Abstract
Integration of a DNA copy of the viral genome into a host chromosome is an essential step in the retrovirus life cycle. The machinery that carries out the integration reaction is a nucleoprotein complex derived from the core of the infecting virion. To successfully integrate into host DNA, the viral DNA within this complex must avoid self-destructive integration into itself, a reaction termed autointegration. We have previously shown [Lee, M. S. and Craigie, R. (1994) Proc. Natl. Acad. Sci. USA 91, 9823–9827] that viral nucleoprotein complexes isolated from Moloney murine leukemia virus-infected cells exhibit a barrier to autointegration. This autointegration barrier could be destroyed by stripping factors from the complexes and subsequently restored by incubation with a host cell extract, but not by incubation with an extract of disrupted virions. We have now used this autointegration barrier reconstitution assay to purify the host factor from uninfected NIH 3T3 fibroblasts. It is a single polypeptide of 89 aa that does not match any previously identified protein. The identity of the protein was confirmed by expressing it in Escherichia coli and demonstrating the activity of the heterologously expressed protein in the reconstitution assay.
Keywords: retrovirus, integration, nucleoprotein complexes
The core of an infecting retrovirus comprises a nucleoprotein complex in which the viral RNA genome is associated with enzymes and structural proteins that are required for reverse transcription and subsequent integration of the viral genome into host DNA (1–3). After reverse transcription, the viral DNA and at least the viral integrase protein (4–7) must remain associated within the nucleoprotein complex until the integration reaction inserts the viral DNA into a chromosome of the host cell. These integration-competent nucleoprotein complexes (INCs) can be isolated from infected cells, and they efficiently integrate their DNA into an exogenously introduced target DNA in vitro (8–12). Analysis of Moloney murine leukemia virus (MoMLV) DNA integration intermediates made in vitro has defined the DNA cutting and joining steps of retroviral DNA integration (13, 14). In the first step, 3′ end processing, two nucleotides are removed from each 3′ end of the initially blunt-ended viral DNA. In the second step, DNA strand transfer, the resulting recessed 3′ ends of the viral DNA are joined to the 5′ ends of the target DNA at the site of integration. The 5′ ends of the viral DNA and the 3′ ends of the target DNA remain unjoined in the resulting integration intermediate. Cellular DNA repair enzymes are likely responsible for degrading the unpaired nucleotides at the 5′ ends of the viral DNA, filling in the single strand gaps and subsequent ligation to complete the integration process.
Although the viral integrase protein is sufficient to carry out 3′ processing and DNA strand transfer in vitro with simple DNA substrates (15–20), these reaction systems lack the full fidelity of the in vivo reaction. For example, many reaction products result from integration of only a single viral DNA end into one strand of target DNA. The high fidelity of in vitro integration reactions with INCs provides a powerful tool to probe aspects of retroviral DNA integration that have not been reproduced with the simplified systems by using integrase protein.
A striking feature of the reactions with INCs isolated from cells infected with MoMLV and HIV type 1 is the strong preference to integrate intermolecularly into another DNA molecule, rather than intramolecularly into their own DNA. This is a biologically important property of the complexes because they must avoid such suicidal autointegration in vivo if they are to successfully integrate into host DNA. In the case of MoMLV INCs we have previously demonstrated the involvement of a host factor in protecting the viral DNA against autointegration (21). The barrier to autointegration can be destroyed by incubating the complexes at high ionic strength. This treatment liberates a trans-acting factor to yield complexes that can only autointegrate, termed autointegration complexes (auto-INCs). These complexes can then be separated from free proteins by sedimentation in a sucrose gradient. The autointegration barrier can be reconstituted by incubating the stripped complexes with an extract of uninfected NIH 3T3 fibroblasts. Here, we have used this autointegration barrier reconstitution assay to purify and identify the host factor that confers the protection of the retroviral DNA against autointegration. This host factor, termed the barrier-to-autointegration factor (BAF), is a single 89-aa polypeptide that does not match any identified protein.
MATERIALS AND METHODS
Autointegration Barrier Reconstitution Assay.
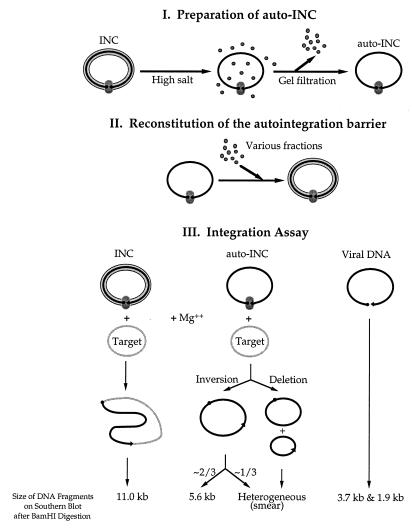
The assay is schematically illustrated in Fig. 1. First, auto-INCs, the substrate for the barrier reconstitution assay, were prepared as previously described (21) except that gel filtration, instead of velocity sedimentation in sucrose gradients, was used to remove free components from the auto-INCs. Briefly, the INCs were prepared from coculture of NIH 3T3 fibroblasts and clone 4, a MoMLV-producing cell line (provided by S. Goff, Columbia University, New York). The autointegration barrier was disrupted by incubating fraction II, containing the INCs, after addition of KCl to 750 mM. The auto-INCs were separated from free components by spin dialysis through a column of Sepharose 4B-CL (Pharmacia) equilibrated with 20 mM Hepes-NaOH, pH 7.5/5 mM MgCl2/6 mM EDTA/1 mM DTT/6% sucrose/750 mM KCl, followed by a second spin dialysis through a column equilibrated with the buffer containing 150 mM KCl. Next, the autointegration barrier was reconstituted onto the purified auto-INC by adding various BAF-containing fractions in standard reaction mixtures containing 20 mM Hepes-NaOH (pH 7.5), 5 mM MgCl2, 6 mM EDTA, 400 mM KCl, 1 mM DTT, 0.04% BSA, 40% Nycodenz, and 10 mM (NH4)2SO4. Inclusion of 10 mM (NH4)2SO4 significantly enhanced the overall signal on the autoradiogram, probably because of increased recovery of the viral DNA during the assay (data not shown). Finally, the reconstitution reaction products were analyzed by the previously described integration assay (21); standard integration reactions (100–200 μl) contained 20 mM Hepes-NaOH (pH 7.5), 200 mM KCl, 10 mM MgCl2, 10 mM DTT, 0.02% BSA, 15% (wt/vol) glycerol, 3 nM øX174 replicative form I DNA as the target. On the autoradiogram, the 11-kb band represents the intermolecular reaction products; the 5.6-kb band and smear are derived from autointegration products and the 3.7-kb and the 1.9-kb bands represent unreacted viral DNA. BAF activity decreases the level of autointegration products and increases the level of intermolecular integration products.
Figure 1.
Outline of the autointegration barrier reconstitution assay. (I) Auto-INC, the substrate for the reconstitution reaction, was prepared by disruption of the autointegration barrier from the INC with high salt treatment, followed by removal of free components through gel filtration. (II) The autointegration barrier was reconstituted onto the purified auto-INC by addition of various BAF-containing fractions. (III) The extent of reconstitution reactions was measured by the integration assay that simultaneously analyzes intermolecular integration and autointegration.
Purification of BAF from Uninfected NIH 3T3 Fibroblasts.
The BAF was purified from uninfected host cells by using the autointegration barrier reconstitution assay. First, cytoplasmic extract (fraction I) was prepared from uninfected NIH 3T3 fibroblasts, followed by high salt treatment and velocity sedimentation in sucrose gradients, as previously described for the INCs (21). The BAF activity in the sucrose gradient top portion (fraction II) was passed through a phosphocellulose P-11 (Whatman) column equilibrated with 20 mM Hepes-NaOH, pH 7.5/1 mM EDTA/150 mM KCl/6% sucrose/1 mM DTT (fraction III) and then a Mono Q anion exchange column equilibrated with 20 mM Tris⋅HCl, pH 7.5/1 mM EDTA/100 mM KCl/6% sucrose/1 mM DTT (fraction IV). The flow-through from the first Mono Q column was fractionated over another Mono Q anion exchange column equilibrated with 20 mM Tris⋅HCl, pH 7.5/1 mM EDTA/50 mM KCl/6% sucrose/1 mM DTT. The pool of peak fractions from the Mono Q column (fraction V) was then fractionated through a Superdex-200 size exclusion column equilibrated with 20 mM Tris⋅HCl, pH 7.5/1 mM EDTA/200 mM KCl/6% sucrose/1 mM DTT. Finally, the pool of peak fractions from the Superdex-200 column (fraction VI) was fractionated through a Phenyl Superose hydrophobic column equilibrated with 50 mM sodium phosphate buffer, pH 7.0/1 mM EDTA/6% sucrose/1 mM DTT/1.7 M (NH4)2SO4.
Partial Amino Acid Sequencing of the 7.0-kDa Polypeptides.
The protein fraction containing the 7.0-kDa band that copurified with the reconstitution activity (Fig. 2) was resolved on an SDS/polyacrylamide gel and transferred to a polyvinylidene difluoride membrane. Initial attempts at N-terminal sequencing revealed the N terminus to be blocked. The protein was then cleaved with endoproteinase Lys-C, which hydrolyzes at the carboxylic side of lysine residues. The resulting peptides were separated by reverse-phase chromatography and subjected to Edman degradation for amino acid sequencing.
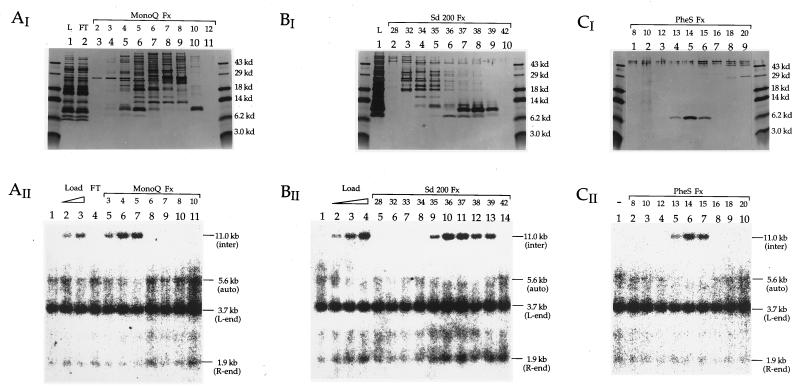
Figure 2.
Purification of the barrier-to-autointegration factor (BAF). (AI, BI, and CI) The silver-stained SDS/polyacrylamide gels (16% run in Tricine buffer). (AII, BII, and CII) The autoradiograms of the Southern blots showing the BAF activity as described in Fig. 1. AI and AII for the Mono Q column; BI and BII for the Superdex-200 column; CI and CII for the Phenyl superose column. Fx, column fractions; FT, flow-through from the column; L, sample load to the column. The load to the Mono Q column (AII, lanes 2 and 3) as well as the load to the Superdex-200 (BII, lanes 2–4) was titrated by 2-fold increments in the activity assay. The BAF activity is represented by conversion of the 5.6-kb band and smear (autointegration products) to the 11.0-kb band (intermolecular integration products) (AII, BII, and CII).
Cloning and DNA Sequencing of BAF-Encoding cDNA.
The internal peptide sequences (underlined in Fig. 3) were used to design primers for amplification of the BAF-encoding DNA sequence from Poly(A)+ mRNA by employing “rapid amplification of cDNA ends” technique (22). As the template for amplification, mRNA was prepared from NIH 3T3 fibroblasts (mRNA isolation kit from Stratagene).
Figure 3.
Nucleotide sequence and deduced amino acid sequence of the ORF of the BAF-encoding cDNA. The four internal peptide sequences that were obtained by partial amino acid sequencing are underlined.
To amplify the 3′ end of cDNA, a pool of minus-strand DNA was generated from the mRNA by using MoMLV reverse transcriptase (Stratagene), and a 50-base [(GA)6-adaptor20-dT18] (5′-GAGAGAGAGAGAGAGAGAGAACTAGTCTCGAGTTTTTTTTTTTTTTTTTT-3′) as a primer. Then, PCR was performed with Taq polymerase (Stratagene) by using the pool of minus-strand cDNA as templates. The primers for this PCR were a pool of 20-base degenerate oligonucleotides derived from one of the specific internal amino acid sequence obtained by Edman degradation (Phe Val Ala Glu Pro Met Gly Glu) and the adaptor20. PCR products were resolved by agarose gel electrophoresis and ≈630-bp DNA fragments were cloned into the TA cloning vector, pCRTM2.1 (Invitrogen), followed by DNA sequencing of the insert.
To amplify the 5′ end of the cDNA, a specific minus-strand cDNA was synthesized by MoMLV reverse transcriptase by using a 20-base specific DNA sequence, 5′-ATCACACCATTCTCGAAGGC-3′, obtained above, as a primer. After removal of excess primers (Promega Wizard PCR preps DNA purification system), oligo-dA tails of ≈20–30 bases were added to the 3′ ends of the minus-strand DNA with terminal deoxynucleotidyl transferase (Bethesda Research Laboratories). The plus-strand DNA was first synthesized by using the specific minus-strand cDNA as a template and the 38-base [adapter20-dT18] as a primer. The product was then amplified by PCR using, as primers, an internal 20-base specific DNA sequence, 5′-AAAGCCCCTTTCCTCCAGCC-3′, for minus strand and the adaptor20 for plus strand. PCR products were resolved by agarose gel electrophoresis, and ≈240-bp DNA fragments were cloned into the TA cloning vector, followed by DNA sequencing of the insert. Based on the sizes of cDNA fragments, the length of untranslated region was estimated to be ≈400 bases for 5′ side and ≈100 bases for 3′ side of the coding sequence.
To finally confirm the DNA sequence of the ORF of the BAF gene, part of the BAF cDNA including ORF was amplified in four separate PCRs by using a pool of minus-strand DNA (as described above for amplification of 3′ end of the cDNA) as the template and four different pairs of specific DNA sequences outside the ORF as primers, followed by cloning into the TA cloning vector and DNA sequencing of the insert.
Expression of Functional BAF in Escherichia coli.
DNA fragments including the ORF were amplified by PCR in the presence of a 28-base 5′-NdeI linker (5′-GGGGCATATGACAACCTCCCAAAAGCAC-3′) and a 30-base 3′-BamHI linker (5′-CCCCGGATCCCTACAAGAAGGCATCACACCA-3′) as primers, and then cloned into the NdeI and BamHI sites of pET-15b vector (Novagen), followed by verification of DNA sequence of the insert. The pET-15b recombinant plasmid containing the ORF of BAF was transformed into a high-stringency expression host, BL21 (DE3) pLysS (Novagen), and the hexahistidine-tagged murine BAF protein was overexpressed under the control of T7 RNA polymerase after isopropyl β-d-thiogalactoside (IPTG) induction. Cells harvested 2.5 hr after IPTG induction were lysed with 20 mM Hepes-NaOH, pH 7.5/150 mM KCl/2 mM EDTA/0.01% lysozyme/0.1% Triton X-100. The BAF in the insoluble portion was extracted with guanidine hydrochloride and enriched by chromatography on a nickel chelate affinity column (23). The hexahistidine tag was cleaved off with thrombin, which was then removed by adsorption to a benzamidine sepharose column (23). The BAF free of the hexahistidine tag was further purified by reverse-phase chromatography on a Protein C4 column (Rainin) and assayed for activity in the autointegration barrier reconstitution assay.
RESULTS
Purification of BAF.
We have used the previously described autointegration barrier reconstitution assay (Fig. 1) to purify the factor that protects the viral DNA against autointegration from cytoplasmic extracts of NIH 3T3 cells. The substrate for reconstitution was INCs that had been stripped with high salt and then separated from free proteins by gel filtration. These stripped complexes were incubated with the various protein fractions and then assayed for integration in a reaction mixture that contained øx174RF DNA as an intermolecular target. The reaction products were cut with a restriction enzyme, BamHI, and visualized by Southern blotting after gel electrophoresis. The intermolecular integration products into øx174RF DNA result in a single band of 11 kb, whereas the autointegration products are seen as a smear together with a band of 5.6 kb.
Preliminary experiments established that more activity was present in cytoplasmic extracts than in nuclear extracts of NIH 3T3 (data not shown). We therefore used the previously described cytoplasmic extracts of NIH 3T3 cells as the starting material for purification of the host factor. Reconstitution activity of the protein was monitored in parallel with SDS/PAGE, followed by silver staining (Fig. 2). Activity is evidenced by the appearance of the 11-kb band resulting from intermolecular integration into øx174 target DNA and a decrease in intensity of the 5.6-kb band and smear that result from autointegration.
Comparison of panels I and II through each of the purification steps reveals that reconstitution activity copurifies with a single protein band of approximately 7.0 kDa in SDS/PAGE (Fig. 2). Peak fractions for both the 7.0-kDa polypeptide and the BAF activity coeluted from the Mono Q column with 100 mM KCl (Fig. 2A, I and II: fraction 5), from the Superdex-200 column at the calibrated elution position for an 18-kDa globular protein (Fig. 2B, I and II: fraction 36), and from the Phenyl Superose with 1.1 M (NH4)2SO4 (Fig. 2C, I and II: fraction 14), respectively. Approximately 2 ng of BAF (estimated by Coomassie blue staining, data not shown) saturated 100 μl autointegration reconstitution reactions (Fig. 2BII, lane 4).
Partial Amino Acid Sequencing and Cloning of BAF-Encoding cDNA.
Initial attempts to obtain N-terminal sequence of the 7.0- kDa protein by Edman degradation revealed the N terminus to be blocked. The protein therefore was digested with endoproteinase Lys-C, and Edman degradation was performed on internal peptides after separation by reverse-phase chromatography. Four of the peptides gave unambiguous sequence (underlined in Fig. 3). These peptide sequences were used to design primers for amplification of the BAF-encoding transcript from poly(A)+ mRNA as described in Materials and Methods.
DNA sequencing of the cloned cDNA generated from four independent PCRs revealed an ORF for 89 aa that contained all four internal peptide sequences obtained by Edman degradation of the peptides. Three of the four were identical in DNA sequences (shown in Fig. 3) and the fourth contained a single base substitution (data not shown). The molecular mass of the polypeptide calculated from the deduced amino acid sequence is 10.1 kDa. That these polypeptides of the native form eluted from the Superdex-200 column at the calibrated elution position for an 18-kDa globular protein (Fig. 2 BI and BII) suggests that they are likely to exist as a dimer in the solution.
Expression of Functional BAF in Escherichia coli.
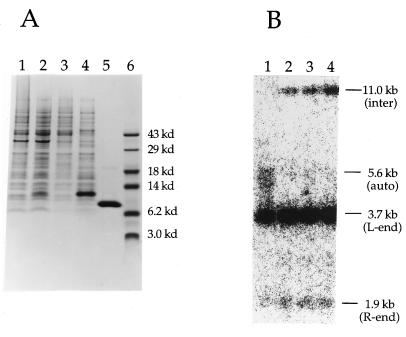
To confirm that the cloned cDNA encodes the functional protein, the ORF was expressed in the E. coli expression vector pET-15b (Novagen) (compare lanes 1 and 2 in Fig. 4). The recombinant protein partitioned to the insoluble fraction of conventional lysates of E. coli cells and was absent in the soluble fraction (Fig. 4A, lane 3). It was extracted from the insoluble pellet with 6 M guanidine hydrochloride (Fig. 4A, lane 4) and further purified as described in Materials and Methods to near homogeneity (Fig. 4A, lane 5). Although expressed in insoluble form, the purified protein remains soluble after removal of guanidine. The purified recombinant protein was found to be active in the autointegration barrier reconstitution assay (Fig. 4B), confirming the identity of the protein and demonstrating that the N-terminal modification is not required for reconstitution activity. Although not carefully quantitated, the specific activity of the recombinant protein appeared to be comparable to that obtained from NIH 3T3 cells (compare Figs. 2BII, lane 4 and 4B, lane 2). We conclude that BAF is necessary and sufficient for reconstitution of the autointegration barrier.
Figure 4.
Expression of functional BAF in Escherichia coli. (A) Overexpression, solubilization, and purification of the murine BAF. E. coli cells with the pET-15b recombinant plasmid containing BAF-ORF were harvested before (lane 1) or after (lanes 2–5) IPTG induction. Lanes: 1 and 2, whole cell lysates in 4% SDS; 3, soluble lysates; 4, guanidine hydrochloride-soluble portion of the insoluble fraction; 5, the purified BAF after removal of the His-tag and reverse-phase chromatography; 6, molecular mass standards. Samples were analyzed by SDS/PAGE, followed by Coomassie blue staining. (B) The recombinant BAF is active in the autointegration barrier reconstitution assay. Lanes: 1, no addition; 2–4, the purified BAF in 2, 10, and 50 ng in each reconstitution reaction.
DISCUSSION
We have used the previously described autointegration barrier reconstitution assay (21) to purify the host factor that confers protection against autointegration. It is a single polypeptide of 89 aa that does not match any previously identified protein. However, a search of DNA sequence databases reveals that a wide variety of species, including humans, zebrafish, and C. elegans express a transcript that can encode a protein with a high degree of sequence identity (data not shown). This high degree of sequence identity among species suggests that BAF plays an important role for the host cell.
How might BAF block autointegration and therefore promote intermolecular integration? One simple possibility is that BAF coats the viral DNA, thereby rendering it inaccessible as a target. Preliminary studies (Fig. 5) demonstrate that BAF does indeed bind DNA, but suggest that a mechanism other than simple coating may be involved in blocking autointegration. Linearized double-stranded øX174 DNA that has been incubated with BAF at a stoichiometry of one BAF dimer per 75 bp can be pelleted by low-speed centrifugation (Fig. 5A, lane 4). An even lower stoichiometry of BAF to DNA (less than one dimer per 300 bp) prevents DNA entering agarose gels in the absence of SDS (data not shown). To explain these observations, we propose that BAF makes intermolecular bridges between DNA segments resulting in an open mesh in which DNA molecules are bridged by BAF. Although DNA associated with BAF forms a large network, it is a competent target for intermolecular integration at the lower range of BAF:DNA stoichiometry (Fig. 5B). However, it becomes refractory to integration as the BAF:DNA stoichiometry is increased (Fig. 5B). Interestingly, naked DNA is highly preferred as a target when both BAF-associated and naked DNA are present in the reaction mixture (Fig. 5C). This phenomenon is observed even when the BAF-associated DNA would be used as a target in the absence of naked DNA (compare lanes 4 of Fig. 5 B and C). The DNA within the BAF-associated mesh (Fig. 5D) is presumably relatively inaccessible to the INC compared with naked DNA.
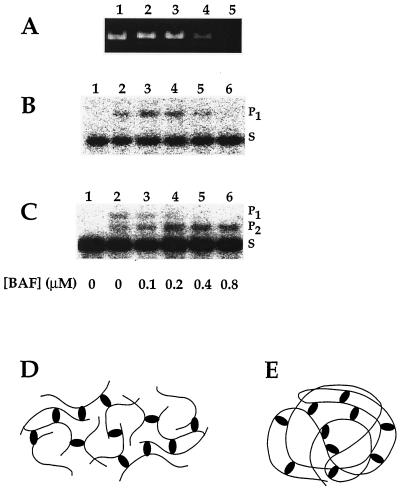
Figure 5.
DNA bridging by BAF and its potential role in blocking autointegration. Linearized double-stranded øX174 DNA (PstI digest of øX174 replicative form I DNA, 30 μM in nucleotides) was preincubated with various concentrations of BAF (0–0.8 μM of monomers) for 10 min on ice. (A) Aggregation of DNA by BAF. One aliquot of each preincubation mixture was centrifuged at 15,000 × g for 10 min. After addition of SDS, the DNA remaining in the supernatants was analyzed by agarose gel electrophoresis and stained with ethidium bromide. (B) Binding of BAF can make a target DNA unavailable for integration. INCs were added to a second set of preincubation mixture aliquots (lanes 2–6) and assayed for their integration activity into the preincubated target DNA. Intermolecular integration reaction products (P1, ≈14 kbp) were separated from unreacted substrate (S, ≈9 kbp) by agarose gel electrophoresis and visualized by Southern blotting and hybridization. (C) Naked DNA is a preferred target for integration. Linear 2.7-kbp DNA (BstNI digest of øX174 replicative form I DNA, 30 μM in nucleotides) was added to a third set of aliquots of linear 5.4-kbp øX174 DNA that had been preincubated with the various concentrations of BAF. Integration reactions were initiated by addition of INCs (lanes 2–6) and analyzed as described in B except that P2 (≈12 kbp) represents integration into the naked target DNA. Lanes 1 for B and C are INCs without addition of target DNA. (D) Intermolecular aggregation of DNA by BAF. Bridging of the DNA by BAF results in the formation of a network. (E) Intramolecular bridging by BAF may compact the viral DNA.
We suggest that the DNA-bridging property of BAF may explain how it prevents autointegration. Intramolecular bridging of DNA segments within the viral DNA (Fig. 5E) would compact it and therefore make it less accessible as an autointegration target. Because only one copy of viral DNA is present in each complex, only intramolecular bridging of the viral DNA should occur. Although we find this model attractive, we cannot completely exclude other possible mechanisms, such as DNA coating, for how BAF blocks autointegration. Is there enough BAF available to coat the viral DNA? Western blotting experiments (data not shown) and assays for BAF activity (21) indicate that BAF is not present in virions. It is therefore likely to be recruited in the cytoplasm of the infected cell. Because there are approximately 105 molecules of BAF per NIH 3T3 cell as judged by Western blotting of whole-cell lysates (data not shown), coating is theoretically possible. However, the total quantity of BAF in the cell is not very informative in the absence of information on how it is compartmentalized. The idea that the viral DNA is organized in an open, mesh-like structure is consistent with observations that the DNA in the INC can be readily cleaved by endonucleases without loss of integration activity (unpublished data). DNA bridging by BAF may provide a means to compact the viral DNA while leaving it relatively accessible to macromolecules that are small enough to penetrate the mesh.
Host-encoded proteins are involved in many DNA recombination reactions. The best understood systems are some of the prokaryotic site-specific recombination and DNA transposition reactions (24). An archetype for the participation of host factors in DNA recombination is the phage lambda Integration Host Factor. This protein is involved in bringing binding sites for lambda integrase into their proper configuration by introducing a sharp bend in the DNA substrate. Another DNA-bending protein, HU, is recruited by several site-specific recombination and transposition systems.
The role of host factors in retroviral DNA integration is less well understood. Although purified retroviral integrases carry out the key DNA cutting and joining steps of retroviral DNA integration in vitro (15–20), these reactions lack the fidelity of those mediated by INCs. In particular, many of the integration products result from joining of only a single viral DNA end to one strand of the target DNA; the authentic reaction inserts a pair of viral DNA ends, one into each strand of the double-stranded target DNA. One possible explanation for the deficiencies of these in vitro systems is that required host factors are not included. Several host proteins, including INI 1 (25) and HMG 1 (26), have been suggested to be involved in retroviral DNA integration. These proteins stimulate in vitro integration reactions with purified integrase by a factor of severalfold. Although such stimulatory effects may be relevant to function in vivo, caution is required in reaching this conclusion; we have observed similar degrees of stimulation in such in vitro assays with E. coli protein HU and ribonuclease A (unpublished data), proteins that are clearly not involved in retroviral DNA integration.
Another report, essentially using the previously described salt stripping and reconstitution assay (21), implicates HMGI(Y) in the function of HIV type 1 INCs (27). Because their assay measures only intermolecular integration activity of the salt stripped complexes, which is partially restored by incubation with HMGI(Y), it is not clear whether this phenomenon is related to the BAF activity reported here. We note that the extent of stimulation by HMGI(Y) appears to never approach that of crude extract, suggesting the possible involvement of other factors. Furthermore, the concentration of HMGI(Y) included in their reconstitution reactions is approximately 1,000 times greater than that of BAF required for our reconstitution reactions.
Host proteins appear to play both direct and regulatory roles in DNA recombination reactions. Activities such as promoting DNA bending can facilitate the assembly of the appropriate nucleoprotein complexes. They also enable the establishment of sophisticated regulatory mechanisms. Many questions remain to be answered concerning the role of BAF in retroviral DNA integration. How does BAF block autointegration and therefore promote intermolecular integration? At what stage of the virus life cycle is BAF recruited into the INC? Future work will investigate its mechanism of action in retroviral DNA integration and its function for the host cell. An understanding of the molecular mechanism of the defense against autointegration may also suggest novel strategies to block retroviral replication.
Acknowledgments
We thank Regis Krah for helpful suggestions for cloning. We also acknowledge the Wistar Sequencing Facility for partial amino acid sequencing. This work was supported in part by the National Institutes of Health AIDS Targeted Antiviral Program.
ABBREVIATIONS
- INC
integration-competent nucleoprotein complex
- auto-INC
autointegration complex
- MoMLV
Moloney murine leukemia virus
- BAF
barrier-to-autointegration factor
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF037080).
References
- 1.Varmus H, Brown P O. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 53–108. [Google Scholar]
- 2.Goff S P. Annu Rev Genet. 1992;26:527–544. doi: 10.1146/annurev.ge.26.120192.002523. [DOI] [PubMed] [Google Scholar]
- 3.Katz R A, Skalka A M. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 4.Andrake M D, Skalka A M. J Biol Chem. 1996;271:19633–19636. doi: 10.1074/jbc.271.33.19633. [DOI] [PubMed] [Google Scholar]
- 5.Schwartzberg P, Colicelli J, Goff S P. Cell. 1984;37:1043–1052. doi: 10.1016/0092-8674(84)90439-2. [DOI] [PubMed] [Google Scholar]
- 6.Donehower L A, Varmus H E. Proc Natl Acad Sci USA. 1984;81:6461–6465. doi: 10.1073/pnas.81.20.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panganiban A T, Temin H M. Proc Natl Acad Sci USA. 1984;81:7885–7889. doi: 10.1073/pnas.81.24.7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown P O, Bowerman B, Varmus H E, Bishop J M. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 9.Bowerman B, Brown P O, Bishop J M, Varmus H E. Genes Dev. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- 10.Ellison V, Abrams H, Roe T, Lifson J, Brown P. J Virol. 1990;64:2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farnet C M, Haseltine W A. Proc Natl Acad Sci USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee Y M, Coffin J M. Mol Cell Biol. 1991;11:1419–1430. doi: 10.1128/mcb.11.3.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujiwara T, Mizuuchi K. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 14.Brown P O, Bowerman B, Varmus H E, Bishop J M. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katzman M, Katz R A, Skalka A M, Leis J. J Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craigie R, Fujiwara T, Bushman F. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 17.Katz R A, Merkel G, Kulkosky J, Leis J, Skalka A M. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 18.Sherman P A, Fyfe J A. Proc Natl Acad Sci USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bushman F D, Fujiwara T, Craigie R. Science. 1990;249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 20.Bushman F D, Craigie R. Proc Natl Acad Sci USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee M S, Craigie R. Proc Natl Acad Sci USA. 1994;91:9823–9827. doi: 10.1073/pnas.91.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craigie R, Hickman A B, Engelman A. In: HIV: A Practical Approach. Karn J, editor. Vol. 2. New York: Oxford Univ. Press; 1997. pp. 53–71. [Google Scholar]
- 24.Nash H A. In: Regulation of Gene Expression in Escherichia coli. Lin E C C, Lynch A S, editors. Austin, TX: R. G. Landes Company; 1996. pp. 149–179. [Google Scholar]
- 25.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 26.Aiyar A, Hindmarsh P, Skalka A M, Leis J. J Virol. 1996;70:3571–3580. doi: 10.1128/jvi.70.6.3571-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farnet C M, Bushman F D. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]