorectal tumors. 3, 4, 5
In the present study, there was no statistical difference in the frequency of genetic alterations found in category 4-a (high-grade adenoma/dysplasia) and category 4-b (well-differentiated intramucosal adenocarcinoma). This finding may indicate that there are no fundamental genetic differences among them. The genetic alteration frequencies reported by Kikuchi-Yanoshita and colleagues, 22 however, are discordant with ours, in that they found higher rates of allelic loss at chromosome 17p and of p53 gene mutations in intramucosal carcinomas than in severe dysplastic adenomas. Although the reasons for these conflicting pieces of evidence remain unclear, the apparent discrepancy may be due to differences in the criteria for histological diagnosis of these lesions. Conversely, this leads to the necessity of establishing a histological classification scheme for colorectal neoplasia that is based on a common consensus. Although we should recognize the existence of intramucosal carcinoma of the colorectum, over-diagnosis of “borderline neoplastic lesions” should be avoided. It is obvious that most adenomas do not become malignant, because the cumulative incidence rate of adenomas at 3 years post-diagnosis is 16%, and the lifetime risk of colorectal carcinoma is estimated to be about 5%. 23
In the present study, we showed that the 17p allelic loss and Ki-ras gene mutations are the most informative and effective genetic markers used to differentiate category 3 tumors from category 4 tumors. Although p53 gene mutations may also be useful for this purpose, 24 multiple PCR analyses are necessary to determine the specific mutation for each patient. On a practical level, multiple PCR-SSCPs represent a technically demanding technique. 25 Carrying out multiple PCR amplifications of the individual exons followed by electrophoretic analyses and sequence determinations is a lengthy and expensive process. 26 Therefore, the other genetic markers used to analyze allelic losses at each chromosomal locus may involve less complex methods to examine the malignant potential of tumor cells. For example, three different markers were necessary to determine an allelic loss at chromosome 18q in the present study, resulting in multiple PCR assays. In contrast, 17p allelic loss, which carries a high informative content, can be examined by a single PCR-LOH assay. 17p allelic loss, which is often associated with p53 gene mutations, is considered to be a late event in the malignant transformation of colorectal adenoma to cancer. 3, 4, 5 Thus, its detection is thought to provide useful information for the clinical management of colorectal neoplasms. Also, Ki-ras gene mutations can be analyzed by performing single PCR-SSCP and direct sequencing. However, there are some problems in assessing the malignant potential of adenomatous cells by Ki-ras gene mutations. Previous studies have shown that Ki-ras gene mutations are associated with various factors, such as large tumor size, a high grade of dysplasia, and old age. 3, 4, 5, 26, 27, 28 These findings do not necessarily imply that occurrence of a Ki-ras gene mutation predicts the malignant propensity of colorectal adenomas. However, a recent study has indicated that Ki-ras gene mutations are associated with the histological features of adenoma progression (high-grade dysplasia) rather than with adenoma growth. 28 Ki-ras gene mutations may, in part, drive the histological progression of adenomas toward high grades of dysplasia. 28 Although it is difficult to classify a given tumor into a particular category by a single genetic marker alone, we believe that 17p allelic loss and Ki-ras gene mutations are simple, useful, and reliable markers in assessing the malignant potential of colorectal adenomas.
In the present study, the absolute frequencies of genetic alterations in category 5 tumors were somewhat higher than those in category 4 tumors, but these differences did not reach statistical significance, possibly due to the small number of the cases tested. On the other hand, the frequency of DNA multiploidy between category 4 and category 5 tumors increased significantly (P < 0.05). This finding may also support the validity of the modified classification scheme. In addition, the cumulative number of genetic changes, including allelic loss of 5q, 17p, 18q, 1p, 8p, and 22q, and Ki-ras and p53 gene mutations, increased along with increases in the histological grades according to the modified Vienna classification criteria. The great majority of advanced carcinomas had multiple associated genetic changes. Our current results indicate that whereas approximately 50% of category 5 tumors showed three or more genetic changes, 80% of category 4 tumors revealed two or fewer genetic changes. These results suggest that accumulations of cancer-related gene mutations provide a basis for the histological diagnosis of colorectal tumors. Although there is a limit to the accuracy of a single genetic marker in separating a given tumor into a particular category, the assessment of a cumulative collection of genetic alterations in tumor cells may be more useful. The utility of this type of genetic analysis is strongly suggested by the hypothesis that colonic carcinogenesis is driven by sequential rounds of mutation followed by clonal expansion. Thus, the ultimate histological and biological features of the adenoma are the result of accumulation of a series of allelic losses or mutations. 3, 4, 5
MSI was a rare finding in sporadic colorectal adenomas, compared with sporadic colorectal carcinomas. Recently, two types of genetic instabilities have been proposed: chromosomal instability and microsatellite instability. 7, 8 Although most colorectal tumors show a CIN genotype, sporadic MSI-positive colorectal carcinomas seem to have some distinguishing characteristics: a preferred localization on the right side of the colon, a high frequency of mucinous and poorly-differentiated histology, low rates of loss of heterozygosity, a low incidence of alterations in p53 and Ki-ras genes, and a high frequency of diploidy. 9, 29, 30 In the present study, two of three high MSI-positive adenomas were found on the right side of the colon. In addition, two of three adenomas with high MSI-positivity showed infrequent LOH, no mutations of Ki-ras and p53 genes, and a DNA diploid state. This genetic profile of sporadic colorectal MSI-positive adenomas resembles that of sporadic colorectal MSI-positive carcinomas. 9 It is interesting to note that one adenoma with high MSI had multiple genetic alterations. This finding contrasts with the genetic characteristics of sporadic MSI-positive colorectal carcinomas, which show low rates of LOH, and a low frequency of Ki-ras or p53 gene mutations. Although a tumor having a genetic profile like this adenoma is not a precursor of sporadic carcinomas, our results suggest the possibility that at least some MSI-positive adenomas may predict future colon cancers. It is widely accepted that carcinomas develop primarily from adenomas through several transitional stages in their carcinogenesis (the so-called LOH pathway, or CIN pathway). 3, 4, 5, 7, 8 The present study indicated that allelic loss at each chromosomal locus and mutations of Ki-ras and p53 genes occur gradually along with an increase in histological grade, suggesting the LOH pathway for the carcinogenesis. These findings imply that a tumor classified according to the modified Vienna scheme will show a LOH genetic type. Our results showed that the frequency of MSI in sporadic colorectal adenomas did not correlate with tumor grade. This finding suggests that MSI-positive adenomas represent a specific subtype of sporadic colorectal adenomas. Furthermore, we propose that such MSI-positive tumors should not be included in a modified Vienna classification scheme, given that this classification is based on the hypothesis that genetic alterations are correlated with histological tumor grade.
Recently, pathologists have referred to serrated adenoma, which is described as a rare entity among colorectal tumors. 31, 32 This rare neoplasm shares the architectural features of a hyperplastic polyp, but it shows distinct cytological atypia. 31 In addition, frequent Ki-ras mutations, MSI, and 1p allelic loss have been observed in serrated adenoma. 31, 33 Therefore, serrated adenoma has been proposed to be a distinct entity in colorectal neoplasia. It is characterized by a new histogenetic pathway to produce hyperplastic aberrant crypt foci, hyperplastic polyps, mixed polyps, and a serrated appearance. 31 On the other hand, there are contrasting data showing no significant differences in genetic alterations between serrated adenoma and conventional adenoma. 34 It therefore may be unlikely that serrated adenoma represents a truly distinct entity in colorectal neoplasias. In the present study, we do not include serrated adenoma as a separate classification. Whether a given tumor is included in the modified classification according to specific genetic alterations is of interest to the pathologist. Further investigations regarding serrated adenoma would be recommended in the near future to investigate its relationship to the modified Vienna classification scheme.
In conclusion, we have demonstrated that allelic losses at multiple chromosomal loci and mutations of Ki-ras and p53 genes are correlated with a proposed modified Vienna classification scheme. In addition, the cumulative number of genetic changes provides useful information to predict the potential propensity of colorectal tumors to transform in progressive steps. We conclude that this modified classification scheme for colorectal neoplasia, which is based on genetic data, will help to create a common consensus between Western and Japanese pathologists.
Address reprint requests to Tamotsu Sugai, M.D., Division of Pathology, Central Clinical Laboratory, Iwate Medical University, 19–1 Morioka, 020-8505, Japan. E-mail: tsugai@cocoa.ocn.ne.jp.
References
- 1.Schlemper RJ, Riddell RH, Kato Y, Borchard F, Cooper HS, Dawsey SM, Dixon MF, Fenoglio-Preoser CM, Flejou J-F, Geboes K, Hattori T, Hirota T, Itabashi M, Iwafuchi M, Iwashita A, Kim YI, Kirchner T, Klimpfinger M, Koike M, Lauwers GY, Lewin KJ, Oberhuber G, Offner G, Price AB, Ribio CA, Shimizu M, Shimoda T, Sipponen P, Solcia E, Stolte M, Watanabe H, Yamabe H: The Vienna classification of gastrointestinal epithelial neoplasia. Gut 2000, 47:251-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlemper RJ, Itabashi M, Kato Y, Lewin KJ, Riddell RH, Shimoda T, Sipponen P, Stolte M, Watanabe H: Differences in the diagnostic criteria used by Japanese and Western pathologists to diagnose colorectal carcinoma. Cancer 1998, 82:60-69 [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AMM, Bos JL: Genetic alterations during colorectal tumor development. N Engl J Med 1988, 319:525-532 [DOI] [PubMed] [Google Scholar]
- 4.Fearon ER, Vogelstein B: A genetic model for colorectal tumorigenesis. Cell 1990, 61:759-767 [DOI] [PubMed] [Google Scholar]
- 5.Miyaki M, Seki M, Okamoto M, Yamanaka A, Maeda Y, Tanaka K, Kikuchi R, Iwama T, Ikeuchi T, Tonomura A, Nakamura Y, White R, Miki Y, Utsunomiya J, Koike M: Genetic changes and histolopathological types in colorectal tumors from patients with familial adenomatous polyposis. Cancer Res 1990, 50:7166-7173 [PubMed] [Google Scholar]
- 6.Laurent-Puig, Blons H, Cugnenc P-H: Sequence of molecular genetic events in colorectal tumorigenesis. Eur J Cancer Prev 1999, 8:539-547 [PubMed] [Google Scholar]
- 7.Lengauer C, Kinzler KW, Vogelstein B: Genetic instabilities in human cancers. Nature 1998, 396:643-649 [DOI] [PubMed] [Google Scholar]
- 8.Lengauer C, Kinzler KW, Vogelstein B: Genetic instability in colorectal cancers. Nature 1997, 386:623-627 [DOI] [PubMed] [Google Scholar]
- 9.Sugai T, Habano W, Nakamura S, Sato H, Uesugi N, Takahashi H, Jiao Y-F, Yoshida T, Itoh C: Genetic alterations in DNA diploid, aneuploid, and multiploid colorectal carcinomas identified by the crypt isolation technique. Int J Cancer 2000, 88:614-619 [DOI] [PubMed] [Google Scholar]
- 10.Muto T, Bussey HJ, Morson BC: The evolution of cancer of the colon and rectum. Cancer 1975, 36:2251-2270 [DOI] [PubMed] [Google Scholar]
- 11.Watanabe T, Muto T: Colorectal carcinogenesis based on molecular biology of early colorectal cancer, with special reference to nonpolypoid (superficial) lesions. World J Surg 2000, 24:1091-1097 [DOI] [PubMed] [Google Scholar]
- 12.Arai T, Kino I: Morphometrical and cell kinetic studies of normal human colorectal mucosa: comparison between the proximal and the distal large intestine. Acta Pathol Jpn 1989, 39:725-730 [DOI] [PubMed] [Google Scholar]
- 13.Nakamura S, Goto J, Kitayama M, Kino I: Application of the crypt isolation technique to flow-cytometric analysis of DNA content in colorectal neoplasms. Gastroenterology 1994, 106:100-107 [DOI] [PubMed] [Google Scholar]
- 14.: Japanese Society for Cancer of the Colon and Rectum: Japanese Classification of Colorectal Carcinoma, first English edition 1997:30-63 Kanehara Co. Tokyo
- 15.Turnbull RB, Kyle K, Watson FR, Spratt J: Cancer of the colon: the influence of the no-touch isolation technique on survival rates. Ann Surg 1967, 166:420-427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Habano W, Sugai T, Nakamura S, Yoshida T: A novel method for gene analysis of colorectal carcinomas using a crypt isolation technique. Lab Invest 1996, 74:933-940 [PubMed] [Google Scholar]
- 17.Sugai T, Habano W, Nakamura S, Yoshida T, Uesugi N, Sasou S, Itoh C, Katoh R: Use of crypt isolation to determine loss of heterozygosity of multiple tumor suppressor genes in colorectal carcinoma. Pathol Res Pract 2000, 196:145-150 [DOI] [PubMed] [Google Scholar]
- 18.Sugai T, Habano W, Nakamura S, Uesugi N, Sasou S, Itoh C: A unique method for mutation analysis of tumor suppressor genes in colorectal carcinomas using a crypt isolation technique. Arch Pathol Lab Med 2000, 124:382-386 [DOI] [PubMed] [Google Scholar]
- 19.Sugai T, Habano W, Nakamura S, Jiao Y-F, Higuchi T, Inomata M, Chiba T: Analysis of Ki-ras gene mutations associated with DNA diploid, aneuploid, and multiploid colorectal carcinomas using a crypt isolation technique. Cytometry 2001, 46:345-350 [DOI] [PubMed] [Google Scholar]
- 20.Halling KC, Harper J, Moskaluk CA, Thibodeau SN, Petroni GR, Yustein AS, Tosi P, Minacci C, Roviello F, Piva P, Hamilton SR, Jackson CE, Powell SM: Origin of microsatellite instability in gastric cancer. Am J Pathol 1999, 155:205-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sommers SC: Large intestine: appendix, colon, and rectum. Rotterdam H Sheahan DG Sommers SC eds. Biopsy Diagnosis of the Digestive Tract 1993, :700-705 Raven Press New York [Google Scholar]
- 22.Kikuchi-Yanoshita R, Kinishi M, Ito S, Seki M, Tanaka K, Maeda Y, Lino H, Fukuyama M, Koike M, Mori T, Sakuraba H, Fukunari H, Iwama T, Miyaki M: Genetic changes of both p53 alleles associated with the conversion from colorectal adenoma to early carcinoma in familial adenomatous polyposis and non-familial adenomatous patients. Cancer Res 1992, 52:3965-3971 [PubMed] [Google Scholar]
- 23.Neugut AI, Jacobson JS, Ahsan H, Santos J, Garbowski GC, Forde KA, Treat MR, Waye J: Incidence and recurrence rates of colorectal adenomas: a prospective study. Gastroenterology 1995, 108:402-408 [DOI] [PubMed] [Google Scholar]
- 24.Hasegawa H, Ueda M, Furukawa K, Watanabe M, Teramoto T, Mukai M, Kitajima M: p53 gene mutations in early colorectal carcinoma, de novo vs. adenoma-carcinoma sequence. Int J Cancer 1995, 64:47-51 [DOI] [PubMed] [Google Scholar]
- 25.Leahy DT, Salman R, Mulcahy H, Sheahan K, O’Donoghue DP, Parfrey NA: Prognostic significance of p53 abnormalities in colorectal carcinoma detected by PCR-SSCP and immunohistochemical analysis. J Pathol 1996, 180:364-370 [DOI] [PubMed] [Google Scholar]
- 26.Morris RG, Curtis LJ, Romanowski P, Hardcastle JD, Jenkins DA, Robinson M, Wyllie AH, Bird CC: Ki-ras mutations in adenomas: a characteristic of cancer-bearing colorectal mucosa. J Pathol 1996, 180:357-363 [DOI] [PubMed] [Google Scholar]
- 27.Boss JL, Fearon ER, Hamilton SR, Verlaan-de Vries M, van Boom JH, van der Eb AJ, Vogelstein B: Prevalence of ras gene mutations in human colorectal cancer. Nature 1987, 327:298-303 [DOI] [PubMed] [Google Scholar]
- 28.Maltzman T, Knoll K, Martinez ME, Gayers T, Stevens BR, Marshall JR, Reid MR, Einspahr J, Hart N, Bhattacharyya AK, Kramer CB, Sampliner R, Alberts DS, Ahnen DJ: Ki-ras proto-oncogene mutations in sporadic colorectal adenomas: relationship to histological and clinical characteristics. Gastroenterology 2001, 121:302-309 [DOI] [PubMed] [Google Scholar]
- 29.Thibodeau SN, Bren G, Schaid D: Microsatellite instability in cancer of the proximal colon. Science 1993, 260:816-819 [DOI] [PubMed] [Google Scholar]
- 30.Samowitz WS, Slattery ML: Microsatellite instability in colorectal adenomas. Gastroenterology 1997, 112:1515-1519 [DOI] [PubMed] [Google Scholar]
- 31.Jass JP: Serrated route to colorectal cancer: back street or super highway. J Pathol 2001, 193:283-285 [DOI] [PubMed] [Google Scholar]
- 32.Makinen MJ, Geoge SM, Jernvall P, Makela J, Vihko P, Karttunen TJ: Colorectal carcinoma associated with serrated adenoma: prevalence, histological features, and prognosis. J Pathol 2001, 193:186-194 [DOI] [PubMed] [Google Scholar]
- 33.Iino H, Lass JR, Simms LA, Young J, Legett B, Ajioka Y, Watanabe H: DNA microsatellite instability in hyperplastic polyps, serrated adenomas, and mixed polyps: a mild mutator pathway for colorectal cancer? J Clin Pathol 1999, 52:5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fogt F, Brien T, Brown CA, Hartmann CJ, Zimmerman RJ, Odze RD: Genetic alterations in serrated adenomas: comparison to conventional adenomas and hyperplastic polyps. Hum Pathol 2002, 33:87-91 [DOI] [PubMed] [Google Scholar]