Abstract
Gene expression profiling using high density oligonucleotide arrays is a powerful method to generate an unbiased survey of a cell’s transcriptional landscape. Increasingly complex biological questions require that this approach be applicable to the small numbers of cells that are obtained from sources such as laser capture microdissection (LCM) of solid tissues. In this report, we demonstrate that two rounds of transcript amplification can generate accurate and reproducible gene expression profiles using high density oligonucleotide microarrays, starting with as little as 10 ng of total RNA. Biased amplification of the 3′ end of transcripts does not have a major impact on the overall transcript profile due to the 3′ bias of probe sets incorporated in the array design. Furthermore, greater than 95% of all genes detected demonstrate less than a twofold difference in expression when independent tissue dissections of identical cell populations are compared. The accuracy and technical reproducibility of the method suggests that expression profiling using transcript amplification and high density oligonucleotide microarrays can be used on a routine basis.
Gene expression profiling using high density oligonucleotide array technology is a powerful and popular approach for characterizing a cell’s transcriptional program. 1 Until recently, utilization of this technology has required relatively large quantities of total cellular RNA (5 to 10 μg) as a labeling substrate. 2, 3 This has generally precluded routine analysis of samples derived from small numbers of cells, such as those obtained by cell sorting, primary cell culture, embryonic dissection, fine needle aspirates, or laser capture microdissection (LCM). 4 To address more sophisticated questions in fields as diverse as developmental biology and clinical oncology, it would be highly desirable to apply global gene expression profiling approaches to these limited cell sources. Several methods have been presented for the amplification and labeling of small quantities of total RNA, which is then suitable for high density oligonucleotide microarray analysis 5, 6, 7 as well as cDNA microarray analysis. 8, 9, 10 However, the overall sensitivity and reproducibility of linear transcript amplification is still under-documented. Furthermore, the protocols used are often tedious and time-consuming.
In an effort to streamline and standardize a process for routinely generating biotin-labeled hybridization targets from limiting cell sources (such as LCM), we have used a commercially available reagent kit. In this report, we compare data generated using this reagent system to that obtained from previously reported, standard protocols. Starting with as little as 10 nanograms of total RNA and using two rounds of linear amplification, a sufficient quantity of labeled target is generated for microarray hybridization. We demonstrate that this procedure is reproducible and results in sensitivity comparable to standard methodologies that routinely employ 200 to 1000 times greater input RNA. The method is easy to use, and reproducibility of expression profiles from duplicate dissections of identical cell populations suggests that the method is reliable for routine microgenomics, addressing complex genomic questions from limiting cell sources.
Materials and Methods
Laser Capture Microdissection
Human tissue specimens were obtained from the Alvin J. Siteman Cancer Center Tissue Procurement Core facility using an IRB-approved protocol. Embedded, frozen tissue specimens were cut as a series of 6-μm thick sections and immediately fixed in 70% ethanol. Subsequently, tissue sections were stained as previously described. 5 In brief, slides were sequentially dipped five times in deionized (DI) water, 10 times in Mayer’s hemotoxylin solution (Sigma, St. Louis, MO), DI water, 1X automation buffer (Biomeda, Foster City, CA), and DI water. Slides were then dehydrated for 60 seconds each in 70% ethanol and 95% ethanol, stained for 15 seconds in alcoholic eosin (Sigma, St. Louis, MO), rinsed with ten dips in 95% ethanol, and placed for 60 seconds in 95% ethanol. Finally, slides were further dehydrated by 10 dips in 100% ethanol, two 60-second washes in 100% ethanol, 10 dips in xylene, and two incubations of 3 minutes each in xylene. Slides were air-dried for 5 minutes and stored in a desiccator for no more than 6 hours before dissection. Areas containing non-malignant ductal epithelial cells were independently isolated from the slides using the PixCell II LCM system (Arcturus, Mountain View, CA). Between 4300 and 6800 laser pulses (30-μm beam diameter; 30 mW power) yielded an estimated average of 15,000 cells for each independent dissection.
Total RNA
Total RNA from human heart and human tumor cell line G-401 was obtained from Ambion (Austin, TX). Total RNA from the human endometrial adenocarcinoma cell line An3CA (American Type Culture Collection (ATCC), HTB-111) was isolated using an affinity resin spin column following the manufacturers’ recommendations (RNeasy, Qiagen, Chatsworth, CA). Breast tissue RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA) followed by further purification with the RNeasy RNA isolation system. RNA was qualitatively assessed by agarose gel electrophoresis or RNA LabChip (Agilent, Palo Alto, CA), quantified by UV absorbance, and diluted to the appropriate working concentration. For microdissected tissue samples, several LCM-caps were pooled into a single tube containing 200 μl of denaturing buffer (GITC) and 1.6 μl of 2-mercaptoethanol (BME). Total RNA was then extracted using a modified protocol of the Stratagene RNA microisolation kit (Stratagene, La Jolla, CA) as previously described. 5 The total RNA obtained from each LCM-dissected tissue was resuspended in 10 μl of Rnase-free water. To assess the quality and concentration of the total RNA, 1 μl was directly analyzed on an RNA LabChip (Agilent) following the manufacturer’s instructions.
Target Synthesis
For biotin-labeled, antisense cRNA target (“cRNA”) synthesis starting from either 2 μg or 10 μg of total cellular RNA, reactions were performed using one of two protocols.
Standard Protocol
Targets were generated using standard protocols supplied by the manufacturer (Affymetrix Inc., Santa Clara, CA) and as previously described. 3
RiboAmp Protocol
Reactions were performed using the RiboAmp RNA amplification kit (Arcturus, Mountain View, CA) following the manufacturer’s protocol for performing one round of amplification with the following modification. After cDNA synthesis and purification, 8 μl of the resulting cDNA was added to the BioArray High Yield in vitro transcription reaction (see below) to generate biotinylated cRNA.
For target synthesis starting from 10 ng of total RNA or LCM extracted RNA, an initial round of amplification was performed before synthesis of biotin-labeled cRNA using one of two protocols.
Standard Two-Round Protocol
In the first method previously described, 4 synthesis of first- and second-strand cDNA was performed using the standard protocol provided by the manufacturer (Affymetrix Inc.). However, instead of proceeding to use the double-stranded cDNA in the biotin-labeled in vitro transcription reaction, the cDNA was resuspended in 8 μl of Rnase-free water and used as a template to transcribe unlabeled RNA using T7 RNA polymerase and the Megascript kit (Ambion). The reaction was incubated for 4 hours at 37°C and the resulting transcribed RNA was purified using RNeasy spin columns (Qiagen). Eluted RNA was precipitated by adding 0.1 volume of 7.5 mol/L ammonium acetate, 0.02 volumes of 5 mg/ml linear acrylamide (Ambion) and 2.5 volumes of 100% ethanol, and resuspended in 10 μl of Rnase-free water. A second round of amplification was initiated by using the in vitro transcribed RNA as template. After annealing RNA with 0.7 μmol/L random hexamers (Pharmacia, Piscataway, NJ) for 10′ at 70°C, the mixture was chilled on ice and extended in a 20-μl reaction containing 4 μl of 5X first-strand reaction buffer, 2 μl of 0.1 mol/L dithiothreitol (DTT), 1 μl of 10 mmol/L dNTPs, and 1 μl of Superscript II (Life Technologies, Rockville, MD). Following a 1-hour incubation at 42°C, 1 μl of 2 units/ml of RNase H was added, incubated for 20 minutes at 37°C, and inactivated at 95°C for 5 minutes. The resulting first-strand cDNA was annealed to 100 pmol of HPLC-purified T7T24 primer (GenSet, La Jolla, CA) for 10′ at 70°C. Then, second-strand cDNA synthesis was performed by adding 90 μl of Rnase-free water, 30 μl of 5X second-strand reaction buffer, 3 μl of 10 mmol/L dNTPs, 10 units DNA ligase, 40 units DNA polymerase and 2 units of RNase H. After incubating the second-strand cDNA reaction for 2 hours at 16°C, 20 units of T4 DNA polymerase were added, followed by incubation at 16°C for 10′. The second-strand cDNA synthesis was stopped by adding 10 μl of 0.5 mol/L EDTA. Double-stranded cDNA was purified by phenol:chloroform:isoamyl alcohol extraction using phase-lock-gel (Eppendorf, Westbury, NY), precipitated with 0.5 volumes of 7.5 mol/L ammonium acetate, 2 μg of glycogen and 2.5 volumes of 100% ethanol, and resuspended in 22 μl of Rnase-free water.
RiboAmp Two-Round Protocol
In the second method, the RiboAmp RNA amplification kit (Arcturus) was used following the protocol for performing first and second rounds of amplification with the following modification. After the second round of cDNA synthesis and purification, 8 μl of the resulting cDNA was added to the BioArray High Yield in vitro transcription reaction (see below) to generate biotinylated cRNA.
Biotin-Labeled cRNA Transcription and Microarray Hybridization
Biotinylated cRNA target was generated from all cDNAs using the Bioarray High Yield transcription kit (Enzo Biochemical, New York, NY) following the manufacturer’s protocol. After a 5-hour incubation at 37°C, the final biotin-labeled cRNA product was purified using RNeasy spin columns (Qiagen) and eluted in 40 μl of Rnase-free water. The concentration of biotin-labeled cRNA was determined by UV absorbance. In all cases, 10 micrograms of each biotinylated cRNA preparation was fragmented, assessed by gel electrophoresis, and placed in a hybridization cocktail containing four biotinylated hybridization controls (BioB, BioC, BioD, and Cre) as recommended by the manufacturer. To initially assess cRNA quality, a subset of samples was first hybridized to GeneChip Test3 microarrays as previously described. 7 Samples were then hybridized to HU-95Av2 GeneChip arrays for 16 hours. Microarrays were washed and stained using the instrument’s standard “Eukaryotic GE Wash 2” protocol, using antibody-mediated signal amplification.
Data Analysis
The images from the scanned chips were processed using Microarray Analysis Suite 4.0 (Affymetrix Inc.). Image data from each microarray was individually scaled to an average intensity of 150. Scaled average difference value (SADV) and absolute call (AC) data were exported to flat text files and used for numerical analysis. Raw image data and numerical data sets used for the analysis described in this study are available at http://bioinformatics.wustl.edu.
Results and Discussion
The purpose of the present study was to compare the sensitivity and reproducibility of a new transcript amplification protocol for expression microarray analysis in two independent laboratories. We first examined the yield and size distribution of biotinylated cRNA starting from different sources and amounts of total RNA and implementing different protocols for transcript amplification. Each sample was performed in duplicate to assess reproducibility. Table 1 summarizes the samples, the methods used, and the resulting yields of biotinylated cRNA. Starting with as little as two micrograms of total cellular RNA, approximately 20 micrograms of biotin-labeled cRNA is generated using the “Standard” Gubler and Hoffman 11 approach to cDNA synthesis, organic extraction, and VanGelder 12 method for T7 polymerase-mediated in vitro transcription (Protocol A). The same amount of input RNA yields three to four times more product (60 to 86 micrograms) when the “Riboamp” method is used (Protocol B). In this procedure, second-strand cDNA synthesis occurs through priming with randomers rather than nicking and strand displacement. The cDNA is then purified using charge-affinity spin chromatography before in vitro transcription. It is likely that the increased efficiency of exogenous primer-mediated second-strand synthesis results in the increased yields. Starting with 10 to 30 nanograms of total RNA and using the “RiboAmp Two-Round” protocol for transcript amplification (Protocol D), 20 to 50 micrograms of biotinylated target was produced. Again, this is three to four times more product than the “Standard Two-Round” method for T7 polymerase-mediated transcript amplification (Protocol C, ref. 5 ). In all cases, the resulting yield of labeled cRNA is more than the requisite 10 micrograms needed to hybridize to the GeneChip microarray.
Table 1.
cRNA Yield and Hybridization Results
| Source | Input | Method | Yield (ug) | 3′/5′ | SF | % Genes = P | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. 1 | No. 2 | No. 1 | No. 2 | No. 1 | No. 2 | No. 1 | No. 2 | |||
| Breast cancer | 10 ug | Standard (A) | 40 | 46 | 3.1 | 3.4 | 1.0 | 1.1 | 55 | 55 |
| An3Ca cell line | 2 ug | Standard (A) | 24 | 22 | 1.0 | 1.1 | 0.8 | 0.6 | 51 | 54 |
| G401 cell line | 2 ug | Standard (A) | 24 | 22 | 1.0 | 1.1 | 1.8 | 1.3 | 54 | 53 |
| NI. heart | 2 ug | Standard (A) | 18 | 20 | 1.3 | 1.2 | 1.4 | 1.4 | 56 | 54 |
| An3Ca cell line | 2 ug | RiboAmp 1 Round (B) | 60 | 56 | 3.1 | 3.5 | 0.7 | 1.2 | 51 | 47 |
| G401 cell line | 2 ug | RiboAmp 1 Round (B) | 86 | 74 | 4.5 | 5.3 | 1.5 | 1.6 | 49 | 46 |
| NI. heart | 2 ug | RiboAmp 1 Round (B) | 72 | 68 | 4.7 | 4.1 | 1.2 | 2.5 | 43 | 49 |
| Breast cancer | 10 ng | Standard 2 Round (C) | 16 | 16 | 4.4 | 4.0 | 5.8 | 4.2 | 36 | 40 |
| Breast cancer | 10 ng | RiboAmp 2 Round (D) | 36 | 24 | 49 | 55 | 3.5 | 2.2 | 40 | 43 |
| An3Ca cell line | 10 ng | RiboAmp 2 Round (D) | 53 | 47 | 43 | 66 | 2.7 | 5.4 | 36 | 35 |
| Normal, heart | 10 ng | RiboAmp 2 Round (D) | 55 | 26 | 57 | 50 | 7.8 | 4.4 | 32 | 35 |
| G401 cell line | 10 ng | RiboAmp 2 Round (D) | 54 | 43 | 66 | 51 | 3.0 | 5.0 | 37 | 36 |
| LCM breast 1 | 30 ng | RiboAmp 2 Round (D) | 40 | 40 | 25 | 33 | 1.1 | 1.7 | 44 | 39 |
| LCM breast 2 | 30 ng | RiboAmp 2 Round (D) | 18 | 18 | 30 | 67 | 2.1 | 5.4 | 43 | 36 |
Source, source of RNA; input, amount of total cellular RNA used for target synthesis; method, method used to generate biotinylated cRNA (see materials and methods); yield, yield of purified cRNA from two, duplicate target synthesis reactions; 3′/5′, ratio of hybridization signals obtained from GeneChip probe pair sets directed at the 3′ or 5′ end of GAPDH transcript; SF, scale factor utilized to generate an average signal intensity of 150 using Microarray Analysis Suite (MAS) 4.0 software; % genes = P, percentage of genes scored detected (“P”) by MAS 4.0.
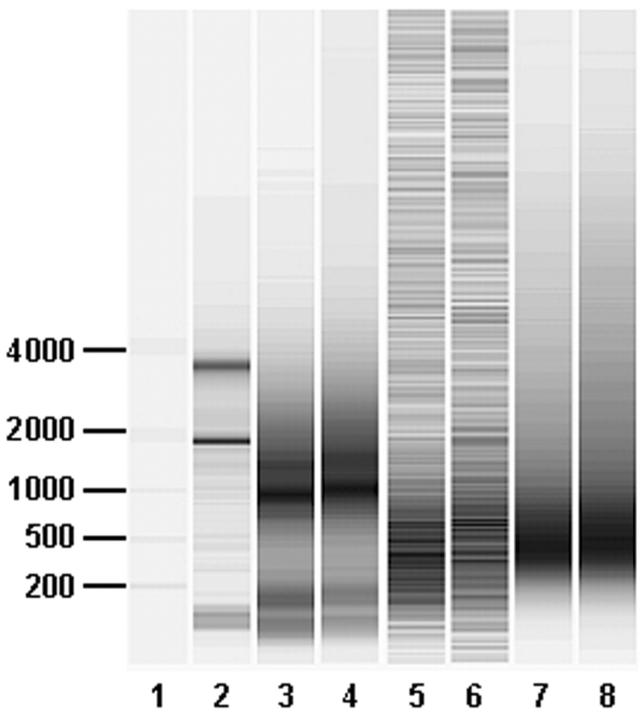
Qualitative electrophoretic analysis of the resulting cRNA targets (Figure 1) revealed that material generated using the “Standard (A)” protocol (lanes 3 and 4) has a mean size distribution of 1000 nucleotides while cRNA generated from the “Riboamp (D)” protocol (lanes 5 and 6) has a mean size distribution of approximately 500 nucleotides. Interestingly, a successive round of amplification of this cRNA (lanes 7 and 8) results in product of similar size distribution. That is, a further round of amplification does not further reduce the mean length of the transcript pool.
Figure 1.
Electrophoretic analysis of aRNA targets. One microliter of each cRNA target was analyzed on an RNA LabChip and Agilent bioanalyzer. Lane 1: Molecular weight ladder with marker sizes indicated in base pairs. Lane 2: Input breast cancer total cellular RNA used for target generation. Lanes 3 and 4: Duplicate targets synthesized using “Standard protocol (A)” and starting with 10 μg of total RNA. Lanes 5 and 6: Duplicate cRNA samples generated from 10 ng of total RNA after the first round of amplification using the “RiboAmp Two-Round (D)” protocol. Lanes 7 and 8: Duplicate aRNA samples generated from 10 ng of total RNA after the second round of amplification using the “RiboAmp Two-Round (D)” protocol. To simultaneously visualize cRNA after both first and second rounds of amplification, the signal intensity in each lane has been scaled to itself so that lane-to-lane quantitative comparisons are not valid. Background bands in lanes 5 and 6 are system noise that is detected due to the low specific signal obtained from the first round amplification cRNA product.
To address whether the shorter mean length of the transcript pool results in loss of specific hybridization signal, we first examined the hybridization signal intensity of probe sets interrogating the 3′ and 5′ ends of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript. Measurement of signal from the 3′ and 5′ ends is a quality control measure for robust, full-length target synthesis. As shown in Table 1 , the GAPDH 3′/5′ ratios range from 1.0 to 3.1 when using the “Standard (A)” protocol and are not much greater when either a single round of the “RiboAmp (B)” protocol or “Standard Two-Round (C)” amplification protocol are used. However, when the “Riboamp Two-Round (D)” protocol is used to make labeled target from 10 ng of RNA, the 3′ and 5′ hybridization signal ratios increase at least tenfold. Given the shorter mean size of Riboamp-generated cRNA targets, it is perhaps not surprising to see a relative decrease in hybridization signal from the 5′ end of the transcript. However, since the majority of probe sets on GeneChip microarrays represent the 3′ end of each transcript, we hypothesized that this effect would not have a major impact on the detection of the majority of other cellular transcripts represented on the arrays. This finding precludes the use of 5′:3′ signal ratios of control transcripts as a reliable measure of target quality when performing the “RiboAmp Two-Round (D)” protocol. Instead, we have used synthetic polyadenylated transcripts as internal controls 5 or simply use the absolute, scaled signal intensity of the control transcript 3′ probe sets and the number of transcripts scored as detected (“P”) to assess overall target quality.
To address the overall method sensitivity, we examined two parameters that reflect signal intensity. Scale factor (SF) is the ratio of a defined target intensity value (in this study, 150) to the average signal intensity of all probe pairs on the microarray. The higher the SF, the lower the overall signal intensity. The number of transcripts detected (% P), is a measure of the cRNA target complexity and reflects not only the intensity of hybridization, but also the number of probe sets that have a detectable, specific hybridization signal. As shown in Table 1 , SF and % P values from targets generated with 2 to 10 μg of RNA using the “Standard (A)” protocol average 1.2 and 54%, respectively. There is no statistical difference (Student’s t-test, P < 0.05) between SF values from targets generated from the “RiboAmp (B)” versus the “Standard (A)” protocols, but a statistical difference between the total number of transcripts called detected (54% vs. 47%). When the “RiboAmp (B)” versus the “RiboAmp Two-Round (D)” protocols are compared there is a statistically significant difference between the scaling factor (1.3 vs. 3.7) and the percentage of transcripts called detected (51% vs. 38%). However, results after two rounds of amplification are comparable whether the “RiboAmp (D)” or the “Standard (C)” protocols are used. A review of several representative probe pair sets with discrepant detection calls revealed that these differences can be primarily attributed to probe pairs directed at more 5′ regions of transcripts whose signal is lost after two rounds of amplification. This observation is consistent with the results obtained from the 3′-specific and 5′-specific signals of control transcripts described above. Complete and selective loss of signal from more 5′-oriented probe pairs, rather than a general diminution of signal across all probe pairs, accounts for lower overall chip signal intensity and correspondingly higher SF. Also, probe pair sets that are more 5′-oriented will have fewer discrete positive probe pair signals and this, in turn, will result in an “A” (non-detected) call from the GeneChip detection algorithm. Given that 200- to 1000-fold less starting RNA is required for the two-round amplification protocol, the modest decrease in performance (in terms of number of transcripts scored detected) seems acceptable. In fact, using the “RiboAmp Two-Round (D)” protocol, it may be advisable to ignore absolute call data and use only signal or fold change calculation data (see below).
We did not perform extensive direct comparisons between one-round amplification methods starting with 2 to 10 micrograms of RNA and two-round amplification methods starting with 10 nanograms of the same RNA. As expected, gene expression correlation between duplicates using the same methodology is higher (r = 0.98 to 0.99) as compared to correlation between duplicates using different methodologies and different amounts of starting material (r = 0.70 to 0.77). We examined several specific transcripts whose relative levels of expression were most discrepant between methods. Transcripts whose levels of expression are most under-represented in the 10-nanogram sample relative to the 10-microgram sample of breast cancer RNA have both high and low level signals in the 10-microgram sample. Conversely, many transcripts with low signal levels in the 10-microgram sample demonstrate very good correlation with signals obtained from the 10-nanogram sample. Transcripts whose level of expression are most over-represented in the 10-nanogram sample relative to the 10-microgram sample were also found to have both high and low level signals in the 10-microgram sample. We also examined the location of probe set features relative to the 3′ end of their corresponding transcript for 20 transcripts that demonstrated the highest correlation between methods and 20 transcripts that were the most discrepant between methods. There is no immediately apparent relationship between the location of probe features relative to the 3′ end of each transcript and relative change in signal intensity between the two methods. For the individual transcripts examined, neither the absolute level of gene expression nor the probe set feature location were well-correlated to the relative change in signal intensity between the two methods. It should also be emphasized that an appropriate experimental design would seldom try to compare data from mixed methodologies in this way, making the significance of these between-method comparisons less relevant.
Technical reproducibility is an essential requirement for successful microarray experiments and so, to address the reproducibility of the amplification methods used in this study, each target synthesis was performed in duplicate from the same starting RNA. As shown in Table 2 , we examined the number of discrepant absolute calls (“A” vs. “P”) between the duplicates. In addition, using the Affymetrix microarray analysis software, we directly compared duplicate arrays to each other and determined the number of genes with expression changes of greater than two-fold. Overall, the percentage of discordant absolute calls (detected, “P” vs. not detected, “A”) is higher than the percentage of transcripts that demonstrate a greater than twofold change in expression. This indicates that, as expected, many of the discrepancies between duplicate samples occur in transcripts whose levels of expression are just within or below the level of confident detection. That is, small (less than twofold) differences in signal intensity are sufficient for the algorithm to alternate between scoring these transcripts “A” or “P.” Between “Standard (A)” and “RiboAmp (B)” single-round amplification protocols, and between “RiboAmp (A)” and “RiboAmp Two-Round (D)” protocols, there is no statistical difference in the percentage of transcripts with discrepant absolute calls or in fold change values of greater than two (t-test, P < 0.05). This data suggests that technical reproducibility is not compromised by starting with 1000-fold less input RNA and using two rounds of transcript amplification.
Table 2.
Reproducibility of Hybridization Results
| Source | Input | Method | “A” vs. “P” | FC > 2 |
|---|---|---|---|---|
| Breast cancer | 10 ug | Standard (A) | 8.1% | 0.3% |
| An3Ca cell line | 2 ug | Standard (A) | 6.8% | 1.5% |
| G401 cell line | 2 ug | Standard (A) | 6.7% | 0.9% |
| NI. heart | 2 ug | Standard (A) | 8.8% | 1.0% |
| An3Ca cell line | 2 ug | RiboAmp 1 Round (B) | 8.1% | 1.9% |
| G401 cell line | 2 ug | RiboAmp 1 Round (B) | 9.5% | 4.3% |
| NI. heart | 2 ug | RiboAmp 1 Round (B) | 10.8% | 2.9% |
| Breast cancer | 10 ng | Standard 2 Round (C) | 11.5% | 1.0% |
| Breast cancer | 10 ng | RiboAmp 2 Round (D) | 11.6% | 1.0% |
| An3Ca cell line | 10 ng | RiboAmp 2 Round (D) | 10.0% | 3.4% |
| Normal, heart | 10 ng | RiboAmp 2 Round (D) | 11.3% | 0.9% |
| G401 cell line | 10 ng | RiboAmp 2 Round (D) | 10.4% | 1.6% |
| LCM breast 1 | 30 ng | RiboAmp 2 Round (D) | 11.8% | 4.6% |
| LCM breast 2 | 30 ng | RiboAmp 2 Round (D) | 14.5% | 4.0% |
Source, source of RNA; input, amount of total cellular RNA used for target synthesis; method, method used to generate biotinylated cRNA (see materials and methods); “A” vs. “P”, percentage of genes in duplicate samples with discrepant absolute calls of detected (“P”) versus not detected (“A”) as determined by MAS 4.0 software; FC > 2, percentage of genes in duplicate samples with apparent expression fold change of greater than 2.0 as determined by MAS 4.0 software. Note that figures in bold italics represent independent RNA samples from duplicate LCM dissections.
Finally, in preparation for studies designed to compare expression profile differences between microdissected cell populations, we assessed the total experiment variability associated with two independent laser capture microdissections, RNA isolations, transcript amplifications, and microarray hybridizations. Two independent isolations of the same population of normal human breast epithelial cells were performed from serial sections in two independent breast biopsy specimens (“LCM Breast 1,” “LCM Breast 2”). As shown in Table 2 , the total experimental variability of this process (as measured by the number of transcripts demonstrating an apparently greater than twofold change between independent dissections of serial sections) is approximately 4.0 to 4.6%. As expected, this variability is higher than that seen from simply performing two independent target syntheses from the same RNA isolation. Technical variability associated with tissue processing, microdissection, and RNA isolation as well as biological microheterogeneity undoubtedly contributes to this increased variability. This measurement provides a useful baseline estimate for the total gene expression variability associated with this experimental approach and suggests that the methodology outlined in this study is still sufficiently robust to reliably measure relatively subtle changes in gene expression profiles from microdissected cell populations. Accordingly, we are now analyzing differences in gene expression between cell populations (ie, normal epithelium, carcinoma in situ, and invasive carcinoma) that have been microdissected from single sections of human breast tumors. In on-going studies, three- to fivefold changes in gene expression initially detected by transcript amplification and GeneChip microarrays have been confirmed by quantitative RT-PCR (Watson MA, manuscript in preparation).
In summary, we have used a commercially available transcript amplification kit to successfully generate biotinylated cRNA targets for hybridization to GeneChip expression microarrays. The protocol may be used with one round of in vitro transcription to generate target from as little as two micrograms of total RNA. Alternatively, two rounds of in vitro transcription may be used to generate target from as little as 10 nanograms of total RNA. Although starting with nanogram quantities of RNA appears to sacrifice some sensitivity (as judged by the total number of transcripts scored as detected, “P”), the method demonstrates comparable reproducibility to existing protocols and performs equally well when directly compared to previously reported methods. 5 In addition, the new method is faster and more convenient, and produces higher yields of labeled cRNA target. The applicability of this method toward obtaining expression profile data from microdissected tissue specimens has been demonstrated here and should provide a useful resource for future studies in “microgenomics.”
Acknowledgments
We thank Chunmei Liu, Kate Hamilton, and Theresa Taylor for technical assistance.
Address reprint requests to Mark A. Watson, M.D., Ph.D., Department of Pathology and Immunology, Box 8118, Washington University School of Medicine, 660 S. Euclid Avenue, St. Louis, MO 63110. E-mail: watsonm@labmed.wustl.edu.
Footnotes
Supported in part by grant number 016-99 from the Mary Kay Ash Charitable Foundation (M.A.W.) and the Alvin J. Sikman Cancer Center.
Veronica Luzzi and Mamatha Mahadevappa should be regarded as joint first authors.
References
- 1.Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, Brown EL: Expression monitoring by hybridization to high-density oligonucleotide arrays. Nature Biotechnol 1996, 14:1675-1680 [DOI] [PubMed] [Google Scholar]
- 2.Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ: High density synthetic oligonucleotide arrays. Nat Genet 1999, 21S:20-24 [DOI] [PubMed] [Google Scholar]
- 3.Mahadevappa M, Warrington JA: A high-density probe array sample preparation method using 10- to 100-fold fewer cells. Nature Biotechnol 1999, 17:1134-1136 [DOI] [PubMed] [Google Scholar]
- 4.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA: Laser capture microdissection. Science 1996, 27:998-1001 [DOI] [PubMed] [Google Scholar]
- 5.Luzzi V, Holtschlag V, Watson MA: Expression profiling of ductal carcinoma in situ by laser capture microdissection and high density oligonucleotide arrays. Am J Pathol 2001, 158:2005-2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohyama H, Zhang X, Kohno Y, Alevizos I, Posner M, Wong DT, Todd R: Laser capture microdissection-generated target sample for high-density oligonucleotide array hybridization. Biotechniques 2000, 29:530-536 [DOI] [PubMed] [Google Scholar]
- 7.Alevizos I, Mahadevappa M, Zhang X, Ohyama H, Kohno Y, Posner M, Gallagher GT, Varvares M, Cohen D, Kim D, Kent R, Donoff RB, Todd R, Yung CM, Warrington JA, Wong DT: Oral cancer in vivo gene expression profiling assisted by laser capture microdissection and microarray analysis. Oncogene 2001, 20:6196-6204 [DOI] [PubMed] [Google Scholar]
- 8.Luo L, Salunga RC, Guo H, Bittner A, Joy KC, Galindo JE, Xiao H, Rogers KE, Wan JS, Jackson MR, Erlander MG: Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat Med 1999, 5:117-122 [DOI] [PubMed] [Google Scholar]
- 9.Wang E, Miller LD, Ohnmacht GA, Liu ET, Marincola FM: High-fidelity mRNA amplification for gene profiling. Nature Biotechnol 2000, 18:457-459 [DOI] [PubMed] [Google Scholar]
- 10.Leethanakul C, Patel V, Gillespie J, Pallente M, Ensley JF, Koontongkaew S, Liotta LA, Emmert-Buck M, Gutkind JS: Distinct pattern of expression of differentiation and growth-related genes in squamous cell carcinomas of the head and neck revealed by the use of laser capture microdissection and cDNA arrays. Oncogene 2000, 19:3220-3224 [DOI] [PubMed] [Google Scholar]
- 11.Gubler U, Hoffman BJ: A simple and very efficient method for generating cDNA libraries. Gene 1983, 25:263-269 [DOI] [PubMed] [Google Scholar]
- 12.Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH: Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci USA 1990, 87:1663-1667 [DOI] [PMC free article] [PubMed] [Google Scholar]