Abstract
DNA amplification in cancer cells frequently involves oncogenes whose increased expression confers a selective advantage on tumor cell growth. In an attempt to identify novel oncogenes involved in hepatocarcinogenesis, representational difference analysis (RDA) was performed using DNA from a primary human hepatocellular carcinoma (HCC) that showed high-level DNA amplifications on chromosomes 1p32 and 11q13 by comparative genomic hybridization. Ten amplification fragments were isolated by RDA, and when used to probe Southern blots of tumor DNA, there was a 5- to 50-fold increase in hybridization intensity relative to normal DNA. The sequence of one amplification product matched that of the EMS1 oncogene, which is located on chromosome 11q13 and is amplified in other cancers. We detected EMS1 amplification in 3 of 17 primary HCC. Overexpression of EMS1 mRNA was observed in 12 of 14 HCC cell lines in the absence of gene amplification or an increased copy-number of the gene. The EMS1 gene encodes cortactin, a cortical actin-associated protein that is a substrate for Src kinase and is involved in cytoskeleton organization. Alterations of the EMS1 gene that lead to overexpression of cortactin may be associated with tumor development in HCC. EMS1 amplification and overexpresion is indicative of unfavorable prognosis in several cancers and may have similar prognostic implications in liver cancer.
Gene amplification is a common alteration in cancer cells, and like other structural changes it is associated with genomic instability and contributes to the process of carcinogenesis. DNA amplification and gene overexpression frequently involve proto-oncogenes that play an important role in the loss of cell cycle control in cancer. 1, 2 In liver cancer, we have reported recurrent DNA copy-number gain on chromosome 11 and the minimal region of gain at 11q13 by comparative genomic hybridization (CGH) analysis of human hepatocellular carcinoma (HCC) cell lines, 3 and others detected amplification of 11q13 in primary HCC. 4, 5, 6, 7, 8 The cyclin D1 gene (CCND1) at 11q13 was found to be amplified or overexpressed in a significant number of HCC, and in certain cases was co-amplified with the FGF3/INT 2 gene. In adjacent non-cancerous liver tissue, cyclin D1 was neither amplified or overexpressed. 9, 10, 11 Chromosome 11q13 amplification is also frequently found in head and neck, oral, lung, gastric, and breast cancer and has clinical relevance as it relates with a poor clinical course of the disease. 12
In an attempt to identify novel oncogenes in liver cancer, we used representational difference analysis (RDA) for detecting sequences that are over-represented in the tumor genome, and we successfully isolated a fragment homologous to the EMSI proto-oncogene. The EMS1 gene encodes cortactin, an actin-binding protein that is a substrate of the src tyrosine kinase. 12, 13, 14 EMS1 has been localized to the 11q13 amplicon that also contains CCND1, two fibroblast growth factor genes (FGF3/INT2 and FGF4/HSTF1) and RIN, a putative effector of the RAS and ABL oncogenes. 12, 15, 16 The isolation of an amplified fragment homologous to EMSI prompted us to examine whether alterations in the gene are present in primary HCC and HCC-derived cell lines.
Materials and Methods
Primary HCC Sample and HCC Cell Lines
All primary tumor DNAs were obtained from surgical resection of HCC tissues from patients from Qidong province, China. The population in this region is exposed to aflatoxin B1 and infected with the hepatitis B virus (HBV). In the majority of cases the tumors were at an advanced stage of the disease and positive for HBV. Each tumor sample was matched with its surrounding non-cancerous liver tissue. DNAs were extracted after diagnosis as hepatocellular carcinoma with or without cirrhosis. HCC cell lines Huh-7, Hlf, Wrl, Hep-40, 7703, Hep-G2, Focus, Hle, Sk-Hep-1, Plc/Prf/5, Huh-1 and Hep-3B were obtained from American Type Culture Collection (Rockville, MD) or kindly provided by Dr. Curtis Harris (Laboratory of Human Carcinogenesis, Center for Cancer Research, National Cancer Institute). They are either negative or positive for hepatitis B virus, and were previously analyzed by CGH. 3
Representational Difference Analysis (RDA)
RDA was performed as previously described in detail by using tumor DNA as tester and normal liver DNA as driver. The RDA products were cloned into pSP72 (Promega, Madison, WI) and characterized as previously described. 17 The sequences of the inserts were used to search the GenBank databases and the human genome database at ENSEMBL (www.ensembl.org).
Southern Blot Analysis
Genomic DNAs extracted from human primary HCC were digested with the BglII restriction enzyme, fractionated in 1% agarose gels, and transferred to a nylon membrane. The membrane was hybridized to the radiolabeled L1–1, L1–2, L3–2, and L4–7 RDA fragments in Rapid-Hyb buffer (Amersham, Piscataway, NJ). The blot was washed for 30 minutes at 62°C in 0.1% SSC/0.1% SDS, exposed to a PhosphorImager plate, analyzed by Software ImageQuant Version 3.3 (Molecular Dynamics, Sunnyvale, CA), and then stripped and rehybridized to a radiolabeled b-actin cDNA probe.
Northern Blot Analysis
RNA was extracted from HCC cell lines and normal liver tissue using TRIzol reagent (Invitrogen/Life Technologies, Carlsbad, CA). Polyadenylated RNA was isolated from total RNA by chromatography using an oligodeoxythymidylic acid column (Bio-Rad). One mg of mRNA for each sample was resolved on 1% agarose-4-morpholinepropanesulfonic acid/formaldehyde gels and transferred to a nylon membrane. The blot was hybridized to the radiolabeled L3–2 RDA fragment, derived from the EMS1 gene, and then rehybridized to a GAPDH cDNA probe. The same procedures for hybridization, membrane washing, detection, and analysis used in Southern blot analysis were also used in Northern blot analysis.
PCR Analysis of EMS1 Gene Amplification
Genomic DNAs from 17 samples of paired normal and HCC tumor tissues were used for PCR analysis. The genes for EMS1 and tyrosine hydroxylase (TH, an internal control), both located on chromosome 11, were amplified in a multiplex PCR reaction. 18 The EMS1 gene primers (5′-TCCCCTGATGCCCAGGTC and 5′-TCCCAATCCAGAGACCCG) amplified a 111-bp product, and the TH gene primers (5′-GCCCCAGCTGCATCCTAC and 5′-CTTGGCAGACACCTGGGG) amplified a 188-bp product. After PCR, 10 μl of each sample were analyzed by agarose gel electrophoresis, and the products visualized by staining with ethidium bromide. The images of the UV-illuminated gels were captured using a digital camera and the EMSI:TH band ratios were determined by computerized densitometry (Laboragerate GmbH, Germany).
Molecular Cytogenetics Analysis
For both CGH and reverse chromosome painting (RCP) analyses, chromosomes were prepared from methotrexate-synchronized normal peripheral lymphocyte cultures. The CGH and RCP protocols, with minor modifications, were previously reported, 3, 17 using genomic DNA from the 94–25 primary HCC and the adjacent normal liver tissue DNA. A P1 genomic clone containing the EMS1 gene was labeled with biotin and used for fluorescence in situ hybridization (FISH) of chromosomes prepared from HCC cell lines. The conditions of hybridization, the detection of hybridization signals, digital-image acquisition, processing and analysis, and direct fluorescent signal localization on banded chromosomes were performed as previously described. 19, 20
Results
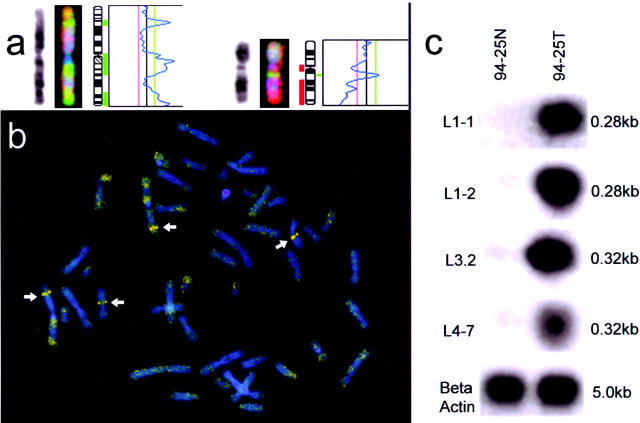
Twenty primary HCC DNA samples were screened for cross-contamination with normal tissue by PCR amplification of exon 7 of p53 gene followed by HaeIII digestion. One of the samples, designated 94–25, showed a homozygous point mutation in the p53 gene that was absent in the adjacent non-tumor tissue, and was selected for further analysis. The CGH profile of this tumor consisted of DNA copy number gains on chromosomes 1p/q, 7,8q, 11q, 17q, 20p/q, 22q, Xq and Y and losses on 8p, 9q, 11q, and 17p. Two discrete regions of gain at 1p32 and 11q13 were regions of high-level DNA amplification as depicted by RCP, which detects a minimum 20-fold amplification 21 (Figure 1a and b) . We proceeded with RDA, using the tumor DNA as tester DNA, and normal DNA as driver. Difference products were observed after the second round of hybridization/selection in the form of multiple diffuse bands. After the third round of subtractive hybridization, two distinct DNA bands were recovered and cloned into the pSP72 vector. One hundred individual clones from each ligation were randomly picked and plasmid DNA was prepared for restriction enzyme analysis and DNA sequencing. From 20 individual clones, 10 were examined for DNA amplification by Southern blot analysis. All fragments tested showed a 5- to 50-fold amplification in DNA from the 94–25 tumor compared with DNA from normal tissue (Figure 1c) .
Figure 1.
Isolation of genomic DNA fragments amplified in HCC by representational difference analysis. Comparative genomic hybridization profile for chromosomes 1 and 11 in hepatocellular carcinoma 94–25 (a), and detection of high-level DNA amplification on chromosomes 1 and 11 of the same tumor by reverse chromosome painting (b). Southern blot analysis of amplified fragments obtained by RDA (c). Four RDA clones (L1–1, L1–2, L3–2, and LA-7) were radiolabeled and hybridized to Southern blots of BglII-digested genomic DNA from the 94–25 liver tumor (94–25T) and from the surrounding normal tissue (94–25N). The blots were re-hybridized to a b-actin probe as a control. The sizes of the bands detected by the probes are indicated.
BLAST searches of the GenBank database showed that the sequences of two of the amplification clones were present in human BAC clones that mapped to the 1p32 region (data not shown). The sequences of three clones were found in BAC clones localized to chromosome 11q13.1, one of the regions of amplification detected in the 94–25 tumor and corresponding to the intense fluorescent band on chromosome 11 detected by both CGH and RCP. The 0.32-kb insert of one amplified fragment, designated L3–2, was 99% identical to nt 122865–123190 of BAC CMB9–66H19 (GenBank AP000487) and contains exon 13 of the EMS1 gene (nt 1126–1195 of the cDNA; GenBank M98343), which has been mapped to chromosome 11q13. 13 The sequences of two clones, L1–1 and L1–2, were found in BAC clones (RP11–826F13 and RP11–736L3, respectively) that have been assigned just telomeric to the EMS1 gene in the draft human genome sequence.
As the EMS1 gene is amplified and overexpressed in several human cancers, 12 we examined 17 primary tumors and HCC cell lines for alterations in the EMS1 gene. By PCR analysis of genomic DNA from tumors, we observed a 2.5- to 3-fold increase in the EMS1 PCR product in two primary tumors as compared to an internal control and normal DNA counterparts (Figure 2a) . As EMS-1 was also amplified in primary tumor used for RDA, the total number of tumors with amplification is three. None of 12 HCC cell lines exhibited an increase in the EMS1 PCR product (Figure 2b). However, by Northern blotting, overexpression of EMS1 mRNA was detected in 12 of 14 HCC cell lines (Figure 2c) . To determine whether the increased expression in these cell lines is due to an increase in the copy-number of the gene, five cell lines (Hep-40, Hep3B, SK-Hep-1, PCL/PRF/5, 7703, and Focus) that have high levels of EMS1 mRNA and DNA copy-number over-representation of 11q13 by CGH 3 were examined by FISH with an EMS1 genomic probe and a chromosome 11 painting probe. Consistent with PCR data showing a lack of EMS1 gene amplification in these lines, the number of signals on intact or rearranged chromosome 11 were proportional with the ploidy level. Clusters of signals reflecting high-level amplification were not observed in any cell line. With the exception of the SK-Hep-1 line, which had three normal copies of chromosome 11, all of the lines had unbalanced translocations, complex rearrangements, or deletions involving chromosome 11 (Figure 3) . Representative spreads from PCL/PRF/5 and SK-Hep-1 cells after FISH with EMS1 genomic probe and rehybridization with chromosome 11 painting probe are shown in Figure 3 . PCL/PRF/5 cells have a deletion of the short arm of chromosome 11 (Figure 3 a and b) , while all three copies of chromosome 11 are normal in SK-Hep-1 cells (Figure 3 c and d) .
Figure 2.
Detection of amplification and overexpression of the EMSI gene in HCC. PCR analysis of EMSI amplification in DNA from primary HCC (a) and HCC cell lines (b), with tyrosine hydroxylase (TH) as an internal control. Northern blot analysis of EMSI mRNA levels in HCC cell lines and in normal liver (c). DNA from two primary HCC, cases 17 and 21, displayed a 3.2- and 2.9-fold amplification, respectively, compared with the normal counterpart (a). None of the HCC cell lines showed amplification (b). There is a very faint expression of EMS1 gene in normal liver cells but higher expression in HCC cell lines (3- to 7-fold increase as compared with normal liver cells). The blot was reprobed with GAPDH cDNA (c).
Figure 3.
Fluorescence in situ hybridization of chromosomes derived from PCL/PRF/5 and SK-Hep-1 cell lines. The chromosomes were hybridized to an EMS1 genomic probe (a and c) and rehybridized to a chromosome 11 painting probe (b and d). The number of fluorescent signals obtained with the EMS1-specific probe corresponds to the number of copies of chromosome 11q.
Discussion
The EMS1 gene was cloned from a human carcinoma cell line with an amplification of 11q13, and overexpression or amplification of EMS1 has been reported in several cancers. 12, 22, 23 The EMS1 gene encodes the human homologue of cortactin, which was first identified as a tyrosine-phosphorylated protein in Src-transformed primary chicken embryo cells. Cortactin binds to F-actin and promotes actin filament assembly through the activation of the Arp2/3 complex. 24 Cortactin also interacts with a number of other proteins via its SH3 domain and may be involved in organizing transmembrane complexes that regulate cell motility and membrane trafficking. 25 In Src-transformed cells and human breast carcinoma cells, cortactin is present in complexes that are associated with sites of increased extracellular matrix degradation. 25 Since cortactin overexpression does not affect the growth rate of carcinoma cells, 26 dysregulation of the EMS1 gene may lead to increased tumor cell motility and invasiveness. The results presented in this report show that overexpression of EMSI is common in HCC cell lines and that amplification of the gene occurs in a subgroup of primary liver tumors. This suggests that disruption of actin cytoskeleton associated with alterations in the EMS1 gene contributes to the process of hepatocarcinogenesis.
Besides being implicated in cancer development, oncogene amplification has clinical relevance as a prognostic marker for the course of the disease. The best known example is amplification or overexpression of the HER2/ErbB2 oncogene in breast cancer, which has provided not only a prognostic criterion for the recurrence of the disease but also led to the development of the first targeted therapy. 27, 28, 29 The amplification or overexpression of EMSI gene is considered to be a negative prognostic clinical marker in several cancers. 12 In liver cancer, gain on 11q13 is more common in the recurrent HCC than in the initial tumor. 7 In addition there appears to be an association between amplification at 11q13 and virus-related cases and geographical distribution. In one study, 11q13 amplification was preferentially found in hepatitis B virus (HBV)-positive HCCs 5 and in a second one, hepatitis C virus-related cases from Japan had a distinctively higher incidence of 11q13 gain. 6 In another study, HBV DNA integration near the HSTF1/FGF4 gene on 11q13 triggered co-amplification of both DNAs. 30 Possibly, in certain HCCs, due to regional amplification of 11q13, EMS1 is coamplified with other oncogenes and does not drive the amplification. The up-regulation of EMS-1 in the majority of the HCC cell lines suggests that mechanisms other than amplification may activate the gene. Recently it has been demonstrated that cortactin promotes bone metastasis of breast cancer cells, implicating EMS1 amplification and overexpression in the late stages of tumor development. 26 Further studies of the EMS1 gene in liver cancer will reveal whether it may serve as a useful molecular marker that correlates with the clinicopathological features and prognosis of the disease.
Address reprint requests to Nicholas C. Popescu, Ph.D., Laboratory of Experimental Carcinogenesis, National Cancer Institute, Building 37, Room 4128B, 37 Convent Drive MSC 4258, Bethesda, MD 20892-4258. E-mail: popescun@mail.nih.gov.
Footnotes
Current address for Bao-Zhu Yuan is the Toxicology and Molecular Biology Branch, Health Effects Laboratory Division, National Institute for Occupational Safety and Health, Morgantown, WV 26505.
References
- 1.Alitalo K, Schwab M: Oncogene amplification in tumor cells. Adv Cancer Res 1986, 47:235-281 [DOI] [PubMed] [Google Scholar]
- 2.Bishop JM: The molecular genetics of cancer. Science 1987, 235:305-311 [DOI] [PubMed] [Google Scholar]
- 3.Zimonjic DB, Keck CL, Thorgeirsson SS, Popescu NC: Novel recurrent genetic imbalances in human hepatocellular carcinoma cell lines identified by comparative genomic hybridization. Hepatology 1999, 29:1208-1214 [DOI] [PubMed] [Google Scholar]
- 4.Sakakura C, Hagiwara A, Taniguchi H, Yamaguchi T, Yamagishi H, Takahashi T, Kyama K, Nakamura Y, Abe T, Inazawa J: Chromosomal aberrations in human hepatocellular carcinomas associated with hepatitis C virus infection detected by comparative genomic hybridization. Br J Cancer 1999, 80:2034-2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kusano N, Shiraishi K, Kubo K, Oga A, Okita K, Sasaki K: Genetic aberrations detected by comparative genomic hybridization in hepatocellular carcinomas: their relationship to clinicopathological features. Hepatology 1999, 29:1858-1862 [DOI] [PubMed] [Google Scholar]
- 6.Wong N, Lai P, Pang E, Fung LF, Sheng Z, Wong V, Wang W, Hayashi Y, Perlman E, Lau JW, Johnson PJ: Genomic aberrations in human hepatocellular carcinomas of differing etiologies. Clin Cancer Res 2000, 6:4000-4009 [PubMed] [Google Scholar]
- 7.Chen YJ, Yeh SH, Chen JT, Wu CC, Hsu MT, Tsai SF, Chen PJ, Lin CH: Chromosomal changes and clonality relationship between primary and recurrent hepatocellular carcinoma. Gastroenterology 2000, 119:431-440 [DOI] [PubMed] [Google Scholar]
- 8.Balsara BR, Pei J, De Rienzo A, Simon D, Tosolini A, Lu YY, Shen FM, Fan X, Lin WY, Buetow KH, London WT, Testa JR: Human hepatocellular carcinoma is characterized by a highly consistent pattern of genomic imbalances, including frequent loss of 16q23.1–24.1. Genes Chromosomes Cancer 2001, 30:245-253 [DOI] [PubMed] [Google Scholar]
- 9.Tanigami A, Tokino T, Takita KI, Ueda M, Kasumi F, Nakamura Y: Detailed analysis of an amplified region at chromosome 11q13 in malignant tumors. Genomics 1993, 13:21-24 [DOI] [PubMed] [Google Scholar]
- 10.Zhang YJ, Jiang W, Chen CJ, Lee CS, Kahn SM, Santella RM, Weinstein IB: Amplification and overexpression of cyclin D1 in human hepatocellular carcinoma. Biochem Biophys Res Commun 1993, 196:1010-1016 [DOI] [PubMed] [Google Scholar]
- 11.Nishida N, Fukuda Y, Komeda T, Kita R, Sando T, Furukawa M, Amenomori M, Shibagaki I, Nakao K, Ikenaga K: Amplification and overexpression of the cyclin D1 gene in aggressive human hepatocellular carcinoma. Cancer Res 1994, 54:3107-3110 [PubMed] [Google Scholar]
- 12.Schuuring E: The involvement of the chromosome 11q13 region in human malignancies: cyclinD1 and EMS1 are two new candidate oncogenes: a review. Gene 1995, 159:83-96 [DOI] [PubMed] [Google Scholar]
- 13.Schuuring E, Verhoeven E, Litvinov S, Michalides RJ: The product of the EMS1 gene, amplified and overexpressed in human carcinomas, is homologous to a v-src substrate and is located in cell-substratum contact sites. Mol Cell Biol 1993, 13:2891-2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuuring E, van Damme H, Schurring-Scholtes E, Verhoeven E, Michalides R, Geelen E, de Boer C, Brok H, van Buuren C, Kluin P: Characterization of the EMS1 gene and its product, human cortactin. Cell Adhesion Commun 1998, 6:185-209 [DOI] [PubMed] [Google Scholar]
- 15.Lese CM, Rossie KM, Appel BN, Reddy JK, Johnson JT, Myers EN, Gollin SM: Visualization of INT2 and HST1 amplification in oral squamous cell carcinoma. Genes Chromosomes Cancer 1995, 12:288-295 [DOI] [PubMed] [Google Scholar]
- 16.Shuster MI, Han L, Le Beau MM, Davis E, Sawicki M, Lese CM, Park NH, Colicelli J, Gollin SM: A consistent pattern of RIN1 rearrangements in oral squamous cell carcinoma cell lines supports a breakage-fusion-bridge cycle model for 11q13 amplification. Genes Chromosomes Cancer 2000, 28:153-163 [PubMed] [Google Scholar]
- 17.Yuan BZ, Miller MJ, Keck CL, Zimonjic D, Thorgeirsson SS, Popescu NC: Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homogous to rat RhoGAP. Cancer Res 1998, 58:2196-2199 [PubMed] [Google Scholar]
- 18.Rodrigo JP, García LA, Ramos S, Lazo PS, Suárez C: EMS1 gene amplification correlates with poor prognosis in squamous cell carcinomas of the head and neck. Clin Cancer Res 2000, 6:3177-3182 [PubMed] [Google Scholar]
- 19.Zimonjic DB, Keck-Waggoner CL, Yuan BZ, Kraus MH, Popescu NC: Profile of genetic alterations and tumorigenicity of human breast cancer cells. Int J Oncol 2000, 16:221-230 [DOI] [PubMed] [Google Scholar]
- 20.Zimonjic DB, Rezanka L, Di Paolo JA, Popescu NC: Refined localization of the erbB-3 proto-oncogene by direct visualization of FISH signals on LUT-inverted and contrast-enhanced digital images of DAPI-banded chromosomes. Cancer Genet Cytogenet 1995, 80:100-102 [DOI] [PubMed] [Google Scholar]
- 21.Joos S, Scherthan H, Speicher MR, Schlegel J, Cremer T, Lichter P: Detection of amplified sequences by reverse chromosome painting using genomic tumor DNA as probe. Hum Genet 1993, 90:584-589 [DOI] [PubMed] [Google Scholar]
- 22.Campbell DH, deFazio A, Sutherland RL, Daly RJ: Expression and tyrosine phosphorylation of EMS1 in human breast cancer cell lines. Int J Cancer 1998, 68:485-492 [DOI] [PubMed] [Google Scholar]
- 23.Van Damme H, Brok H, Schuuring-Scholtes E, Schuuring E: The redistribution of cortactin into cell-matrix contact sites in human carcinoma cells with 11q13 amplification is associated with both overexpression and post-translational modification. J Biol Chem 1997, 272:7374-7380 [DOI] [PubMed] [Google Scholar]
- 24.Uruno T, Liu J, Zhang P, Egile C, Mueller SC, Zhan X: Activation of Arp2/3 complex-mediated actin polymerization of cortactin. Nat Cell Biol 2001, 3:259-266 [DOI] [PubMed] [Google Scholar]
- 25.Weed SA, Parsons JT: Cortactin: coupling membrane dynamics to cortical actin assembly. Oncogene 2001, 20:6418-6434 [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Tondravi M, Liu J, Smith E, Haudenschild CC, Kaczmarek M, Zhan X: Cortactin potentiates bone metastasis of breast cancer cells. Cancer Res 2001, 61:6906-6911 [PubMed] [Google Scholar]
- 27.Kraus MH, Popescu NC, Amsbaugh SC, King CR: Overexpression of the EGF receptor-related proto-oncogene Erbb-2 in human mammary tumor cell lines by different molecular mechanisms. EMBO J 1987, 6:605-610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL: Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987, 235:177-182 [DOI] [PubMed] [Google Scholar]
- 29.Pegram MD, Lipton A, Hayes DF, Weber BL, Baselga JM, Tripathy D, Baly D, Baughman SA, Twaddell T, Glaspy JA, Slamon DJ: Phase II study of receptor-enhanced chemosensitivity using recombinant humanized anti-p185HER2/neu monoclonal antibody plus cisplatin in patients with HER2/neu-overexpressing metastatic breast cancer refractory to chemotherapy treatment. J Clin Oncol 1998, 16:2659-2671 [DOI] [PubMed] [Google Scholar]
- 30.Hatada I, Tokino T, Ochiya T, Matsubara K: Co-amplification of integrated hepatitis B virus DNA and transforming gene hst-1 in a hepatocellular carcinoma. Oncogene 1988, 3:537-540 [PubMed] [Google Scholar]