Abstract
Excellent electrophoretic separations of a variety of biological molecules can be accomplished by using uncharged, triblock copolymers as the “gel” media. These copolymers form uncrosslinked, lyotropic liquid crystalline phases of large micelles between which molecules must travel. Unlike crosslinked hydrogels in common use, these alternative media have highly ordered internal structures. Pluronic F127, representative of the copolymer class, contains poly(ethylene oxide) (EO) and poly(propylene oxide) (PO) units with an approximate molecular formula (EO)106(PO)70(EO)106. Concentrated (18–30%) solutions of Pluronic F127 are freely flowing liquids at low temperature (0–5°C) but form gel-like, cubic liquid crystals of large, spherical micelles when warmed. The utility of these media is illustrated by separations of linear, double-stranded DNA up to 3,000 bp long by conventional electrophoresis, and of single-stranded DNAs from 4 to 60 nt long by capillary electrophoresis. Extraordinary separations of supercoiled DNAs were also obtained by capillary electrophoresis. The versatility, availability, and ease of use of Pluronic polymers offer major advantages over conventional media for preparative and high performance analytical separations of nucleic acids and other biomolecules. Mechanisms of molecular transport and separation operating in polymer liquid crystals must differ in fundamental ways from those in crosslinked gels. Lyotropic polymer liquid crystals are unique systems for elucidating mechanisms of macromolecule migration in ordered, dense media, and provide opportunities in separations science.
Electrophoresis is an essential separation method of biochemistry and molecular biology that distinguishes between proteins or nucleic acids differing only slightly in size, charge, conformation, or degree of association. Gene mapping, DNA sequencing, and transcription factor identification stand out among recent advances uniquely attributable to this method.
Conventional electrophoresis is usually conducted on crosslinked hydrogels such as polyacrylamide “chemical” gels, formed by solution polymerization of simple monomers; or agarose “physical” gels formed by hydrogen bonding between pre-existing polymer chains. These completely interconnected polymer networks provide mechanical support and quench convection, and separate molecules by sieving according to size and shape. Structural details such as pore shape, size distribution, and connectivity strongly influence separations. The structures of both gel classes have been studied, and there is a basic understanding of how globular proteins (1–5) and semi-rigid polymers such as DNA (6–9) migrate through gel networks, but understanding of the influence of gel structure on separations is still incomplete.
Capillary electrophoresis (CE) with high-voltage gradients offers much higher resolution than conventional electrophoresis or HPLC (10–13) and has been proposed to speed DNA sequencing for the Human Genome Project (14, 15). CE is often performed in concentrated solutions of uncrosslinked, hydrophilic linear polymers such as polyacrylamide, agarose, and poly(ethylene oxide) (e.g., see refs. 16–20 and references therein). Replacement of polymer solutions between runs enhances reproducibility and avoids problems associated with the physical and chemical instabilities of crosslinked gels under the conditions of CE.
We introduce here a distinctly different class of electrophoresis media consisting of aqueous, lyotropic liquid crystalline solutions of uncrosslinked, triblock Pluronic copolymers (21–25). Pluronics are (EO)x (PO)y (EO)x copolymers, which are available with different length poly(ethylene oxide) or (EO)x, and poly(propylene oxide) or (PO)y blocks. Above some critical concentration and temperature, Pluronic polymers associate into micelles as the hydrophobic effect drives (PO)y segments into a nearly water-free central core surrounded by hydrated (EO)x tails (21, 23, 26). Liquid crystalline phases form at high solution concentrations to minimize the volume excluded by spherical or columnar micelles (21–25).
The suitability of Pluronic liquid crystals as electrophoresis media was investigated by using Pluronic F127 (BASF Performance Chemicals, Mt. Olive, NJ). Aqueous solutions of Pluronic F127 [approximate molecular formula (EO)106(PO)70(EO)106, Mr 13,000] at low temperatures (0–5°C) and concentrations below about 32% are free flowing liquids; but at room temperature and concentrations above 17%, the polymers self organize into a transparent, liquid crystalline gel of spherical micelles that fill space by packing on a locally cubic lattice (21, 24, 25). Pre-chilled polymer solutions can be readily introduced into capillary tubes or conventional gel templates, then allowed to gel at room temperature. We show that such gels are suitable for conventional and capillary electrophoresis and in certain circumstances provide separations superior to conventional media.
Pluronic gels are versatile media which are easy and safe to use and have an ordered structure, on the length scale of several nanometers, which is distinct from the random structures of conventional media. Excellent separations of different nucleic acid forms were obtained by electrophoresis in gels of Pluronic F127. Members within other molecule classes can be separated in Pluronic gels, including proteins and small amphiphilic molecules. Mechanisms of molecular sieving through ordered arrays of large micelles must differ fundamentally from sieving through random, interconnected polymer networks or entangled polymer chains. Because micelle structures and phase transitions of liquid crystal-forming polymers depend intimately on polymer composition, concentration, and temperature, many options are available for tuning the medium to optimize separations. Micellar polymer liquid crystals offer unique challenges to understanding mechanisms of molecular migration in ordered media and opportunities for developments in separations science.
MATERIALS AND METHODS
Materials.
Pluronic F127 (manufactured by BASF Performance Chemicals), buffer salts, “123-bp ladder” double-stranded DNA standard, and oligonucleotide standards were obtained from Sigma. A “supercoiled DNA ladder” and additional linear DNA standards were obtained from Promega. Restriction endonucleases were obtained from Promega and used according to the supplier’s instructions.
Conventional Electrophoresis.
Pluronic solutions of the stated concentrations (wt/wt) were prepared on ice or in a cold room (5°C) in TBE buffer (90 mM Tris/90 mM boric acid/3 mM EDTA, pH 8.3) containing 0.5 μg/ml ethidium bromide. Cold solutions were pipetted into 13-cm-long by 5-mm i.d. tubes, which were placed in a rack and allowed to warm to ambient temperature (21–23°C) for about 10 min before installation in the electrophoresis apparatus. The liquid crystalline phase formation (gelation) occurred within a few minutes of warming. No “curing” was required before use. Care was taken to assure flat gel tops. In some cases it was useful to push a small amount of gel out of the tube by light pressure from a pipet bulb applied to the opposite end, slice the gel top even with the tube, then force the gel back into the tube by using slight suction on the opposite end. A similar procedure can be used to recover samples from band slices after electrophoresis.
Electrophoresis was performed at room temperature with an applied voltage of 80 or 100 V (6–8 V/cm) for 3–6 hr, depending on the Pluronic concentration. The buffer in both trays was TBE containing 0.5 μg/ml ethidium bromide. The conductivity of a 20% Pluronic solution is about half that of a 6% polyacrylamide gel, allowing application of higher voltage gradients than those typically used for tube gels, without significant heating. A short polyacrylamide plug was cast in the tube bottom if electrophoresis was to continue beyond 4–5 hr to prevent excessive loss of the gel as it dissolved from the end.
Pluronic gels are stable at high temperatures, but effects of temperature on separations have not been examined. Electrophoresis under conditions similar to the above was also performed in Pluronic gels cast as slabs, but the best band shapes were obtained in tube gels. Band irregularities in slab gels arose from distortions of wells during sample application as the Pluronic transitioned from the liquid crystalline to fluid liquid phase on dilution. We believe this technical problem can be overcome to allow routine use of the slab gel format.
PAGE was performed in tube gels as above except that the applied voltage gradient was reduced to 3 V/cm to minimize heating and consequent band cupping. Gels of the indicated acrylamide concentrations were prepared with a 29:1 ratio of acrylamide/bisacrylamide. Electrophoresis in agarose was performed by using a horizontal minigel apparatus (gel thickness ≈4 mm, length 8 cm) and a voltage gradient of 5 V/cm. Tray and gels buffers were TBE containing ethidium bromide, as above.
DNA-bound ethidium fluorescence was visualized by 302 nm illumination (Hoefer) and photographed on Polaroid 665P/N film through UV and red filters. Densitometry was performed on the film negatives.
CE.
Initial studies of oligonucleotide separations were performed at ambient temperature by using a homemade CE system (27) kindly made available by William Cooper (Florida State University). Separations of poly(U) and supercoiled plasmids shown in figures were performed by using a Beckman P/ACE 5010 instrument at the cited temperatures. Electrophoresis was performed in 75 μm i.d., Celect-N columns (Supelco, Inc.) that have a surface coating reported to be exceptionally stable and to nearly eliminate electroosmosis (28). No electroosmosis was noted in CE runs on Pluronic gels. The total capillary length was 38.8 cm (34.8 cm effective length), unless otherwise stated. The UV absorbance detector was set at 254 nm.
The electrophoresis buffer was TBE (without ethidium bromide). Pluronic F127 solutions in TBE buffer were prepared and stored in the cold room (5°C). Capillaries were filled with Pluronic solution with modest hand pressure applied by a syringe and equilibrated to room temperature for 5–10 min before use. Electrophoresis was performed in the reversed polarity mode. Samples were injected electrokinetically. Sample concentrations for both conventional and capillary electrophoresis were typically 1–2 mg/ml in 0.1× TBE buffer. After each electrophoretic run the capillary was put in the cold room and flushed with 1× TBE buffer, with hand pressure applied by syringe, before refilling.
Restriction Endonuclease Digestion of DNA in 10% Pluronic Solution.
Restriction endonuclease buffer concentrate (supplied by Promega) was added to plasmid pGEM3Zf in 50 mM Tris⋅HCl/0.1 mM Na2EDTA (pH 8.0). The plasmid samples were then diluted either with an equal volume of water or with chilled 20% Pluronic F127 in water. Restriction endonucleases (AvaII, EcoRI, HaeIII, HindIII, HpaII, PstI, or ScaI) were added to individual aliquots and incubated for 60 min at 37°C. Samples were applied directly to 1.4% agarose gels and electrophoresed as described above.
RESULTS
Conventional Electrophoresis.
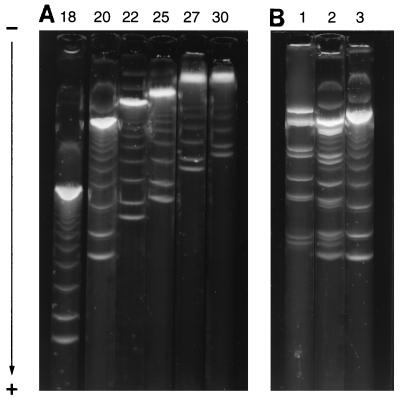
Double-stranded DNA fragments from n = 1 (123 bp) to n = 11 (1,353 bp) of a standard Nx123 bp ladder, and 10 fragments from 222 to 2,977 bp derived from the pGEM3Zf plasmid, were separated in 20% Pluronic F127 gels (Figs. 1 and 2). Increasing the Pluronic concentration progressively decreased the absolute mobilities and size resolving range, but ladder fragments to n = 6 (738 bp) were still separated in 30% gels (Fig. 1).
Figure 1.
Electrophoretic separation of double-stranded DNA fragments in Pluronic F127 gels. (A) Nx123 bp DNA ladder electrophoresed in (left to right): 18, 20, 22, 25, 27, and 30% Pluronic gel. (B) Electrophoresis of DNA restriction fragments in a 20% Pluronic gel. Lanes: 1, restriction endonuclease fragments of pGEM3Zf plasmid; 2, mixture of Nx123 ladder and pGEM fragments; 3, Nx123 bp ladder. The pGEM fragment lengths are 2,977, 1,799, 1,509, 724, 676, 517, 396, 325, 244 and 222 bp.
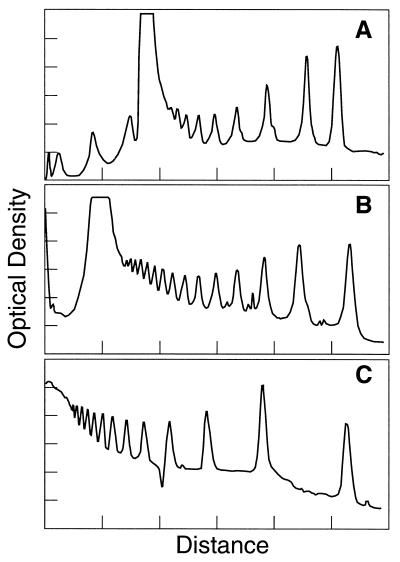
Figure 2.
Comparison of electrophoretic separations of Nx123 bp DNA ladder fragments in (A) 20% Pluronic, (B) 2% agarose, and (C) 6% polyacrylamide gels. Resolution between peaks, Ri,i+1 in the order R2,3, R3,4, R4,5, R5,6, and R6,7 were (6.1, 5.5, 3.7, 2.4, 1.8) for the Pluronic gel, (4.0, 3.8, 3.4, 2.8, 2.2) for the agarose gel, and (4.9, 3.1, 2.5, 1.7, 1.1) for the polyacrylamide gel, where Ri,i+1 = (di+1 − di)/[1/2(wi+1 + wi)], with di or i+1 = migration distance, and wi or i+1 = peak width at half-height.
DNA transport in Pluronic gels is facile in comparison to polyacrylamide gels of the same polymer concentration, as DNA standards >100 bp barely migrated in 20% polyacrylamide gels (data not shown). The effective DNA size separation range of 20% Pluronic gels, up to about 1,000 bp, is more comparable to that obtained in 6–8% polyacrylamide gels or 2–3% agarose gels (Fig. 2), although the absolute mobility of the 123-bp fragment in 20% Pluronic F127 was about 60% of its mobility in 6% polyacrylamide. The low absolute mobilities of small DNA fragments in Pluronic media are not a practical disadvantage because the low conductivities of Pluronic media allow application of higher voltage gradients (see Materials and Methods). In comparison to 6% polyacrylamide gels or 2% agarose gels, the resolution of Nx123 bp ladder fragments obtained in 20% Pluronic F127 was superior from 246 to 615 bp (n = 2–5), approximately equivalent from 625 to 861 bp, and poor above 1,000 bp (see legend to Fig. 2).
DNA mobilities in 20% Pluronic are nearly independent of length above 3,000 bp, as nearly identical mobilities were observed for linear, 3-kb pGEM DNA and 48-kb λ phage DNA (data not shown). Lack of size discrimination above 3 kb is correlated with unusually high mobilities of long, linear DNAs in Pluronic gels. Although the absolute mobilities of short DNA fragments were smaller in 20% Pluronic than 6% polyacrylamide gels, the absolute mobilities of fragments larger than 984 bp were higher in the Pluronic gel (Fig. 2). λ DNA (48 kb) migrated with almost the same absolute mobility in 20% Pluronic and 2% agarose gels (data not shown).
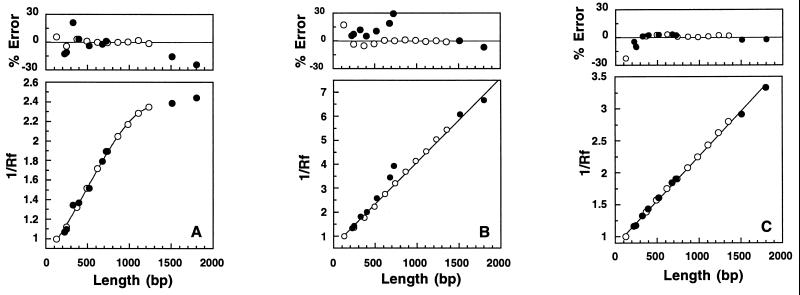
Transport of moderate length double-stranded DNA in agarose and polyacrylamide gels is described to a first approximation by the biased reptation model when the fragments are longer than the gel pore dimensions (6–9). The inverse dependence of mobility on DNA length predicted for migration by biased reptation was observed in 2% agarose and 5% polyacrylamide gels over a range from 123 to 1,800 bp (Fig. 3 B and C). Linear least-squares fits of these data were very good (R2 > 0.999). By contrast, the dependence of inverse mobility of the Nx123 ladder fragments in 20% Pluronic gels was linear only over a range of about 246–738 bp (Fig. 3A), and a third-order polynomial was required to fit data for ladder fragments from 123 to 1,353 bp with the same precision. In this sense the Pluronic gels are more size selective than the polyacrylamide or agarose gels, and the biased reptation model describes the migration of DNA fragments only over a narrow size range.
Figure 3.
(Lower) Dependencies of inverse mobilities on DNA length observed for electrophoresis in gels of (A) 20% Pluronic, (B) 5% polyacrylamide, and (C) 2% agarose. Open circles (○) represent Nx123 bp ladder fragments, and filled circles (•) represent pGEM fragments. Mobilities were normalized to the mobility of the 123-bp fragment. Solid lines for polyacrylamide and agarose gels in frames B and C are linear least-squares fits to all points. Solid line for Pluronic gel in A is a best-fit third order polynomial to data for the Nx123 bp fragments only. R2 ≥ 0.999 for all fits. (Upper) Errors (%) in the lengths estimated for Nx123 bp fragments (○) and pGEM fragments (•). Mobilities of the Nx123 ladder fragments only were least-squares fit to straight lines (2% agarose, 5% polyacrylamide) or a third-order polynomial (20% Pluronic). Best-fit equations (R2 ≥ 0.999) were used to calculate estimated lengths of the pGEM and Nx123 fragments from mobilities.
Calibration curves obtained by least-squares fits to mobilities of Nx123 ladder fragments were used to estimate the sizes of seven pGEM fragments (222–724 bp) from their mobilities in each medium. Examination of errors in these estimates revealed anomalies in fragment mobilities in Pluronic and polyacrylamide gels, but not in agarose gels (Fig. 3 Upper). All the pGEM fragments <1,000 bp migrated slower than expected in 5% polyacrylamide gels. Largest errors (>20%) were observed for 676 and 724 bp fragments. Analogous results were obtained in 7.5 or 10% polyacrylamide gels. In the 20% Pluronic gel a comparably large error (>25%) was obtained only for the 325-bp fragment.
Anomalous migration of pGEM fragments may be caused by sequence-dependent DNA curvature or bending (29, 30), which is detected in polyacrylamide but not agarose gels; a phenomenon attributed to the “tightness” of polyacrylamide (9). The 325-bp fragment migrated slower than expected in both Pluronic and polyacrylamide gels, but the effect was largest in Pluronic gels. This 325-bp sequence (positions 2,864–3,189) is contained within the 724-bp fragment (sequence positions 2,494 to 19) that migrated most anomalously in polyacrylamide gels, suggesting that Pluronic gels are particularly sensitive to curvature in small DNA fragments. Studies of other ladder standards supported this suggestion as “10-bp ladder” and “25-bp ladder” DNA standards (Promega) exhibited a smooth dependence of mobility on length when electrophoresed in agarose gels, but not in polyacrylamide gels or Pluronic gels (data not shown).
CE.
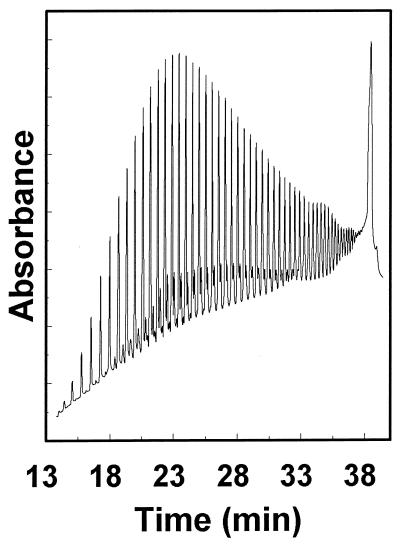
CE separations of oligonucleotides on Pluronic gels were investigated by using several standards including dT4–16 even, dT12–18, dT19–24, dN20–60, even, and a “poly(U)” sample (Sigma). Optimal separations were achieved in 25% gels. Excellent resolution was obtained for oligomers differing by 1 nt from 4 to about 40 nt, and oligonucleotides up to 60 nt long were separated in the poly(U) sample (Fig. 4). It should be noted that each major component in the poly(U) sample apparently was associated with two minor components that were also resolved in the region of the smaller oligonucleotides. One source of minor components may be terminally dephosphorylated chains. The presence of these contaminants degraded the apparent resolution of larger oligomers. The resolution between oligomers differing by 1 nt up to 24 nt long in the dT12–18 and dT19–24 standards ranged from 2.8 to 3.5, about twice that calculated from several published electropherograms of separations that used capillaries filled with linear polymer solutions or polyacrylamide gels (11, 17, 31).
Figure 4.
Single nucleotide resolution of components of poly(U) (Sigma) by capillary electrophoresis in Pluronic F127. The size range of distinguishable components is estimated as approximately 5–60 nt based on comparison with the mobilities of dTn samples of known lengths. Electrophoresis was performed in 25% Pluronic in TBE buffer at 25°C with an applied voltage of 18 kV (500 V/cm).
Different forms of double stranded DNA were also effectively separated by CE in Pluronic F127. Linear DNA fragments of the 123-bp ladder up to 861 bp were well resolved in less than 30 min (data not shown). CE of a supercoiled DNA ladder standard in 25% Pluronic gels yielded remarkable separations of the nine plasmids from 2 to 10 kb long, differing by 1 kb in length. Conventional electrophoresis on 1% agarose revealed the expected ladder of nine bands of nearly equal intensity; excepting the fourth band, the 5-kb plasmid supplied in higher abundance than the others. Electrophoresis in a short capillary of 25% Pluronic revealed roughly nine components as broad, poorly resolved bands (Fig. 5 Inset). The most intense band, presumably the 5-kb plasmid, was the sixth band detected, not the fourth, raising the surprising possibility that large supercoiled plasmids migrated faster than small ones. A separate experiment confirmed that the 3-kb pGEM3Zf plasmid migrated slower than larger plasmids on Pluronic gels.
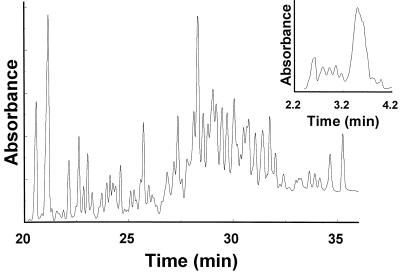
Figure 5.
Separation of supercoiled DNA plasmids by capillary electrophoresis in Pluronic gels. The standard (Promega) contains nine plasmid DNAs from 2 to 10 kb in length, differing by 1 kb. The 5-kb plasmid was supplied at about 3–4 times the abundance of other species. Optimal resolution (shown) was obtained in 30% Pluronic F127 (25°C; gradient, 500 V/cm; column length, 36 cm). (Inset) Low-resolution electrophoresis in 25% Pluronic F127 (25°C; gradient, 300 V/cm; column length, 7 cm).
Progressive increases of the capillary column length and Pluronic concentration caused progressive separation of the broad bands into multiple components. Optimized conditions revealed about 68 different components (Fig. 5), as the broad bands were resolved into sharp multiplets. The origins of this multiplet structure were not established independently. At least part of this complexity may arise because supercoiled plasmids consist of a Boltzmann distribution of topoisomers differing by units of one in linking number (number of superhelical turns). Topoisomers of small plasmids can be resolved by electrophoresis on agarose gels under optimal conditions (32).
DISCUSSION
We have found that Pluronic F127 gels are effective, versatile, and convenient media for electrophoresis. The most diverse applications of Pluronic and related media may be in capillary electrophoresis. Pluronic F127 offers practical advantages over other polymer media as technical problems limit applications of replaceable solutions of linear, random coil polymers to automated separations (17). Highly viscous solutions at optimal concentrations for oligonucleotide separations cannot be loaded directly into capillaries because of large back pressures. In situ polymerization of linear polyacrylamide used to solve this problem is not conveniently automated or highly reproducible (17). By contrast, the low viscosities of chilled, 20–30% solutions of Pluronic F127 (≈1 poise at 0°C; Y.L. and R.L.R., unpublished data) allowed easy hand-filling of capillaries. Capillary electrophoresis in Pluronic gels should be particularly useful for rapid assessment of purity, conformational properties, and interactions of oligonucleotides; capabilities pertinent to development of therapeutic oligonucleotides (33, 34) and PCR primers. Applications to rapid DNA sequencing, characterization of restriction endonuclease digests and PCR products, and peptide–protein separations are also feasible.
Gels of Pluronic F127 are also useful media for traditional electrophoresis. Excellent separations were obtained for double-stranded DNA fragments up to about 800 bp, which is sufficient for many applications such as characterization and recovery of PCR products or restriction fragments for subcloning. (Potential users should note that sequence-dependent DNA conformational variations may influence mobilities in Pluronic gels, as has been observed for polyacrylamide gels.) Preliminary studies have demonstrated separations of members within several other classes of molecules as varied as supercoiled plasmids, single-stranded oligonucleotides, native proteins, proteins unfolded in SDS, and even small dyes and mononucleotides.
Pluronic gels are quickly cast with pre-prepared, chilled solutions, without concerns for reproducibility of polymerization or dangers associated with hot agarose solutions or the toxicity of acrylamide. The latter attributes may be particularly beneficial in large volume operations where operator safety and waste disposal are major concerns.
Preparative electrophoresis is an attractive application of Pluronic gels. DNA and other substances are easily recovered because the gels are not crosslinked and liquify on chilling or dilution. Liquified samples can be applied directly to other gels for further analysis. Several restriction endonucleases are active in 10% Pluronic F127 (data not shown), hence the polymer is unlikely to interfere with enzyme-mediated manipulations of recovered DNA or proteins.
The well-defined structures of Pluronic liquid crystals provide a matrix so distinctly different from interconnected polymer networks that mechanisms of sieving of certain molecular classes on the two media must be fundamentally different. For example, DNA electrophoresis in crosslinked gel networks is commonly viewed as biased reptation, by which the chain is confined by the network to move in snake-like fashion within a “tube” determined by the path of the leading segment, while retaining an overall random coil configuration (6–9). Very long DNA fragments tend to stretch as some chain segments become “hooked” or otherwise trapped by the medium, ultimately causing loss of the dependence of mobility on length (6–8).
Biased reptation is not the only option available to long DNA migrating through Pluronic liquid crystals. Pluronic F127 micelles in the liquid crystalline state in a 20% solution (25°C) have an aggregate Mr of about 700,000, and micelles pack with an average intermicellar distance of 18 nm (24). Because the hard sphere diameter of the (PO)70 micelle core is about 9 nm, the minimum gap between cores in 20% gels is also about 9 nm. This gap is occupied by hydrated, uncrosslinked (EO)106 chains that extend from the micelle core surface. One can imagine these as molecular strands of a splayed soft brush. There are other spaces in interstices between micelles, presumably with low densities of EO chain segments. Thus DNA molecules must travel around large, regularly arranged spheres; through space occupied by uncrosslinked (EO)x chains (Fig. 6). “Reptation in a tube,” “hooking,” and related concepts are not appropriate because chain segments may escape a potential tube or hook by sliding around the micelle. The ease of sliding will depend on the resistance offered by the EO brush, relative to the force driving the DNA. Large variations in local electrical fields caused by the low dielectric susceptibility of micelle cores may also affect DNA motion. Both factors could cause significant effects of field strength on separations. We have not explored field effects on separations of linear DNAs, but strong effects of field and Pluronic concentration on separations of supercoiled plasmids were observed and will be described elsewhere.
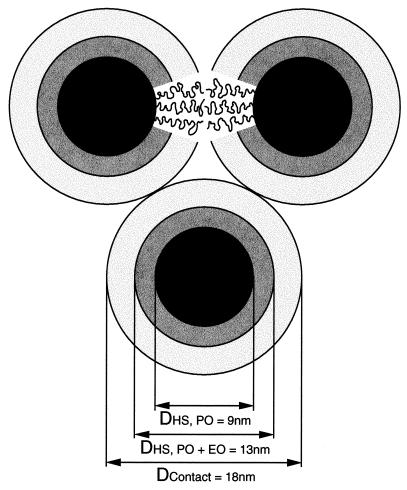
Figure 6.
Schematic of Pluronic F127 micelles in the liquid crystalline phase (20% solution). The central, black region represents the approximate hard-core dimensions of the poly(propylene oxide) or (PO)70 micelle core. The gray region represents the approximate hard-core dimensions of the poly(ethylene oxide) or (EO)106 micelle brush if hypothetically flattened onto the core surface. White plus gray regions represent the dimensions of the hydrated (EO)106 chains. Individual (EO)106 chains (curly lines) will be more extended than an unperturbed random coil because of crowding near the micelle core surface. Dimensions of the interstitial region depend on the packing geometry and degree of extension of (EO)106 chains into this region. Hard-core radii were calculated from the approximate molecular formula based on an aggregation number, n = 54 (24), and an assumed density of 1.0 g/cm3. The inter-micelle distance of 18 nm shown is based on small angle neutron diffraction studies (24).
The ability of a single medium to distinguish between molecules in diverse size classes from mononucleotides to supercoiled DNA plasmids is unique to Pluronic gels. Several models for separation appear to apply for these gels. We suggest that it is useful to view this structure as tripartite, consisting of interbrush, intrabrush, and core regions (Fig. 6). Highly charged macromolecules like single- and double-stranded DNA should avoid hydrophobic micelle cores and separate by sieving through the hydrated, (EO)x-rich inter- and intrabrush domains. Charged organic dyes and other amphiphilic molecules may separate by partitioning between hydrophobic and hydrophilic environments in a manner analogous to reverse phase chromatography.
Small oligonucleotides and much longer double-stranded DNA fragments were found to have roughly comparable electrophoretic mobilities in 20% Pluronic F127. This behavior can be rationalized in terms of differential access to inter- and intrabrush regions. The intrabrush region, defined by the spaces between individual (EO)x chains emerging from the micelle core, is envisioned as crowded and accessible only to relatively small or long, thin molecules such as single-stranded oligonucleotides. “Fatter” species such as double-stranded DNA and proteins should be largely restricted to migrate through interbrush regions defined by interstitial spaces and brushtips between neighboring micelles. Migration through the latter regions would require significant distortion of the uncrosslinked brush.
Liquid crystalline phases of nonionic polymers, as a class, should be useful for electrophoretic separations of diverse types of molecular ions. Various Pluronic and structurally related polymers are commercially available. Differences in lengths of EO and PO blocks define the micelle characteristics and liquid crystal-forming properties. Many undergo transitions between two or more liquid crystalline phases as functions of concentration and temperature (24). This diversity at the levels of molecular, micellar, and liquid crystalline phase structure presents rich opportunities for tuning media for optimal separations within different classes of molecular ions. The ordered structures of these phases also provide new challenges to those interested in mechanisms of macromolecular transport in dense media.
Acknowledgments
We thank Dr. Leonore C. Witchey-Lakshmanan (Schering-Plough Research Institute, Kenilworth, NJ) for bringing our attention to the properties of Pluronic polymers. We also thank Dr. William Cooper (Florida State University) for use of capillary electrophoresis equipment, and David Grassian, Mark Bacchus, Franz Kettwig, and Samuel Davis (Florida State University) for technical assistance. This work was supported by National Science Foundation Grant BES-9521381 and the Florida State University Center for Materials Research and Technology.
ABBREVIATIONS
- CE
capillary electrophoresis
- EO
poly(ethylene oxide)
- PO
poly(propylene oxide)
References
- 1.Ogston A G. Trans Faraday Soc. 1958;54:1754–1757. [Google Scholar]
- 2.Giddings J C, Kucera E, Russell C P, Myers M N. J Phys Chem. 1968;72:4397–4408. [Google Scholar]
- 3.Rodbard D. In: Methods of Protein Separation. Catsimpoolas N, editor. New York: Plenum; 1976. pp. 145–179. [Google Scholar]
- 4.Slater G W, Guo H L. Electrophoresis. 1996;17:977–988. doi: 10.1002/elps.1150170604. [DOI] [PubMed] [Google Scholar]
- 5.Slater G W, Guo H L. Electrophoresis. 1996;17:1407–1415. doi: 10.1002/elps.1150170903. [DOI] [PubMed] [Google Scholar]
- 6.Slater G W, Rousseau J, Noolandi J, Turmel C, Lalande M. Biopolymers. 1988;27:509–524. doi: 10.1002/bip.360270311. [DOI] [PubMed] [Google Scholar]
- 7.Slater G W, Noolandi J. Biopolymers. 1989;28:1781–1791. doi: 10.1002/bip.360281011. [DOI] [PubMed] [Google Scholar]
- 8.Deutsch J M, Madden T L. J Chem Phys. 1989;90:2476–2485. [Google Scholar]
- 9.Zimm B H, Levene S D. Q Rev Biophys. 1992;25:171–204. doi: 10.1017/s0033583500004662. [DOI] [PubMed] [Google Scholar]
- 10.Cohen A S, Najarian D R, Paulus A, Guttman A, Smith J A, Karger B L. Proc Natl Acad Sci USA. 1988;85:9660–9663. doi: 10.1073/pnas.85.24.9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baba Y, Matsuura T, Wakamoto K, Tsuhako M. J Chromatogr. 1991;558:273–284. [Google Scholar]
- 12.Karger B L. Curr Opin Biotechnol. 1992;3:59–64. doi: 10.1016/0958-1669(92)90127-5. [DOI] [PubMed] [Google Scholar]
- 13.Karger B L, Chu Y H, Foret F. Annu Rev Biophys Biomol Struct. 1995;24:579–610. doi: 10.1146/annurev.bb.24.060195.003051. [DOI] [PubMed] [Google Scholar]
- 14.Fung E N, Yeung E S. Anal Chem. 1995;76:1913–1919. [Google Scholar]
- 15.Carrilho E, Ruiz-Martinez M C, Berka J, Smirnov I, Goetzinger W, Miller A W, Brady D, Karger B L. Anal Chem. 1996;68:3305–3313. doi: 10.1021/ac960411r. [DOI] [PubMed] [Google Scholar]
- 16.Heiger D N, Cohen A S, Karger B L. J Chromatogr. 1990;516:33–48. doi: 10.1016/s0021-9673(01)90202-x. [DOI] [PubMed] [Google Scholar]
- 17.Chiari M, Nesi M, Fazio M, Righetti P G. Electrophoresis. 1992;13:690–697. doi: 10.1002/elps.11501301147. [DOI] [PubMed] [Google Scholar]
- 18.Barron A E, Soane D S, Blanch H W. J Chromatogr A. 1993;652:3–16. doi: 10.1016/0021-9673(93)80639-P. [DOI] [PubMed] [Google Scholar]
- 19.Chrambach A, Aldroubi A. Electrophoresis. 1993;14:18–22. doi: 10.1002/elps.1150140104. [DOI] [PubMed] [Google Scholar]
- 20.Tietz D, Aldroubi A, Pulyaeva H, Guszczynski T, Garner M M, Chrambach A. Electrophoresis. 1992;13:614–616. doi: 10.1002/elps.11501301124. [DOI] [PubMed] [Google Scholar]
- 21.Mortensen K, Brown W, Norden B. Phys Rev Lett. 1992;68:2340–2343. doi: 10.1103/PhysRevLett.68.2340. [DOI] [PubMed] [Google Scholar]
- 22.Malmsten M, Lindman B. Macromolecules. 1992;25:5440–5445. [Google Scholar]
- 23.Linse P. J Phys Chem. 1993;97:13896–13902. [Google Scholar]
- 24.Wanka G, Hoffmann H, Ulbricht W. Macromolecules. 1994;27:4145–4149. [Google Scholar]
- 25.Mortensen K, Talmon Y. Macromolecules. 1995;28:8829–8834. [Google Scholar]
- 26.Linse P, Malmsten M. Macromolecules. 1992;25:5434–5439. [Google Scholar]
- 27.Sullivan L R. M.S. Thesis. Tallahassee: The Florida State University; 1997. [Google Scholar]
- 28.Huang M, Bigelow M, Byers M. Am Lab. 1996;October:32–36. [Google Scholar]
- 29.Koo H S, Crothers D M. Proc Natl Acad Sci USA. 1988;85:1763–1767. doi: 10.1073/pnas.85.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagerman P J. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- 31.Auriola S, Jaaskelainen I, Mikko R, Urtti A. Anal Chem. 1996;68:3907–3911. [Google Scholar]
- 32.Pulleyblank D E, Shure M, Tang D, Vinograd J, Vosberg H P. Proc Natl Acad Sci USA. 1975;72:4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gelfi C, Perego M, Morelli S, Nicolin A, Righetti P G. Antisense Nucleic Acid Drug Dev. 1996;6:47–53. doi: 10.1089/oli.1.1996.6.47. [DOI] [PubMed] [Google Scholar]
- 34.Christoffersen R E. Nat Biotechnol. 1997;15:483–484. doi: 10.1038/nbt0697-483. [DOI] [PubMed] [Google Scholar]