Summary
Sp/KLF family of factors regulates gene expression by binding to the CACCC/GC/GT boxes in the DNA through their highly conserved three zinc finger domains. To investigate the role of this family of factors in erythroid differentiation and globin gene expression, we first measured the expression levels of selected Sp/KLF factors in primary cells of fetal and adult stages of erythroid development. This quantitative analysis revealed that their expression levels vary significantly in cells of either stages of the erythroid development. Significant difference in their expression levels was observed between fetal and adult erythroid cells for some Sp/KLF factors. Functional studies using RNA interference revealed that the silencing of Sp1 and KLF8 resulted in elevated level of γ globin expression in K562 cells. In addition, K562 cells become visibly red after Sp1 knockdown. Benzidine staining revealed significant hemoglobinization of these cells, indicating erythroid differentiation. Moreover, the expression of PU.1, ETS1 and Notch1 is significantly down-regulated in the cells that underwent erythroid differentiation following Sp1 knockdown. Overexpression of PU.1 or ETS1 efficiently blocked the erythroid differentiation caused by Sp1 knockdown in K562 cells. The expression of c-Kit, however, was significantly upregulated. These data indicate that Sp1 may play an important role in erythroid differentiation.
Keywords: RNAi, Sp/KLF factors, globin expression, erythroid differentiation
Introduction
The Specificity protein (Sp)/Krüppel-like factor (KLF) transcription factors (Sp/KLF factors hereafter) are a family of highly conserved transcription factors that are characterized by the presence of three highly homologous Cys2/His2 type zinc-fingers near their C-termini. So far 9 members in the Sp subgroup and 16 members in the KLF subgroup have been identified in mammalian cells 1. By regulating gene expression through binding to the CACCC/GC/GT boxes (CACCC box hereafter), which is one of the most common DNA elements in the regulatory regions of many cellular and viral genes, this family of factors regulates diverse cellular processes including cell growth and differentiation, and is essential for early embryonic development 2; 3; 4; 5. Currently, however, for most of the KLF factors, their exact function in vivo and their target-specificity remains largely unknown. This is mainly because that they all have similar DNA binding specificity and affinity to the CACCC boxes at least under in vitro conditions and many of them are co-expressed in the same tissues 2.
KLF1 (EKLF) is the only KLF factor that is specifically expressed in erythroid cells. KLF1 interacts with the CACCC boxes of β globin promoter and is required for β globin expression but not for murine embryonic (Ey, βh1) and human ε and γ globin gene expression 6; 7; 8. Currently, the roles of this family of factors in erythropoiesis are not established. We have carried out extensive studies to identify the KLF factors that may play a role in globin expression and erythroid differentiation. Quantitative analysis of KLF factor expression in primary fetal and adult erythroid tissues showed that their expression levels vary significantly in both tissues. In addition, some KLF factors are differentially expressed in a developmental stage-specific manner. Functional studies of selected KLF factors using RNAi showed that the silencing of Sp1 resulted in increased expression of γ globin and erythroid differentiation of K562 cells. Furthermore, gene expression analysis revealed that the expression of c-Kit is significantly upregulated, whereas the expression of PU.1, ETS1 and Notch1 genes is significantly down-regulated in K562 cells that underwent erythroid differentiation following Sp1 knockdown. The results indicate that Sp1 may negatively control erythroid differentiation.
Results
Expression of KLF factors in primary erythroid cells
The expression of KLF factors in primary erythroid cells that express γ and β globin has not been completely analyzed. To help functional studies on the role of KLF factors in globin gene expression, it is necessary to know the status of their expression in γ and β globin expressing tissues. Therefore, we analyzed their expression profile in fetal and adult erythroid cells derived from purified human fetal liver and adult peripheral blood hematopoietic stem cells and progenitors. As shown in figure 1, this analysis revealed that most of the KLF factors are expressed, albeit at variable levels in these cells. The general expression profile of KLF factors in both types of cells is as following: Sp1, Sp2, Sp3, KLF1 and KLF13 are expressed at relatively high levels; Sp4, KLF3, KLF6, KLF9, KLF10, KLF11 and KLF16 have moderate expression levels; the expression levels of KLF2, KLF5, KLF8 and KLF15 are comparatively low; and there are only detectable levels of KLF4, KLF7, KLF12 and KLF14. When their expression levels in fetal and adult erythroid cells were compared, however, it was found that there are more transcripts of Sp1, Sp2, Sp3, Sp4, KLF1, KLF3, KLF8, KLF9 and KLF13 in adult than in fetal liver erythroid cells. In contrast, KLF2, KLF4, KLF5, KLF7, KLF10, KLF12, KLF15 and KLF16 are expressed higher in fetal liver than in adult erythroid cells. Expression analysis showed that the expression levels of these factors in fetal liver cells are similar to that in K562 cells, a erythroid cell line that expresses γ globin mRNA (data not shown).
Figure 1.
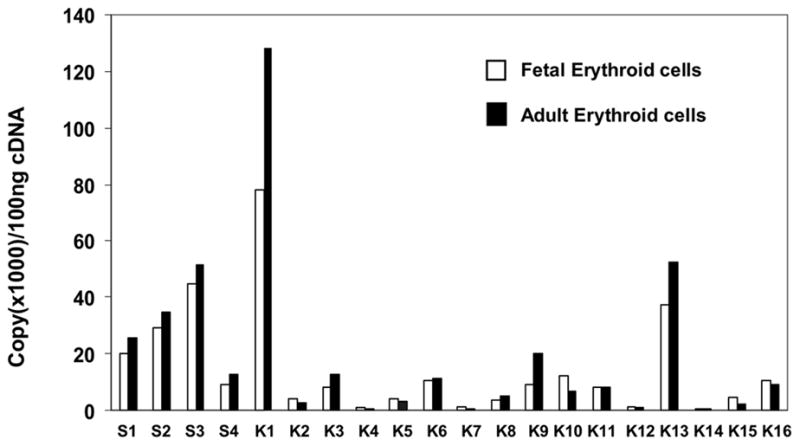
KLF factor expression profile in primary human fetal and adult erythroid cells. Total RNAs from 6–7 day phase II cultures of purified human fetal liver and adult peripheral blood hematopoietic stem cells and progenitors were analyzed for Sp/KLF factor expression using real-time RT-PCR. S1-S4 and K1-K16 denote Sp1-4 and KLF1-16, respectively. The expression level of each factor is presented by copy number per 100 ng cDNA that was transcribed by RT-PCR from total RNA. The amplification of RNAs from human fetal cells and human adult erythroid cells is as indicated.
Development of a lentiviral RNAi system and a reporter system for systematic silencing of KLF factors
We have developed a reporter system (Figure 2A) for quick identification of effective siRNAs for gene silencing. Since the reporter and the KLF factor are transcribed as a single mRNA, the targeting of the KLF factor by RNAi results in degradation of the entire transcript. By transiently co-transfecting the plasmid expressing siRNA targeting a specific KLF factor and the reporter that expresses the KLF factor bicistronically with the luciferase gene, it is possible to determine the efficiency of a specific siRNA sequence in target gene silencing by measuring luciferase activity. Based on the expression profile data and previous functional studies on KLF factors using gene knockout and transgenic mice, 15 Sp/KLF factors were selected as the target genes for functional studies using a systematic RNAi approach. These include Sp1, KLF1-8, KLF10, and KLF12-16. We screened 132 siRNAs using this reporter system in order to identify effective siRNAs for the 16 selected factors. The knockdown efficiency of each individual siRNA was measured using luciferase assay 48 hours following transient transfection. As summarized in figure 3, the knockdown efficiency ranges from 0% to 94%.
Figure 2.
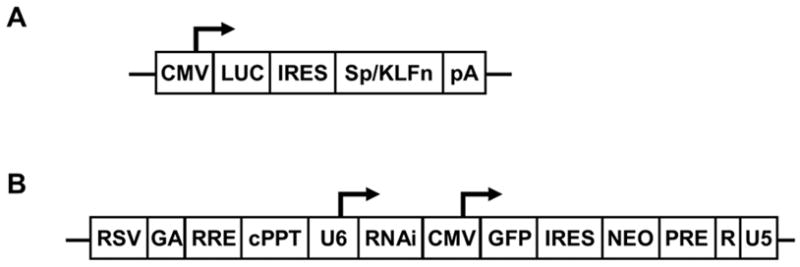
Lentiviral siRNA expression and reporter system. (a) Schematic presentation of the reporter system that was used for the screening of effective siRNA targeting KLF factors. The luciferase reporter gene and the target KLF gene were expressed bicistronically under the CMV promoter. (b) Lentiviral vectors for siRNA expression. It expresses the siRNA under the human U6 promoter. It also expresses the GFP and Neo genes under a CMV promoter bicistronically. Transduced cells can be monitored by GFP expression and selected by GFP sorting or G418 resistance.
Figure 3.
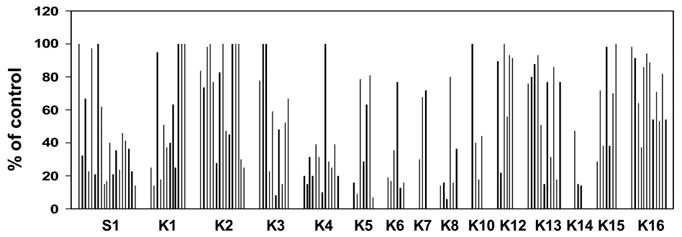
Identification of effective siRNA using transient reporter assay. A total of 132 siRNAs targeting 15 Sp/KLF factors were screened for RNAi efficiency by using a reporter system that we have developed. COS cells were transiently transfected with lentiviral vector expressing siRNA targeting the indicated KLF factors and their corresponding reporters. Data are presented as percentage of the control lentiviral vector. K1-K16 is abbreviation of KLF1-KLF16. Each graph represents the efficiency of an individual siRNA, with a range of 0% to 94%.
To achieve efficient endogenous expression of siRNA in mammalian cells for studying the role of candidate Sp/KLF factors in γ globin gene expression using RNAi, we have developed a lentiviral vector system for efficient delivery and expression of siRNA in the cells (Figure 2B). In addition to expressing the siRNA under the human small nuclear RNA U6 promoter, this lentiviral system also expresses GFP and Neo bicistronically, thus allowing monitoring transduction efficiency and selecting transduced cells. siRNAs that are able to inhibit the luciferase activity more than 70% under our reporter assay condition are selected as effective siRNA for further endogenous gene knockdown study. The numbers of the siRNA that are screened for each factor and their effectiveness in target reporter silencing are summarized in figure 3. According to the results of luciferase reporter assay, some of the KLF factors are easier to knockdown, e.g. KLF4, KLF6, KLF8, while other factors, such as KLF1, KLF2, KLF12, KLF13, KLF15 and KLF16, are difficult to silence.
Knockdown of endogenous KLF factors in K562 cells
Effective siRNAs in the reporter assay (>70% knockdown efficiency) were used to silence their respective endogenous genes in K562 cells. Near 100% transduction of K562 cells was achieved under our lentiviral production and transduction conditions as judged by GFP expression (data not shown). K562 cells were transduced with lentivirus expressing siRNA targeting a specific factor or targeting the luciferase gene as a control. The knockdown of endogenous Sp/KLF factors at the mRNA level is measured using semi-quantitative RT-PCR compared to the expression level of the target gene in K562 cells following transduction with control lentivirus expressing siRNA targeting the luciferase gene. These assays identified siRNAs that are able to silence the expression of endogenous KLF factors (Figure 4A). The target genes that are significantly silenced include Sp1, KLF2, KLF3, KLF4, KLF5, KLF7, KLF8, KLF10 and KLF13. The efficiency of endogenous gene silencing ranges from around 60% up to more than 95% as quantified by densitometry (Figure 4B). Although effective in reporter silencing, the siRNAs targeting KLF1, KLF6, KLF12, KLF14 KLF15 and KLF16 were unable to silence their respective endogenous genes.
Figure 4.
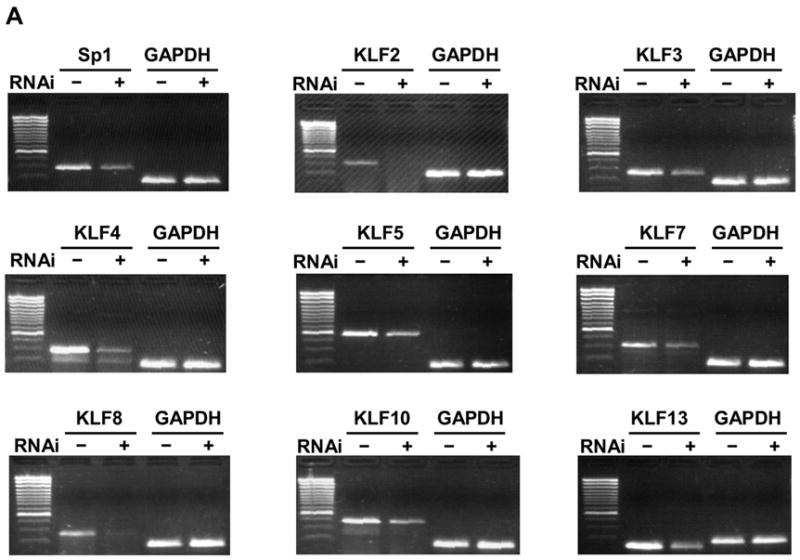
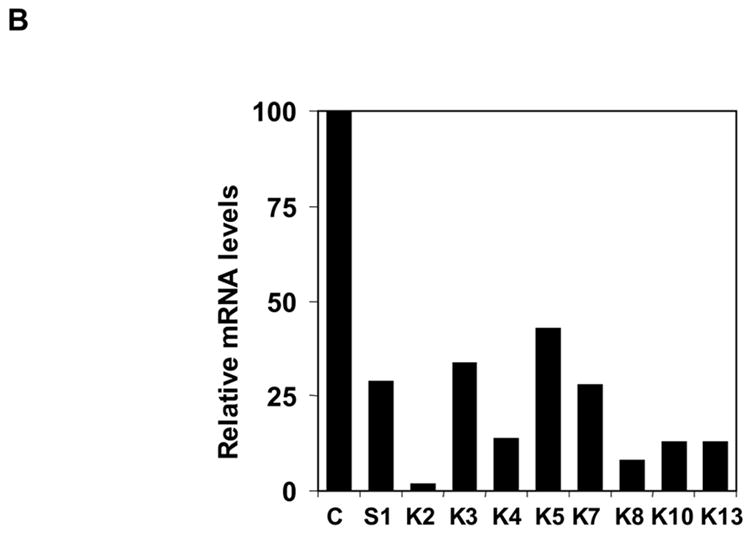
Knockdown of Endogenous Sp/KLF factors by RNAi. (a) Endogenous knockdown of Sp/KLF factors in K562 cells. Lentiviral vector expressing siRNA targeting Sp/KLF factors were used to transduce K562 cells. Total RNA were isolated 2 weeks later. The expression of endogenous Sp/KLF factors following transduction with lentiviruses expressing siRNA targeting a KLF factor or control siRNA targeting luciferase was determined using RT-PCR with GAPDH as an internal control. (b) Quantification of the data shown in (a). The data are presented as the percentage of mRNA level in cells transduced with lentiviruses expressing siRNA targeting the KLF factor and cells transduced with lentiviruses expressing luciferase control siRNA after normalization with the GAPDH internal control.
Changes in globin gene expression following knockdown of Sp/KLF factors in K562 cells
Since K562 cells express γ globin mRNA and we are interested in the functional role of Sp/KLF factors in γ globin expression, we next determined the effect of the knockdown of these Sp/KLF factors on endogenous γ globin expression in K562 cells. As shown in figure 5, the knockdown of Sp1 and KLF8 resulted in significant increase in γ globin expression in K562 cells. In contrast, no significant change in γ globin expression was observed after the knockdown of KLF2, KLF3, KLF4, KLF5, KLF7, KLF10 and KLF13 (data not shown). RNase protection analysis (Figure 5A) and quantification using PhosphoImage and ImageQuant software showed that γ globin expression increased 6.8 fold and 4.3 fold following Sp1 and KLF8 knockdown, respectively (Figure 5B).
Figure 5.
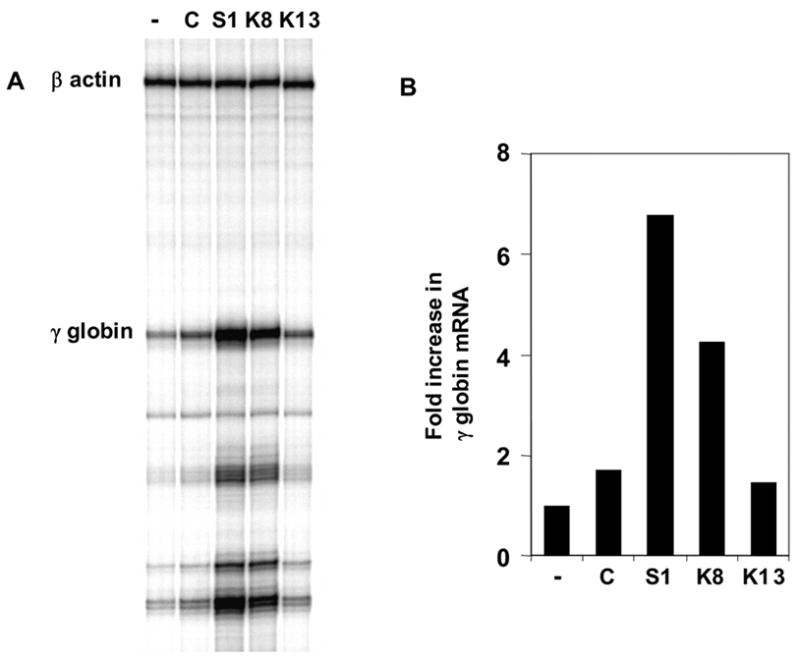
Changes in expression levels of γ globin following KLF factor knockdown. (a) RNase protection analysis of γ globin expression in K562 cells following siRNA knockdown of KLF factors. Human β-actin was used as internal control. (b) Quantification of the data in (a) using PhosphoImage and ImageQuant software. -, C, S1, K8 and K13 denote untransduced, luciferase control, Sp1, KLF8 and KLF13 siRNA transduced cells, respectively.
Although it was reported that β globin is not detectable in K562 cells by Northern blot analysis 9, a very low level of β globin mRNA was detected in K562 cells using RT-PCR. Therefore, we also examined the changes of β globin expression in K562 cells following the knockdown of the KLF factors. The knockdown of Sp1, KLF3, KLF4 and KLF7 resulted in significant increases of β globin in K562 cells, although the level of β globin is still very low compared with that of γ globin (data not shown). The strongest up-regulation of β globin expression is observed when KLF7 is silenced (data not shown). Furthermore, the expression of α globin was also increased in Sp1 knockdown cells (Figure 7).
Figure 7.
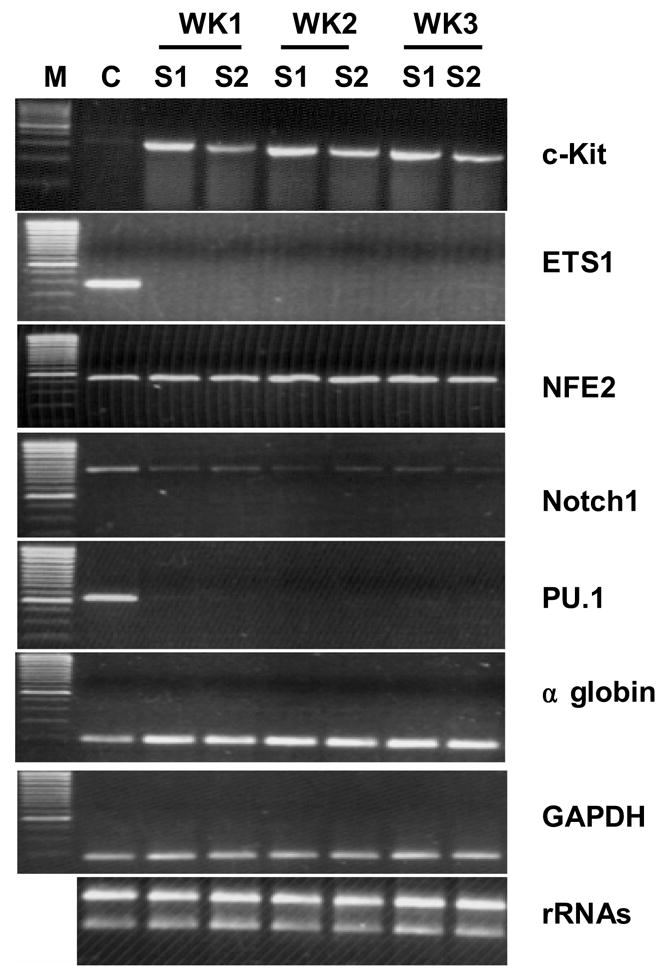
The expression of Key erythroid regulators and globins in K562 cells that underwent erythroid differentiation following Sp1 knockdown. C-Kit is significantly upregulated, whereas ETS1, Notch1 and PU.1 are significantly down-regulated. NF-E2 expression is slight upregulated. The expression level of α globin is increased. GAPDH is the internal control of the RT-PCR. rRNA is the control of quality and quantity of the isolated RNA. M, Marker; WK, week; C, S1 and S2, K562 cells transduced with lentiviruses expressing siRNAs targeting luciferase control, Sp1 siRNA-1 and Sp1 siRNA-2, respectively.
Erythroid differentiation of K562 cells following the Sp1 knockdown
Sp1 transcript levels were reduced to about 30% (Figure 4B) as quantified by densitometer of semi-quantitative RT-PCR results (Figure 4A). The knockdown of Sp1 protein was confirmed using Western blot analysis (Figure 6A). Visual inspection found that K562 cell pellet turned into red color starting around 10 days post lentiviral delivery of Sp1 siRNA, indicating erythroid differentiation of K562 cells following Sp1 knockdown (Figure 6B). The red color intensity increases at 2–3 weeks after transduction with lentiviruses expressing Sp1 siRNAs. The use of two siRNAs targeting different regions of Sp1 with different silencing efficiency demonstrated that redness of the cells correlates with the degree of Sp1 knockdown by RNAi. To further establish that the knockdown of Sp1 resulted in erythroid differentiation of K562 cells, we next carried out benzidine staining of the cells. This assay showed that 29% and 16% of the cells were benzidine positive following transduction with lentiviruses expressing Sp1 siRNA-1 and Sp1 siRNA-2 respectively, demonstrating significant hemoglobinization of the cells when Sp1 is silenced (Figure 6C). Similar to the redness of the cells, the numbers of the benzidine staining positive cells also correlate with the degree of Sp1 knockdown in these cells. The knockdown of KLF2, KLF3, KLF4, KLF5, KLF7, KLF10 and KLF13 showed no changes in either γ globin expression or cell phenotype. In addition, although the knockdown of KLF8 also resulted in changes in γ globin gene expression in K562 cells, no erythroid differentiation is observed (data not shown), indicating a specific role of Sp1 in regulation of erythroid differentiation of K562 cells.
Figure 6.
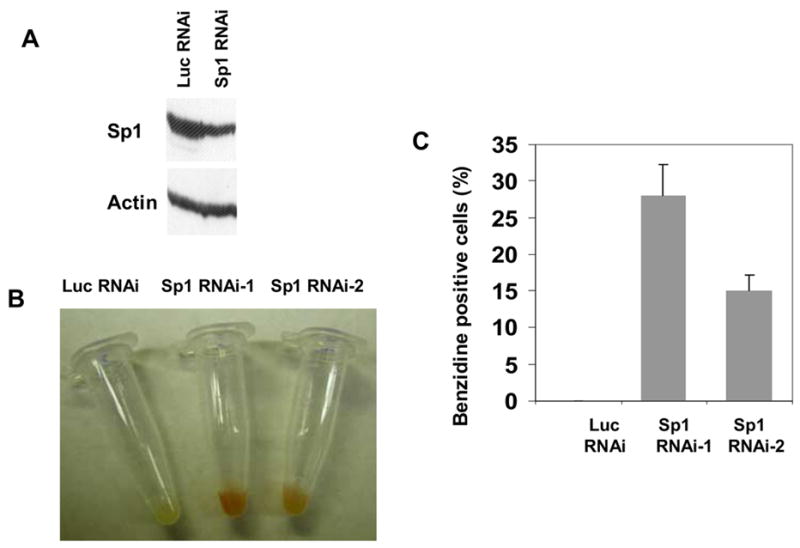
Endogenous knockdown of Sp1 in K562 cells and the phenotypic changes of the cells. (a) Western Blot analysis using anti-Sp1 antibody showed a decrease of Sp1 protein in K562 cells transduced with lentivirus expressing Sp1 siRNA. The decrease of Sp1 protein is comparable to that of Sp1 transcripts as demonstrated by RT-PCR. (b) and (c) Phenotypic change of K562 cells from white to red when Sp1 is silenced. The redness is correlated with the effectiveness of the Sp1 siRNA. Sp1 siRNA-1 is more potent than Sp1 siRNA-2 in Sp1 knockdown, therefore, the redness of K562 cells transduced with the former are darker than that transduced with the latter.
Changes in the expression of key factors of erythroid differentiation following Sp1 knockdown
The results that Sp1 knockdown resulted in a significant increase in the globin expression and hemoglobinization suggest that Sp1 may play an inhibitory role in erythroid differentiation of the cells. Sp1 is expressed at relatively high levels in fetal and adult erythroid cells (Figure 1), and K562 cells (data not shown) comparing with other Sp/KLF factors. A number of factors have been shown to play a key role in regulating erythroid differentiation. To help understand the molecular basis of Sp1 inhibition of erythroid differentiation, we next examined the expression of several of these factors in K562 cells that underwent erythroid differentiation following Sp1 knockdown. These analyses showed that expression levels of Notch1, PU.1 and ETS1 are significantly reduced, while c-Kit is significantly increased and NF-E2 is marginally increased when Sp1 is silenced in K562 cells (Figure 7). We next examined whether the down-regulation of ETS1 and PU.1 is required for Sp1 knockdown induced-erythroid differentiation. K562 cells were first transduced with lentiviruses that express ETS1 or PU.1. Then these cells were transduced with lentiviruses that express Sp1 siRNA. These studies showed that overexpression of these two factors efficiently blocked the Sp1 knockdown-induced erythroid differentiation (Figure 8).
Figure 8.
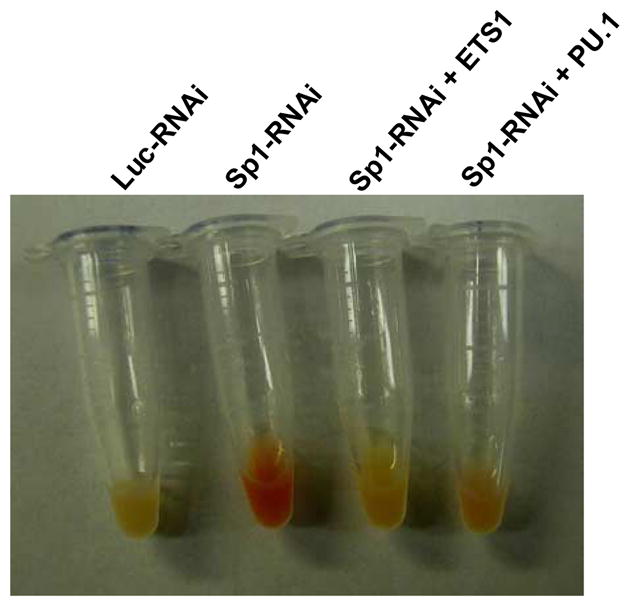
Overexpression of PU.1 or TES1 blocks Sp1 knockdown induced-erythroid differentiation of K562 cells. K562 cells were first transduced with lentiviruses that express ETS1 or PU.1. Then these cells were transduced with lentiviruses that express Sp1 siRNA.
Discussion
The efficient silencing of target genes by RNAi in mammalian cells relies on the identification of effective siRNA. As it is difficult to predict which siRNA will be effective in target gene silencing, several siRNAs per target have to be screened for effectiveness in specific target gene silencing. It is very time consuming to screen a large number of siRNAs for effectiveness in silencing endogenous genes directly since this requires establishing stable cell lines or generating lentiviruses for each siRNA. By overcoming this rate-limiting step, the reporter system that we have developed will make it possible to identify effective siRNAs quickly and easily. Out of the 132 siRNAs that we have tested, 44 are effective in reporter silencing. Our studies found that the efficiency of silencing the reporter gene correlates with that of the endogenous KLF gene (data not shown). Therefore, this reporter system allows quick selection of effective siRNAs using transient transfection assay. However, it is also found that although very effective in gene silencing under reporter assay conditions, some siRNAs showed no or minimal activity in silencing their respective endogenous gene expression. For example, effective siRNAs are identified for KLF6 and KLF14 at a relatively high ratio using the reporter system. Nevertheless, all of them failed to silence their endogenous target gene expression to significant levels. This discrepancy may be due to that the secondary structure of endogenous transcripts may differ from that of the reporter, thus preventing siRNA from accessing its endogenous targets. The structure of the mRNA has been shown to significantly influence the effectiveness of siRNA 10. This may also be because the differences in assay conditions. For example, the reporter assay used a target to siRNA ratio of 1 to 20. The expression levels of siRNA may not be sufficient for endogenous target silencing since the copy numbers of lentivirus in the transduced cells is generally low.
Sp/KLF factors bind to similar GC/CACCC boxes on DNA. How they regulate tissue- and developmental stage-specific expression of globin genes is of great interest and is under extensive investigations. Our functional studies using systematic RNAi revealed that once KLF8 is silenced, the expression of γ globin is elevated. It has been reported that KLF8 inhibits γ globin promoter activity through the CACCC box using a transient transfection assay 11. KLF8 has been shown to repress transcription by binding the CACCC box in association with the transcription repressor CtBP 12. Together with our results using RNAi, it is suggested the KLF8 may play a negative role in γ globin expression. No changes in γ globin expression and cell functions are observed following the knockdown of KLF2, KLF3, KLF4, KLF5, KLF7, KLF10 and KLF13 in K562 cells. Our RNAi studies in K562 cells together with the studies using KLF2 knockout mice carrying a human β globin transgene 13 suggest that KLF2 is not required for γ globin expression. The expression level of KLF13 is relatively high in both primary fetal and adult erythroid cells (Figure 1). KLF13 was cloned as a candidate trans-activator of γ globin expression 14. KLF13 activates γ globin promoter activity under transient transfection assays 14; 15. However, no change in endogenous γ globin gene expression was observed when KLF13 was silenced in K562 cells using RNAi. This result demonstrated that KLF13 is not required for endogenous γ globin gene expression in K562 cells.
Our quantitative real time RT-PCR analysis showed the expression levels of selected Sp factors and all KLF factors in primary human fetal and adult erythroid cells. The expression level of KLF1 is the highest of all the family members analyzed. In addition, KLF1 expression in adult erythroid cells is higher than that in the fetal cells. This is consistent with the fact that KLF1 is a erythroid specific factor and it is specifically required for adult β globin gene expression 16. Sp1 is expressed at relatively high levels in both fetal and adult erythroid cells (Figure 1). Sp1 is also expressed at similarly high levels in K562 cells (data not shown). The knockdown of Sp1 in K562 cells resulted in erythroid differentiation since the cells become red in color, benzidine staining positive and showed significant increases in the expression of both α and β chains (Figure 5 and Figure 7).
Currently several approaches that are both complimentary and redundant have been considered as standard practice in functional studies using RNAi to rule out the possibility of off-target and side effects of RNAi. These include BLAST analysis, the use of control siRNA, the use of two or more siRNAs targeting different regions of the same gene and overexpression rescue. We have taken the following steps to establish that the change in the erythroid differentiation of K562 cells is due to specific silencing of Sp1 rather than the consequence of off-target silencing of other genes or side effects. First, a BLAST analysis against the sequence database showed that the Sp1 siRNA sequences are specific to Sp1. This significantly eliminated the possibility of potential off-target silencing of unintended targeted genes. Secondly, Luciferase control siRNA was included in all RNAi experiments to rule out the possibility that the phenotype is caused by other experimental procedures such as viral infection instead of by the silencing of Sp1. We chose the siRNAs targeting the firefly luciferase as our control. Since the siRNA targeting the luciferase has been validated against its target 17, it can efficiently enter the RNAi pathway but would not be expected to affect cellular gene expression in mammalian cells. Therefore, it serves as a better control than scrambled siRNAs, which may not have the biological activity and may not enter the RISC complex. Thirdly, a second siRNA from a different region of the Sp1 was used to confirm the phenotype. Since it is extremely unlikely that different RNAi triggers produce the same off-target effect, the same results from two independent siRNAs targeting different regions in Sp1 provided strong evidence that the phenotype is caused by specific silencing of Sp1. Moreover, the second siRNA (Sp1 RNAi-2) is less potent than the first siRNA (Sp1 RNAi-1) in Sp1 knockdown. The degree of Sp1 knockdown correlated with the degree of hemoglobinization (figure 6C). This does-response relationship provided strong supports for our conclusion that the reduced level of Sp1 by RNAi is the cause of erythroid differentiation of K562 cells. Therefore, we concluded that the erythroid differentiation of K562 cells following Sp1 knockdown is not a result of off-target effect or side effects.
A number of factors have been shown to play a key role in regulation of erythroid differentiation. In mouse and human erythroleukemia cell culture or in primary human erythroblasts, the expression levels of ETS1 and PU.1, and the DNA binding activity of ETS1 are down-regulated during erythroid differentiation 18; 19. PU.1 transgenic mice were reported to develop erythroleukemia 20. PU.1 interacts with GATA1 and inhibits GATA1 function at multiple levels including DNA binding and transcription activity 21; 22; 23 It has been reported that the balance between the activity of GATA1 and GATA2 regulates erythroid differentiation and this balance is determined by the function of transcription factors PU.1 and Notch1 23; 24; 25. The receptor tyrosine kinase, c-kit plays important roles in hematopoiesis. The expression of c-Kit has been reported to be regulated by Sp1 26; 27. C-kit expression increased during early erythroid differentiation 28. NFE2 is a hematopoietic-specific activator and is implicated in the control of α and β globin expression 29; 30. The expression levels of NFE2 increased along erythroid differentiation of K562 cells 31; 32. In addition, NFE2 has been found to be sumoylated in K562 cells and mouse fetal liver; and this sumoylation of NFE2 is believed to activate β globin gene 33.
To understand the mechanisms underlying the Sp1 knockdown-induced erythroid differentiation, we examined the expression levels of c-Kit, PU.1, ETS1, Notch1 and NFE2 in K562 cells transduced with lentiviruses expressing Sp1 siRNA or control luciferase siRNA. This analysis found that the knockdown of Sp1 in K562 cells resulted in a significant decrease in ETS1, Notch1 and PU.1 expression and a significant increase in c-Kit expression (Figure 7). A slight increase in NFE2 expression was also observed (Figure 7). In addition, overexpression of ETS1, Notch1 and PU.1 was reported to interfere with or block erythroid differentiation in mouse and human erythroleukemia cells 18; 19; 23; 24. Therefore, we also examined whether the overexpression of ETS1 or PU.1 can also block Sp1 knockdown-induced erythroid differentiation of K562 cells. K562 cells were first transduced with lentiviruses that express ETS1 or PU.1. Then these cells were transduced with lentiviruses that express Sp1 siRNA. These studies showed that overexpression of these two factors efficiently blocked the Sp1 knockdown-induced erythroid differentiation (Figure 8). These results reveal a functional interaction between Sp1 and ETS1 or PU.1, and the down-regulation of ETS1 and PU.1 is required for the erythroid differentiation following Sp1 knockdown. At present, it remains to be determined whether these factors are direct targets of Sp1 regulation or the changes in their expression levels is merely a consequence of the erythroid differentiation resulted from Sp1 knockdown.
It has been reported by several previous studies that Erk and p38 MAPK signaling pathways play a role in globin expression and in erythroid cell differentiation 34; 35; 36. Therefore, we also examined whether PI3K, Erk and p38 MAPK signaling pathways are involved in the changes of globin expression and erythroid differentiation following Sp1 knockdown in K562 cells. Western blot analysis using antibodies against non-phosphorylated and phosphorylated forms of these proteins revealed no changes in the expression levels and phosphorylation status of PI3K, Erk and p38 in K562 cells that underwent erythroid differentiation following Sp1 knockdown (data not shown).
Materials and Methods
Profiling of KLF factor expression in primary erythroid cells at different stages of development
Real-time RT-PCR was carried out to measure the expression levels of KLF factors in fetal and adult human primary erythroid cells. RNA was extracted from 6–7 day phase II culture of purified human fetal liver and adult peripheral blood hematopoietic stem cells and progenitors using TRIZOL reagent according to the manufacturer’s protocol (Invitrogen). The adult peripheral blood CD34+ cells were provided by Dr. Shelly Heimfeld at the Fred Hutchinson Cancer Research Center. The cells were provided to us anonymously and without identifiers. All the experiments involving the use of human tissues are approved by IRB of University of Washington. The human fetal liver hematopoietic stem cells and progenitors were isolated from 7–16 week fetal livers. Two-phase liquid culture was done as described 37. Real-time RT-PCR analysis was performed with SYBR green chemistry using Lightcycler (Roche Applied Science). Two μg of RNA from each cell type were reverse transcribed with oligo(dT)20 using standard protocol (Invitrogen). The cDNA at a concentration of 40 ng/μl was analyzed for the expression of KLF factors using real time PCR with specific primers for each KLF factor (primer sequences are available upon request). Standard curves for each KLF factor were generated by amplifying 3-fold serial dilutions of known quantities of plasmid DNA or PCR fragment of each factor. Human β-actin was used as internal control for normalization. Hematopoietic tissues of fetal origin were obtained from the Birth defects Research Laboratory of University of Washington. This laboratory is organized to provide fetal tissues to investigators studying fetuses. Fetal material is obtained from induced abortions and spontaneous abortions. Maternal consent for use of the fetal tissues for research is always obtained. The procedures followed by the Laboratory of Human Embryology have been reviewed and approved by the appropriate Committees of the University of Washington.
Screening for effective siRNA using a reporter system
The reporter vector for quick identification of effective siRNA was made by inserting the firefly luciferase (Luc) gene from pGL3-Basic (Promega) into pIRESneo3 (Clontech) generating a CMV-Luc-IRES-Neo expression plasmid (pLIN). Each target KLF cDNA was then cloned into pLIN (replacing the Neo gene). This reporter was named pLI-KLFn, which expresses the specific target KLF gene bicistronically with the luciferase reporter (Figure 2A). COS cells were then co-transfected with the reporter pLI-hKLFn together with the lentiviral vector expressing siRNA targeting the KLF factor (Figure 2B, see below for construction), at a ratio of 1 to 20 using Fugene 6 (Roche). Luciferase assay (Promega) was carried out 48 hours after transfection to identify effective siRNAs. Sequences of effective siRNAs are 5′-TTGCTGCCATTGGTACTGCTGC for Sp1-1; 5′-TAGTCCTGTCAGAACTTGCTGG for Sp1-2; 5′-TGCTTTCGGTAGTGGCGCGTGA for KLF2; 5′-GGCCTCTTGCTTGACAGGAACTGGGTCAA for KLF3; 5′-GCTGCGGCGGAATGTACACCGGG for KLF4; 5′-GTGCCTCTTCATATGCAGGGCC for KLF5; 5′-GTCTCTTCATGTGGAGGGCAAG for KLF7; 5′-GGTGTCATGGCGACGGCGATGC for KLF8; 5′-GAACGACTGTTGTCACAACAG for KLF10 and 5′-ACTTTGCATCAAGTTCTCTCTC for KLF13.
Construction of a lentiviral system for RNAi
The lentiviral system is the third generation of SIN (self-inactivating) virus 38. The lentiviral vector for RNAi was constructed by inserting a U6 promoter driven siRNA expression cassette and a CMV-GFP-IRES-NEO cassette into the lentiviral vector as shown in figure 2.
Lentiviral production and transduction of K562 cells
Lentiviral production was carried out as described 38. Briefly, Human kidney 293D cells (5 × 106) were plated on 10 cm plates. The cells were transfected the following day with the transfer vector expressing the specific siRNA together with packaging constructs. Culture medium was changed 16 hours after adding DNA. Viral supernatants were collected 48 hours later after transfection. The viral supernatant was used for transduction of K562 cells immediately or stored at −70°C for later use. Transduction efficiency is 100% as indicated by GFP expression 48 hours post transduction.
Knockdown of endogenous Sp/KLF factors
Total RNA was isolated two weeks after lentiviral transduction of K562 cells. The expression of the endogenous Sp/KLF factor following transduction with lentiviruses expressing KLF-specific siRNA or luciferase control siRNA was analyzed using semi-quantitative RT-PCR. GAPDH was used as a control for RNA quantity and quality. The knockdown of endogenous Sp1 at the protein level was further confirmed by Western Blot analysis using anti-Sp1 antibody (SC-420, Santa Cruz Biotechnology).
Analysis of the effect of the knockdown of a specific KLF factor on the expression of globin and other genes
The expression levels of endogenous globin and other genes following RNAi knockdown of a KLF factor were examined by semi-quantitative RT-PCR with rRNAs as a control for RNA quantity and quality. One μg of total RNA was used for RNase protection analysis as described 39 with human β-actin (Ambion) as an internal control.
Benzidine staining
K562 cells were smeared on glass slides and air-dried. Then it was fixed in 100% Methanol for 5 minutes, stained in 1% Benzidine solution for 5 minutes and followed by incubation in 30% H2O2 for 2 minutes.
Lentiviral expression of PU1 and ETS1
Lentiviral vectors expressing PU1 and ETS1 were constructed by inserting the CMV-PU1 or ETS1-IRES-Neo cassette into the lentiviral vector described above. Then lentivirus was produced and used to transduce K562 cells. The transduced K562 cells were selected with G481. The overexpression of PU1 and ETS1 in K562 cells was confirmed by RT-PCR (data not shown). PU1 or ETS1-overexpressing K562 cells were then transduced with lentiviruses that express siRNA targeting Sp1.
Acknowledgments
This work was supported by grants from National Heart, Lung and Blood Institute, National Institute of Health. We thank Dr. Jeffrey S. Chamberlain (University of Washington) for kindly providing the lentiviral system.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551–6. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Bieker JJ. Kruppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355–8. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 3.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–60. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 4.Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–86. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–8. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 8.Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–22. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 9.Donze D, Townes TM, Bieker JJ. Role of erythroid Kruppel-like factor in human gamma- to beta-globin gene switching. J Biol Chem. 1995;270:1955–9. doi: 10.1074/jbc.270.4.1955. [DOI] [PubMed] [Google Scholar]
- 10.Schubert S, Grunweller A, Erdmann VA, Kurreck J. Local RNA target structure influences siRNA efficacy: systematic analysis of intentionally designed binding regions. J Mol Biol. 2005;348:883–93. doi: 10.1016/j.jmb.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Zhang P, Basu P, Redmond LC, Morris PE, Rupon JW, Ginder GD, Lloyd JA. A functional screen for Kruppel-like factors that regulate the human gamma-globin gene through the CACCC promoter element. Blood Cells Mol Dis. 2005;35:227–35. doi: 10.1016/j.bcmd.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 12.van Vliet J, Turner J, Crossley M. Human Kruppel-like factor 8: a CACCC-box binding protein that associates with CtBP and represses transcription. Nucleic Acids Res. 2000;28:1955–62. doi: 10.1093/nar/28.9.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basu P, Morris PE, Haar JL, Wani MA, Lingrel JB, Gaensler KM, Lloyd JA. KLF2 is essential for primitive erythropoiesis and regulates the human and murine embryonic {beta}-like globin genes in vivo. Blood. 2005 doi: 10.1182/blood-2005-02–0674. prepublished online June 9, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asano H, Li XS, Stamatoyannopoulos G. FKLF-2: a novel Kruppel-like transcriptional factor that activates globin and other erythroid lineage genes. Blood. 2000;95:3578–84. [PubMed] [Google Scholar]
- 15.Song CZ, Keller K, Murata K, Asano H, Stamatoyannopoulos G. Functional interaction between coactivators CBP/p300, PCAF, and transcription factor FKLF2. J Biol Chem. 2002;277:7029–36. doi: 10.1074/jbc.M108826200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bieker JJ. Role of erythroid Kruppel-like factor (EKLF) in erythroid-specific transcription. In: Stamatoyannopoulos G, editor. Molecular biology of hemoglobin switching. Intercept, Andover; United Kingdom: 1995. pp. 231–241. [Google Scholar]
- 17.Chen Y, Stamatoyannopoulos G, Song CZ. Down-regulation of CXCR4 by inducible small interfering RNA inhibits breast cancer cell invasion in vitro. Cancer Res. 2003;63:4801–4. [PubMed] [Google Scholar]
- 18.Rao G, Rekhtman N, Cheng G, Krasikov T, Skoultchi AI. Deregulated expression of the PU.1 transcription factor blocks murine erythroleukemia cell terminal differentiation. Oncogene. 1997;14:123–31. doi: 10.1038/sj.onc.1200807. [DOI] [PubMed] [Google Scholar]
- 19.Marziali G, Perrotti E, Ilari R, Lulli V, Coccia EM, Mazzeo S, Kuhn LC, Testa U, Battistini A. Role of Ets-1 in erythroid differentiation. Blood Cells Mol Dis. 2002;29:553–61. doi: 10.1006/bcmd.2002.0595. [DOI] [PubMed] [Google Scholar]
- 20.Moreau-Gachelin F, Wendling F, Molina T, Denis N, Titeux M, Grimber G, Briand P, Vainchenker W, Tavitian A. Spi-1/PU.1 transgenic mice develop multistep erythroleukemias. Mol Cell Biol. 1996;16:2453–63. doi: 10.1128/mcb.16.5.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stopka T, Amanatullah DF, Papetti M, Skoultchi AI. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. Embo J. 2005;24:3712–23. doi: 10.1038/sj.emboj.7600834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rekhtman N, Choe KS, Matushansky I, Murray S, Stopka T, Skoultchi AI. PU.1 and pRB interact and cooperate to repress GATA-1 and block erythroid differentiation. Mol Cell Biol. 2003;23:7460–74. doi: 10.1128/MCB.23.21.7460-7474.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang P, Zhang X, Iwama A, Yu C, Smith KA, Mueller BU, Narravula S, Torbett BE, Orkin SH, Tenen DG. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 2000;96:2641–8. [PubMed] [Google Scholar]
- 24.Kumano K, Chiba S, Shimizu K, Yamagata T, Hosoya N, Saito T, Takahashi T, Hamada Y, Hirai H. Notch1 inhibits differentiation of hematopoietic cells by sustaining GATA-2 expression. Blood. 2001;98:3283–9. doi: 10.1182/blood.v98.12.3283. [DOI] [PubMed] [Google Scholar]
- 25.Zhang P, Behre G, Pan J, Iwama A, Wara-Aswapati N, Radomska HS, Auron PE, Tenen DG, Sun Z. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci U S A. 1999;96:8705–10. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lecuyer E, Herblot S, Saint-Denis M, Martin R, Begley CG, Porcher C, Orkin SH, Hoang T. The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood. 2002;100:2430–40. doi: 10.1182/blood-2002-02-0568. [DOI] [PubMed] [Google Scholar]
- 27.Park GH, Plummer HK, 3rd, Krystal GW. Selective Sp1 binding is critical for maximal activity of the human c-kit promoter. Blood. 1998;92:4138–49. [PubMed] [Google Scholar]
- 28.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, Valtieri M, Calin GA, Liu CG, Sorrentino A, Croce CM, Peschle C. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005;102:18081–6. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu SJ, Rowan S, Bani MR, Ben-David Y. Retroviral integration within the Fli-2 locus results in inactivation of the erythroid transcription factor NF-E2 in Friend erythroleukemias: evidence that NF-E2 is essential for globin expression. Proc Natl Acad Sci U S A. 1994;91:8398–402. doi: 10.1073/pnas.91.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forsberg EC, Downs KM, Bresnick EH. Direct interaction of NF-E2 with hypersensitive site 2 of the beta-globin locus control region in living cells. Blood. 2000;96:334–9. [PubMed] [Google Scholar]
- 31.Chenais B. Requirement of GATA-1 and p45 NF-E2 expression in butyric acid-induced erythroid differentiation. Biochem Biophys Res Commun. 1998;253:883–6. doi: 10.1006/bbrc.1998.9869. [DOI] [PubMed] [Google Scholar]
- 32.Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. Embo J. 2004;23:2841–52. doi: 10.1038/sj.emboj.7600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shyu YC, Lee TL, Ting CY, Wen SC, Hsieh LJ, Li YC, Hwang JL, Lin CC, Shen CK. Sumoylation of p45/NF-E2: nuclear positioning and transcriptional activation of the mammalian beta-like globin gene locus. Mol Cell Biol. 2005;25:10365–78. doi: 10.1128/MCB.25.23.10365-10378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McElveen RL, Lou TF, Reese K, Xia S, Baliga BS, Pace BS. Erk pathway inhibitor U0126 induces gamma-globin expression in erythroid cells. Cell Mol Biol (Noisy-le-grand) 2005;51:215–27. [PubMed] [Google Scholar]
- 35.Huang HM, Chiou HY, Chang JL. Activin A induces erythroid gene expressions and inhibits mitogenic cytokine-mediated K562 colony formation by activating p38 MAPK. J Cell Biochem. 2006 doi: 10.1002/jcb.20713. [DOI] [PubMed] [Google Scholar]
- 36.Matsuzaki T, Aisaki K, Yamamura Y, Noda M, Ikawa Y. Induction of erythroid differentiation by inhibition of Ras/ERK pathway in a friend murine leukemia cell line. Oncogene. 2000;19:1500–8. doi: 10.1038/sj.onc.1203461. [DOI] [PubMed] [Google Scholar]
- 37.Cao H, Stamatoyannopoulos G, Jung M. Induction of human gamma globin gene expression by histone deacetylase inhibitors. Blood. 2004;103:701–9. doi: 10.1182/blood-2003-02-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S, Kimura E, Fall BM, Reyes M, Angello JC, Welikson R, Hauschka SD, Chamberlain JS. Stable transduction of myogenic cells with lentiviral vectors expressing a minidystrophin. Gene Ther. 2005;12:1099–108. doi: 10.1038/sj.gt.3302505. [DOI] [PubMed] [Google Scholar]
- 39.Navas PA, Peterson KR, Li Q, Skarpidi E, Rohde A, Shaw SE, Clegg CH, Asano H, Stamatoyannopoulos G. Developmental specificity of the interaction between the locus control region and embryonic or fetal globin genes in transgenic mice with an HS3 core deletion. Mol Cell Biol. 1998;18:4188–96. doi: 10.1128/mcb.18.7.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]


