Abstract
Increased acetylation at specific N-terminal lysines of core histones is a hallmark of active chromatin in vivo, yet the structural consequences of acetylation leading to increased gene activity are only poorly defined. We employed a new approach to characterize the effects of histone acetylation: A Drosophila embryo-derived cell-free system for chromatin reconstitution under physiological conditions was programmed with exogenous histones to assemble hyperacetylated or matching control chromatin of high complexity. Hyperacetylated chromatin resembled unmodified chromatin at similar nucleosome density with respect to its sensitivity toward microccal nuclease, its nucleosomal repeat length, and the incorporation of the linker histone H1. In contrast, DNA in acetylated chromatin showed an increased sensitivity toward DNase I and a surprisingly high degree of conformational flexibility upon temperature shift pointing to profound alterations of DNA/histone interactions. This successful reconstitution of accessible and flexible chromatin outside of a nucleus paves the way for a thorough analysis of the causal relationship between histone acetylation and gene function.
The basal unit of eukaryotic chromatin, the nucleosome, consists of an octamer of histones, around which the DNA is wrapped in approximately two helical turns. The flexible N termini of all four core histones protrude from the otherwise very compact nucleosome structure. The amino acid sequences of these exposed “tails” have been highly conserved during evolution, reflecting their importance for chromatin structure and function (1, 2). The many reported roles of histone N termini can be explained by their ability to reach out and to contact other components of chromatin. Conserved basic residues in the tails interact with DNA in isolated nucleosomes leading to a moderate stabilization of the nucleosome structure (reviewed in ref. 2). Within a nucleosomal array the tail domains rearrange from positions on the nucleosomal DNA (3) to contact sites within the linker DNA as well as on neighboring nucleosomes and to promote the folding of the array into higher order fibers (2, 4, 5). They are also important for the regular spacing of nucleosomes in such an array (6). Histone tails contribute to the chromatin-mediated repression of basal and activated transcription (7, 8). They also participate in the establishment of higher order chromatin structures involved in complex biological phenomena, like the silencing of the yeast mating type loci (7), through direct interaction with specific nonhistone proteins (9, 10).
The accessible tails are the preferred targets of posttranslational histone modifications, notably acetylations at the ɛ-amino groups of conserved lysine residues (11). It is assumed that this acetylation affects important properties of the tails, but the causalities between acetylation and alterations in chromatin structure and function have not been determined. Acetylation only moderately affects nucleosome structure and stability at physiological conditions in the absence of additional factors (12–15) but has profound consequences for the folding of the nucleosomal fiber (16, 17). However, it has been reported that hyperacetylated nucleosomes constrain DNA superhelicity differently from unmodified ones (18–21).
Studies on native chromatin and intact nuclei revealed striking correlations between general histone acetylation, transcription potential, DNase I sensitivity (22), and a altered interaction of the linker histone H1 (ref. 23 and references therein). Conversely, inactive, silenced, or heterochromatin domains are characterized by global hypoacetylation (24, 25). However, these correlations do not hold true when particular H4 isoforms are monitored indicating that acetylation of specific lysines rather than bulk acetylation is of functional importance (26, 27). In addition to these domain-wide phenomena, model systems using mononucleosomes show that acetylation facilitates the interaction of transcription factors with nucleosomal binding sites (28, 29). Bulk hyperacetylation of chromatin in vivo can facilitate the remodeling of specific nucleosomes leading to activation of some promoters, but not others (30–33). The recent identification of histone acetyltransferase activity of some transcriptional regulators (reviewed in ref. 34) has fueled hypotheses that histone modification is an essential step toward the establishment of active transcription in chromatin and that acetylation may be targeted specifically to those nucleosomes at promoters that restrict the access of the transcription machinery.
Despite the strong correlations between histone acetylation and gene activity the features that distinguish acetylated chromatin from unmodified domains leading to the activation of nuclear processes, are unknown. We used a cell-free system for chromatin reconstitution derived from Drosophila embryo extracts (35) to reconstitute chromatin with elevated acetylation levels on all four core histones in vitro that allowed to biochemically address a number of open questions regarding the structural consequences of histone acetylation.
MATERIALS AND METHODS
Histone Isolation.
Green monkey kidney CV1 cells were treated with 500 ng/ml trichostatin A (TSA; Wako) for about 20 h to induce hyperacetylation. TSA-treated or untreated cells were harvested on ice. Core histones were isolated as described (20) with the following modifications: Nuclei were extracted with 20 ml of 0.25 M H2SO4 per 0.5 ml of pelleted nuclei for 30 min on ice, the reaction was centrifuged at 15,000 × g for 30 min and the supernatant was dialyzed overnight (Mr cutoff 6,000–8,000) at 4°C against buffer A (10 mM 2-mercaptoethanol/5 mM Na butyrate/0.1 mM phenylmethylsulfonyl fluoride/5 ng/ml TSA). After dialysis, core histones were precipitated with an equal amount of 10% perchloric acid for 30 min on ice, collected by centrifugation at 10,000 × g for 30 min, and resuspended in 10 ml of 0.5 M HCl. The histone solution was concentrated in Filtron 3K ultrafiltration tubes to about 1 ml and finally dialyzed overnight against buffer A. To visualize histones after chromatin assembly, chromatin immobilized on Dynabeads was washed twice with 1 ml EX-80 buffer and then extracted for 1 h at 0°C with 0.5 ml of 2 M NaCl. Histones in the supernatant were precipitated with 0.25% trichloroacetic acid, reextracted with HCl, and then precipitated with 6 vol of acetone. Histones were analyzed on SDS/13% polyacrylamide gels (36) and in 12% Triton-acid-urea gels as described (16).
Chromatin Assembly.
Chromatin assembly extracts (37) from 3–6 hr embryos were depleted of endogenous histones by incubation with 50 μg of immobilized DNA per 250 μl of extract for 30 min at 4°C (38). A standard assembly reaction (37) contained 20 μl of depleted extract, 100 μl EX buffer, 13.3 μl of a 10× buffer containing 0.3 M creatine phosphate, 30 mM MgCl2, 30 mM ATP (pH 8.0), 1 μg/ml creatine kinase, and 10 mM dithiothreitol, 650 ng plasmid DNA and purified core histones at the desired histone:DNA ratio in a final volume of 133 μl and was incubated for 6 h at 26°C. The histone:DNA ratio was usually 3:1 (wt/wt) for assembly without H1 and 1.5:1 for assembly in the presence of H1. The H1:DNA ratio (wt/wt) was generally 0.25. The KCl concentrations were varied as indicated in the figures (according to refs. 6 and 38).
Structural Analysis of Reconstituted Chromatin.
Digestion with micrococcal nuclease (MNase) was as described (37, 38), directly scaled to the volume of the assembly reactions. For analysis of DNase I sensitivity, 20 μl of the standard assembly reaction were digested with 0, 1, 1.5, 2.25, 3.4, 5, 7.6, and 11.3 units of DNase I in 10 μl of EX buffer/1 mM CaCl2 (KCl concentrations adjusted to the assembly conditions) for 1 min at 26°C. For analysis of temperature-dependent DNA flexibility, 20 μl of the standard assembly reaction were incubated for 1 h at 35°C or 3–4 h at 5°C with 0.5 unit of Drosophila topoisomerase I (Pharmacia). Reactions were stopped with 10 μl of 2.5% (wt/vol) Sarkosyl/100 mM EDTA. DNA was isolated and analyzed on 1.2% agarose gels containing 3–5 μM chloroquin in the dark.
RESULTS
Reconstitution of Hyperacetylated Chromatin.
Nuclear histone acetylation levels are the result of a dynamic equilibrium between acetylase and deacetylase activities. Treating tissue culture cells with nanomolar concentrations of a specific deacetylase inhibitor, TSA leads to a general hyperacetylation of nucleosomes (39) and selective activation of some genes (30–33). To recreate hyperacetylated chromatin in vitro we employed a cell-free system derived from Drosophila embryo extracts (35). Chromatin assembly in these extracts usually relies on maternal pools of histones and chromatin assembly factors. Extracts from postblastoderm embryos contain significantly less endogenous histones (40) and if these histones are further depleted from the extract, chromatin assembly will entirely depend on exogenously added histones (38). To assemble chromatin with elevated acetylation levels we programmed the assembly extracts with hyperacetylated histones. Since Drosophila cells do not respond well to TSA (41) we purified bulk histones from either TSA-treated CV1 cells that accumulated hyperacetylated histone isoforms or from untreated cells as controls. In a direct comparison CV1 cells accumulated significantly higher levels of histone acetylation upon TSA treatment than the more commonly used HeLa cells. An acetylation-dependent contamination of the histone preparations by nonhistone proteins was ruled out by SDS/PAGE of the histones (data not shown).
Histone isoforms differing in their acetylation status were resolved on discontinuous Triton/acid/urea gels (42). The isoform composition of the histones derived from either control or TSA-treated cells (“acetylated”) is shown in Fig. 1 (in). The well-resolved isoforms of H4 may be used as a convenient indicator for the degree of acetylation. While control H4 is predominantly unmodified, the bulk of these histones from TSA-treated cells are tetraacetylated. Clear differences in isoform composition can also be identified for the other core histones.
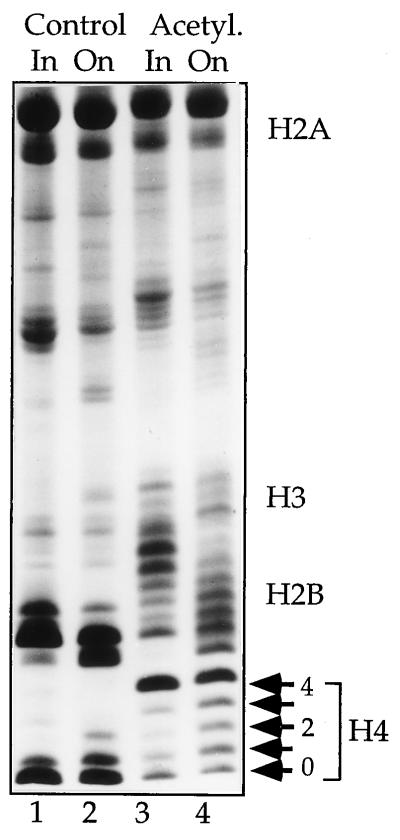
Figure 1.
Acetylation state of histones used. Histone isoforms purified from TSA-treated (Acetyl.) or untreated (Control) CV1 cells were separated on Triton/acid/urea gels. The isoform distribution of input histones (In) is contrasted with the status of histones after incorporation into chromatin (On). For histone H4 the familiar isoform distribution from unacetylated (0) to tetraacetylated H4 (4) is marked to the right side of the gel.
The assembly of long, regular arrays of nucleosomes requires the extended incubation of DNA and histones in a crude Drosophila cytoplasmic extract. To monitor the action of presumed histone acetylases and deacetylases in the extract that might alter the acetylation status of input histones (43, 44) we assembled chromatin for 6 h in the extract and reextracted the histones for Triton/acid/urea gel analysis. In each case the isoform composition of input histones (Fig. 1, In) differed significantly from the composition of the histones assembled into nucleosomes (Fig. 1, On). During assembly, the acetylation status of control histones was moderately increased, while hyperacetylated histones were deacetylated significantly, pointing to the presence of a minor histone acetylase activity and considerable deacetylase activity. As predicted from the insensitivity of Drosophila cells to deacetylase inhibitors, the presumed extract deacetylases could not be inhibited by either TSA or trapoxin (39). Nevertheless, a striking difference in acetylation status remained between control and acetylated chromatin even after extended incubation in the extract (Fig. 1, compare On lanes).
Histone Acetylation Does Not Affect the Nucleosome Repeat Length.
Nucleosome assembly extracts depleted of endogenous histones failed to assemble significant amounts of nucleosomes, as revealed by digestion of chromatin with MNase (Fig. 2). Titration of either control or acetylated histones into the system allowed increasing numbers of nucleosomes to be deposited until extended nucleosomal arrays with regular spacing were reconstituted (Fig. 2). The nominal histone input per DNA (wt/wt) to achieve optimal assembly was empirically determined and was similar for control and acetylated histones (see Fig. 4B). The rate of MNase digestion of the two types of chromatin and the extent of the regular repeat pattern were similar, confirming earlier observations on native chromatin (12, 45) and demonstrating that nucleosome assembly from the two histone sources occurred with equal efficiency.
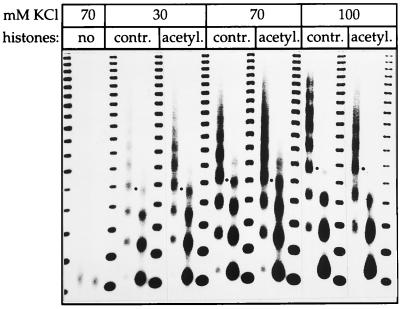
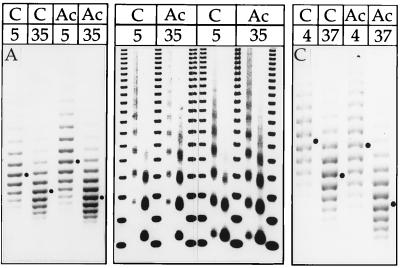
Figure 2.
Analysis of nucleosome spacing. Chromatin was reconstituted at the indicated KCl concentrations from control or acetylated histones and digested with MNase to reveal the NRL. As the KCl concentration is raised the NRL increases to a similar extent for both types of chromatin. Dots marking the position of the tetranucleosomal fragments. A negative print of the ethidium-stained DNA is shown.
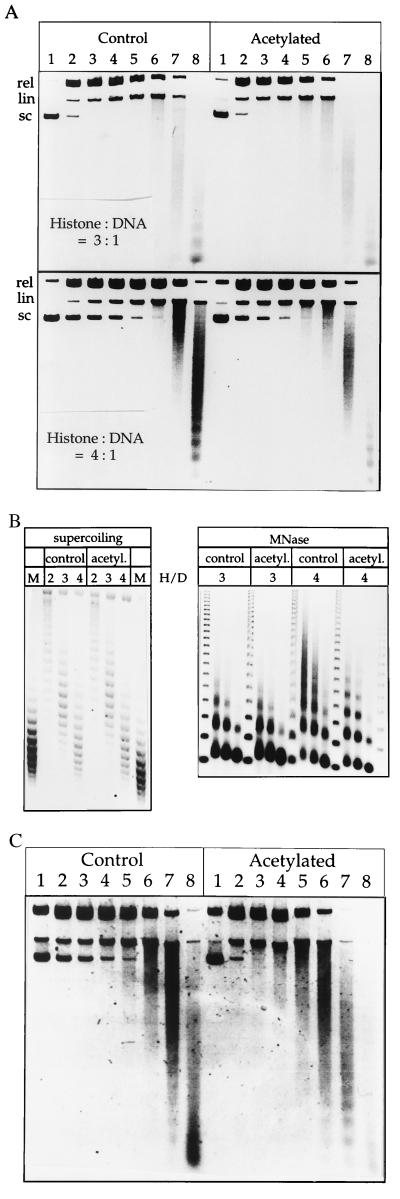
Figure 4.
Acetylated chromatin is DNase I sensitive. (A) DNase I analysis of control and acetylated chromatin. Reconstituted chromatin was digested with increasing concentrations of DNase I (see Materials and Methods), and the deproteinized DNA fragments were analyzed on an agarose gel. A negative print of the ethidium-stained DNA is shown. (B) Left, The superhelical density of the two types of chromatins is identical. Acetylated and control chromatin was reconstituted at histone:DNA (wt/wt) ratios between 2 and 4. Deproteinization leaves an unrestrained supercoil for each nucleosome. Plasmid topoisomers were resolved on an agarose gel in the presence of chloroquin. The plasmid isolated from bacteria, at a superhelical density of 0.05 (69), served as a marker (M) equivalent to ≈31 superhelical turns. An equivalent superhelicity on plasmids in chromatin corresponds to about one nucleosome per 197 bp. Right, DNase I-sensitive chromatin is not sensitive to MNase. The MNase analysis was as in Fig. 2. (C) The preferential DNase I sensitivity of acetylated chromatin is enhanced in the presence of H1. Chromatin was reconstituted from control and acetylated histones in the presence of H1 (see Fig. 2) to an equivalent degree (not shown) and digested with DNase I as in A.
A regular ladder of fragments was obtained from partial digestion of reconstituted chromatin with MNase, indicating a defined nucleosome repeat length (NRL), a hallmark of physiological chromatin. We recently observed that the precise NRL established in reconstituted chromatin is determined by the histone tails, the presence and stoichiometry of histone H1 and the overall cation concentrations during assembly (6). This fueled the hypothesis that acetylation, which neutralizes charges in N-terminal lysines, would impact on the NRL. However, in contrast to this notion, the nucleosome spacing was unaffected by the acetylation status of reconstituted chromatin under several assembly conditions (Fig. 2). While the NRL clearly increased as a function of monovalent cation concentration, it was identical in acetylated and control chromatin.
Histone H1 Incorporation Is Unaffected by Bulk Histone Acetylation.
Earlier analyses of native chromatin indicated that the linker histone H1 interacted differently with acetylated chromatin, suggesting that histone acetylation may affect chromatin structure indirectly through differential association with H1 (23, 46). Since the cell-free reconstitution system allows the efficient incorporation of purified histone H1 into chromatin we addressed the question whether this incorporation was affected by histone acetylation. This can be monitored conveniently since the NRL increases as a function of the amount of incorporated H1 (6, 47). Addition of appropriate amounts of H1 led to the expected increase in NRL, but also to significantly longer nucleosomal arrays (Fig. 3 Left). This improved assembly/alignment of nucleosomes by the linker histone is usually not observed if chromatin is assembled from endogenous histones (6). Importantly, however, the analysis did not reveal any difference in H1 incorporation between acetylated and control chromatin under a variety of conditions (Fig. 3). We conclude that bulk nucleosome acetylation does not affect the affinity of histone H1 for chromatin.
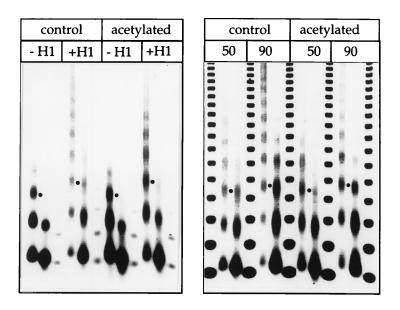
Figure 3.
Incorporation of histone H1 into acetylated and control chromatin. The incorporation of histone H1 can be visualize by a characteristic increase in NRL. Addition of equivalent amounts of H1 to assembly reaction with control or acetylated histones has identical effects on the extent of the MNase pattern and the NRL (see dots marking equivalent fragments). A negative print of the ethidium-stained DNA is shown.
Reconstituted Acetylated Chromatin Is DNase I-Sensitive.
In intact nuclei, chromatin domains with transcription potential are marked by elevated levels of specifically acetylated histone isoforms and by a generally increased sensitivity toward DNase I digestion (12, 22, 45, 48). This domain-wide DNase I sensitivity may be due to the differential packaging of chromatin into higher order chromosomal structures. Isolated or reconstituted mononucleosomes differing in acetylation status show only little difference in DNase I sensitivity, unlikely to explain the DNase I sensitivity observed in nuclei (12, 49). The reconstitution of hyperacetylated chromatin in Drosophila embryo extracts enables a new approach to examine whether histone acetylation is causally involved in DNase I sensitivity.
To compare the DNase I sensitivity of both types of chromatin we needed to assure that nonacetylated and acetylated chromatin was reconstituted to a similar degree. MNase analysis of the two types of chromatin revealed that both samples were assembled to a similar degree when comparable amounts of histones were titrated into the assembly reaction (histones:DNA mass ratios were varied between 2–4; Fig. 4B). As a further control we also analyzed the topology of these chromatin samples. The superhelical density of a plasmid minichromosome is a reflection of the nucleosome density, since the winding of DNA around a nucleosome introduces approximately one superhelical turn into the DNA (50). Whether or not acetylated nucleosomes restrain DNA to the same extent has been debated: Bradbury and coworkers (18) reported that acetylated nucleosomes, reconstituted by salt gradient dialysis, restrained only ≈0.8 superhelical turn each, but Lutter and coworkers (21) failed to detect a corresponding change of minichromosome topology in vivo upon butyrate-induced hyperacetylation of simian virus 40 minichromosomes. Upon assembly of chromatin with increasing amounts of control and acetylated histones the superhelical density on the plasmid increased accordingly and to similar levels for each histone:DNA ratio (Fig. 4B). Remarkably, the acetylated chromatin revealed an increased DNase I accessibility when compared with equivalent control chromatin that became more pronounced as the nucleosomal density on the template increased (Fig. 4A; compare DNase sensitivity at histone DNA ratios of 3 and 4). When the experiment was repeated in the presence of histone H1 the relative difference in DNase I sensitivity between the two types of chromatin was even higher (Fig. 4C). Indirect endlabeling experiments demonstrated that the increased susceptibility of acetylated chromatin to DNase I was indeed due to a global sensitivity and not to local, acetylation-induced DNase I hypersensitivity (data not shown). The increased DNase I sensitivity of hyperacetylated chromatin may be severely underestimated, if according to Norton et al. (18) acetylated chromatin contains significantly more nucleosomes (see Discussion). We conclude (i) that DNase I sensitivity is a feature of acetylated nucleosomal arrays in the absence of the native domain structure characteristic of a nucleus (ii), that the association of histone H1 increases the difference in DNase I sensitivity between acetylated and control chromatin, and (iii) that the relationship between histone acetylation and DNase I sensitivity is a causal one, rather than a simple correlation.
Histone Acetylation Increases the Conformational Flexibility of DNA in Chromatin.
One very sensitive measure of histone/DNA interactions is the degree with which nucleosomes constrain temperature-induced flexibility of associated DNA (51). In covalently closed circular DNA the temperature-dependent untwisting of DNA leads to a corresponding increase in negative superhelicity (ref. 52 and references therein). Nucleosomes prevent the unwinding of associated DNA in vitro and in intact simian virus 40 minichromosomes (53, 54). Therefore, the degree of temperature-dependent unwinding (increase in negative superhelicity) observed in a minichromosome is a measure of the free, unrestrained DNA. To determine the amount of flexible DNA in reconstituted chromatin, each sample was split and incubated with topoisomerase I at two different temperatures (Fig. 5A). Relaxation of control minichromosomes at 35°C leads to accumulation of approximately two additional negative superhelical turns when compared with relaxation at 5°C (Fig. 5A, control). This is due to the unwinding of all unrestrained DNA in long nucleosomal linkers and gaps between nucleosomal arrays. In parallel, chromatin was reconstituted from hyperacetylated histones to a similar degree (compare with topoisomer distribution between the two samples relaxed at 5°C and the parallel MNase analysis, Fig. 5 Center). When the sample was relaxed at 35°C, four additional superhelical turns appeared. Acetylated nucleosomes obviously constrain DNA much less than nonacetylated nucleosomes (see below), a clear demonstration of altered or reduced histone/DNA interactions that may be causal to the observed DNase I sensitivity and transcriptional potential of acetylated chromatin. Increased thermal untwisting of acetylated chromatin was also observed in H1-containing chromatin (Fig. 5C). A temperature increase of 33°C resulted in the appearance of ≈2.5 superhelical turns in control, but 4.5 turns in acetylated chromatin.
Figure 5.
Increased temperature-dependent flexibility of DNA in acetylated chromatin. (A) Left, Control (C) and acetylated (Ac) chromatin was incubated at 5°C or 35°C (indicated by the numbers above the lanes) and relaxed completely with topoisomerase I. Samples were deproteinized and analyzed on chloroquin-containing agarose gels. A negative print of the ethidium-stained DNA is shown. Center, Aliquots of the relaxed chromatin were treated with MNase to visualize the NRL (see Fig. 2). (C) Right, Histone H1-containing chromatin was subjected to temperature-dependent unwinding analogous to the experiment shown in A), except that the temperature was shifted from 4°C to 37°C.
DISCUSSION
To determine the structural effects of acetylation in reconstituted chromatin we needed to compare chromatin with equivalent nucleosome densities and hence control for possible effects of histone acetylation on the efficiency of nucleosome assembly. In addition to the qualitative assessment by comparison of MNase ladders we determined the nucleosome numbers from the superhelical state of the minichromosomes: the winding of DNA around a nucleosome introduces approximately one negative supercoil (50). The direct comparison is complicated by the observation of Bradbury and colleagues (18) that hyperacetylated nucleosomes, assembled by salt gradient dialysis, restrained only 0.8 supercoils. If this also applied to our extract reconstitution, acetylated chromatin with a superhelical density equivalent to that of control chromatin should, on average, contain about six more nucleosomes than control chromatin, leading to severe underestimation of the effects of nucleosome acetylation on chromatin structure. It is, however, unclear whether this reduced supercoiling by acetylated nucleosomes also applies to chromatin reconstituted under more physiological conditions in the embryo extract that contains a wealth of associated nonhistone proteins. Addition of equivalent amounts of either acetylated or control histones to an assembly reaction leads to formation of chromatin with similar superhelical densities, extent of MNase ladders and nucleosome spacing. Lutter et al. (21) failed to see significant changes in minichromosome topology upon hyperacetylation of nucleosomes in vivo. The conservative assumption that the nucleosomal densities of acetylated chromatin is at least as high as the nucleosome density of control chromatin is supported by a determination of nucleosome densities with psoralen crosslinking (K. P. Nightingale, R. Wellinger, J. Sogo, and P.B.B., unpublished results). Under those conditions we find remarkable structural and functional (see below) consequences of nucleosome acetylation.
To appreciate the striking difference in histone/DNA interactions revealed by the thermal untwisting experiments one needs to consider that upon temperature shift, protein-free DNA untwists 0.011°/°C/bp (ref. 51 and references therein). Accordingly, the unwinding of our 6.1-kb plasmid during a temperature shift of 30°C is compensated by the appearance of about six superhelical turns that are fixed in the presence of topoisomerase I (data not shown). The interaction with the histone octamer significantly restricts the flexibility of DNA to untwist. Taking into account the partial dissociation of histones from DNA at elevated temperature, each nucleosome restrains ≈160–180 bp from thermal untwisting (59, 60).
When we analyzed control chromatin for thermal untwisting about two negative supercoils appeared. Therefore, about two-thirds of the plasmid was restrained by chromatin while 1/3 of the DNA, in the gaps between nucleosomal arrays and in long nucleosomal linkers, was flexible to unwind. Remarkably, if acetylated chromatin with similar superhelical densities was subjected to this assay, four superhelical turns were introduced. Acetylated chromatin therefore only constrains one-third of the total DNA. Since the nucleosome spacing was identical in both types of chromatin and acetylated chromatin did not contain less nucleosomes than control chromatin, any difference in the degree of thermal untwisting must be due to altered histone/DNA interactions in the DNA that is usually restrained by nucleosomes. This includes DNA associated with the histone octamer (146–160 bp) as well as part of the adjacent linker DNA (60). A similarly high degree of rotational flexibility has recently been observed in vivo in yeast minichromosomes (61, 62) and transcriptionally active bovine papillomavirus-based minichromosomes in intact mouse cells and in isolated nuclei (42), which may represent structural features of transcriptionally active chromatin (for a discussion see ref. 63).
Acetylation-induced altered histone/histone interactions have been derived from salt-induced nucleosome unfolding experiments and loosened histone/DNA interactions are detected in the premelting region of thermal denaturation profiles (64). Because DNA at the end of nucleosomes melts first in thermal denaturation experiments (65, 66) it may be that DNA at the entry/exit points is released from the nucleosome core particle in acetylated chromatin. Simpson (12) observed that the ends of DNA from hyperacetylated mononucleosomes were somewhat more accessible than those in control nucleosomes. Interestingly, the differential DNase I sensitivity of acetylated chromatin is essentially lost if chromatin is degraded to mononucleosomes (12). This may reflect rearrangement of the histone tails, which somehow must play a role as determinants of DNase I sensitivity, to bind contact sites on the nucleosome core when a nucleosome is taken out of the context of a regular array (2). Paradoxically, the temperature-induced untwisting of chromatin is unaltered if the histone N termini are removed from nucleosomes by trypsin (60, 67), which could be taken as an indication that the modification status of the tails might affect the properties of the nucleosome indirectly through allosteric distortions. It is possible that these effects are mediated by yet unknown proteins present in the assembly extracts that interact with tails in ways modulated by their acetylation status.
Increased levels of acetylation have been correlated with chromatin domains that are, in general, DNase I-sensitive and therefore more accessible than bulk chromatin containing genes with transcription potential (22). It is likely that acetylation at specific N-terminal histone lysines rather than the global net charge neutralization in tails is an important determinant. For example, the presence of an H4 isoform acetylated specifically at Lys-16 at the Drosophila male X chromosome correlates with a decondensed appearance of the entire chromosome and a two-fold increased transcription. This dosage compensation of X-linked genes is vital for male flies (55). By contrast, chromatin-mediated gene silencing, as occurs in yeast mating type regulation, Drosophila position-effect variegation or X-chromosome inactivation in mammalian cells is accompanied by a general hypoacetylation of nucleosomes (1, 22, 24, 56). Histone acetylation therefore appears to be a marker for chromosomal domains or even entire chromosomes (this does not exclude the additional targeting of acetyltransferases to specific regulatory sites; ref. 57). It is, therefore, likely that histone acetylation affects, among other things, the higher order folding of chromatin in nuclei.
In contrast to the rather small effects that acetylation has on the structure of isolated mononucleosomes (12, 13, 58), we have detected clear effects when analyzing nucleosomes in arrays with regular spacing. Under physiological ionic conditions nucleosomal arrays fold into more or less regular structures with an approximate diameter of 30 nm (2). In model experiments, arrays of hyperacetylated nucleosomes do not undergo cation-dependent folding (16, 17), although an earlier analysis of complex, native chromatin failed to reveal an effect of butyrate-induced hyperacetylation on the extent of chromatin folding (68). Chromatin reconstituted under physiological conditions is highly folded but the issue of whether a particular higher order structure is adopted has not been addressed so far. It is possible that histone acetylation modulates histone/DNA and nucleosome/nucleosome interactions to establish a less tightly folded, accessible fiber. There are also functional consequences of acetylation in the cell-free system: we recently observed that a 26-kDa heat shock protein gene was transcribed more efficiently if reconstituted into acetylated vs. control chromatin (K. P. Nightingale, W.A.K., and P.B.B., unpublished observations).
It has previously been proposed that the structural differences observed between acetylated and control chromatin were due to a differential interaction of histone H1 and accordingly different H1-mediated degrees of condensation (23, 46). We show that H1 associates well with acetylated nucleosomes in nucleosomal arrays, extending earlier findings that H1 can interact with equal efficiency with acetylated and control mononucleosomes (58). We did not address the stability of this interaction and therefore cannot exclude that H1 is selectively removed from acetylated chromatin in vivo. The increased DNase I sensitivity and transcription potential of acetylated chromatin were observed in the absence of H1 and are therefore intrinsic to the nucleosomal array. However, the difference between the two types of chromatin in terms of DNase I sensitivity and derepression of transcription were more pronounced in presence of H1 (Fig. 4C and K. P. Nightingale, W.A.K., and P.B.B., unpublished results). These results are consistent with the earlier suggestion, that histone acetylation may alter the capacity of H1 to compact the nucleosomal fiber (23, 46).
Acknowledgments
We thank Dr. K. P. Nightingale for critical reading of the manuscript and hypersensitive site analysis. W.A.K. was supported by a Human Frontier Science Program fellowship.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: MNase, micrococcal nuclease; NRL, nucleosomal repeat length; TSA, trichostatin A
References
- 1.Turner B M, O’Neill L P. Semin Cell Biol. 1995;6:229–236. doi: 10.1006/scel.1995.0031. [DOI] [PubMed] [Google Scholar]
- 2.Hansen, J. C. (1998) ChemTracts: Biochemistry and Molecular Biology, in press.
- 3.Usachenko S I, Bavykin S G, Gavin I M, Bradbury E M. Proc Natl Acad Sci USA. 1994;91:6845–6849. doi: 10.1073/pnas.91.15.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fletcher T M, Hansen J C. J Biol Chem. 1995;270:25359–25362. doi: 10.1074/jbc.270.43.25359. [DOI] [PubMed] [Google Scholar]
- 5.Fletcher T M, Hansen J C. Crit Rev Eukaryotic Gene Expression. 1996;6:149–188. doi: 10.1615/critreveukargeneexpr.v6.i2-3.40. [DOI] [PubMed] [Google Scholar]
- 6.Blank T A, Becker P B. J Mol Biol. 1995;252:305–313. doi: 10.1006/jmbi.1995.0498. [DOI] [PubMed] [Google Scholar]
- 7.Grunstein M, Hecht A, Fisher A G, Wan J, Mann R K, Strahl B S, Laroche T, Gasser S. J Cell Sci Suppl. 1995;19:29–36. doi: 10.1242/jcs.1995.supplement_19.4. [DOI] [PubMed] [Google Scholar]
- 8.Lenfant F, Mann R K, Thomsen B, Ling X, Grunstein M. EMBO J. 1996;15:3974–3985. [PMC free article] [PubMed] [Google Scholar]
- 9.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 10.Edmondson D G, Smith M M, Roth S Y. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 11.Bradbury E M. BioEssays. 1992;14:9–16. doi: 10.1002/bies.950140103. [DOI] [PubMed] [Google Scholar]
- 12.Simpson R T. Cell. 1978;13:691–699. doi: 10.1016/0092-8674(78)90219-2. [DOI] [PubMed] [Google Scholar]
- 13.Ausio J, van Holde K E. Biochemistry. 1986;25:1421–1428. doi: 10.1021/bi00354a035. [DOI] [PubMed] [Google Scholar]
- 14.Imai B S, Yau P, Baldwin J P, Ibel K, May R P, Bradbury E M. J Biol Chem. 1986;261:8784–8792. [PubMed] [Google Scholar]
- 15.Bauer W R, Hayes J J, White J H, Wolffe A P. J Mol Biol. 1994;236:685–690. doi: 10.1006/jmbi.1994.1180. [DOI] [PubMed] [Google Scholar]
- 16.Krajewski W A, Panim V M, Razin S V. Biochem Biophys Res Commun. 1993;196:455–460. doi: 10.1006/bbrc.1993.2271. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Ramirez M, Rocchini C, Ausio J. J Biol Chem. 1995;270:17923–17928. doi: 10.1074/jbc.270.30.17923. [DOI] [PubMed] [Google Scholar]
- 18.Norton V G, Imai B S, Yau P, Bradbury E M. Cell. 1989;57:449–457. doi: 10.1016/0092-8674(89)90920-3. [DOI] [PubMed] [Google Scholar]
- 19.Norton V G, Marvin K W, Yau P, Bradbury E M. J Biol Chem. 1990;265:19848–19852. [PubMed] [Google Scholar]
- 20.Krajewski W A, Luchnik A N. Mol Gen Genet. 1991;230:442–448. doi: 10.1007/BF00280301. [DOI] [PubMed] [Google Scholar]
- 21.Lutter L C, Judis L, Paretti R F. Mol Cell Biol. 1992;12:5004–5014. doi: 10.1128/mcb.12.11.5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perry C A, Annunziato A T. Exp Cell Res. 1991;196:337–345. doi: 10.1016/0014-4827(91)90269-z. [DOI] [PubMed] [Google Scholar]
- 24.Braunstein M, Rose A, B, Holmes S G, Allis D C, Broach J B. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 25.Jeppesen P, Turner B M. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 26.Turner B M. Cell. 1993;75:5–8. [PubMed] [Google Scholar]
- 27.O’Neill L P, Turner B M. EMBO J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee D Y, Hayes J J, Pruss D, Wolffe A P. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 29.Vettese-Dadey M, Grant P A, Hebbes T R, Crane-Robinson C, Allis C D, Workman J L. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 30.Almouzni G, Dimitrov S, Khochbin S, Wolffe A P. Dev Biol. 1994;165:654–669. doi: 10.1006/dbio.1994.1283. [DOI] [PubMed] [Google Scholar]
- 31.Bartsch J, Truss M, Bode J, Beato M. Proc Natl Acad Sci USA. 1996;93:10741–10746. doi: 10.1073/pnas.93.20.10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Lint C, Emiliani S, Ott M, Verdin E. EMBO J. 1996;15:1112–1120. [PMC free article] [PubMed] [Google Scholar]
- 33.Van Lint C, Emiliani S, Verdin E. Gene Expression. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 34.Roth S Y. Nat Genet. 1996;14:3–4. doi: 10.1038/ng0996-3. [DOI] [PubMed] [Google Scholar]
- 35.Becker P B, Wu C. Mol Cell Biol. 1992;12:2241–2249. doi: 10.1128/mcb.12.5.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidmann J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley-Interscience; 1997. [Google Scholar]
- 37.Becker P B, Tsukiyama T, Wu C. Methods Cell Biol. 1994;44:207–223. doi: 10.1016/s0091-679x(08)60915-2. [DOI] [PubMed] [Google Scholar]
- 38.Blank T A, Sandaltzopoulos R, Becker P B. Methods. 1997;12:28–35. doi: 10.1006/meth.1997.0444. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida M, Horinouchi S, Beppu T. BioEssays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 40.Kamakaka R T, Bulger M, Kadonaga J T. Genes Dev. 1993;7:1779–1795. doi: 10.1101/gad.7.9.1779. [DOI] [PubMed] [Google Scholar]
- 41.Munks R J L, Turner B M. Biochim Biophys Acta. 1994;1223:23–28. doi: 10.1016/0167-4889(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 42.Krajewski W A, Luchnik A N. Mol Gen Genet. 1991;231:17–21. doi: 10.1007/BF00293816. [DOI] [PubMed] [Google Scholar]
- 43.Wiegand R C, Brutlag D L. J Biol Chem. 1981;256:4578–4583. [PubMed] [Google Scholar]
- 44.Sobel R E, Cook R G, Allis C D. J Biol Chem. 1994;269:18576–18582. [PubMed] [Google Scholar]
- 45.Mathis D J, Oudet P, Wasylyk B, Chambon P. Nucleic Acids Res. 1978;5:3523–3547. doi: 10.1093/nar/5.10.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ridsdale J A, Hendzel M J, Delcuve G P, Davie J R. J Biol Chem. 1990;265:5150–5156. [PubMed] [Google Scholar]
- 47.Sandaltzopoulos R, Blank T, Becker P B. EMBO J. 1994;13:373–379. doi: 10.1002/j.1460-2075.1994.tb06271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weisbrod S. Nature (London) 1982;297:289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- 49.Marvin K W, Yau P, Bradbury M E. J Biol Chem. 1990;265:19839–19847. [PubMed] [Google Scholar]
- 50.Simpson R T, Thoma F, Brubaker J M. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 51.Morse R H, Cantor C R. Proc Natl Acad Sci USA. 1985;82:4653–4657. doi: 10.1073/pnas.82.14.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drlica K. Microbiol Rev. 1984;48:273–289. doi: 10.1128/mr.48.4.273-289.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keller W, Muller U, Eicken I, Wendel I, Zentgraf H. Cold Spring Harbor Symp Quant Biol. 1978;43:227–244. doi: 10.1101/sqb.1978.042.01.025. [DOI] [PubMed] [Google Scholar]
- 54.Morse R H, Simpson R T. Cell. 1988;54:285–287. doi: 10.1016/0092-8674(88)90190-0. [DOI] [PubMed] [Google Scholar]
- 55.Kelley R L, Kuroda M I. Science. 1995;270:1607–1610. doi: 10.1126/science.270.5242.1607. [DOI] [PubMed] [Google Scholar]
- 56.Bone J R, Lavender J, Richman R, Palmer M J, Turner B M, Kuroda M I. Genes Dev. 1994;8:96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- 57.Roth S Y, Allis C D. Trends Cell Biol. 1996;6:371–375. doi: 10.1016/0962-8924(96)20032-7. [DOI] [PubMed] [Google Scholar]
- 58.Ura K, Wolffe A P, Hayes J J. J Biol Chem. 1994;269:27171–27174. [PubMed] [Google Scholar]
- 59.Ambrose C, McLaughlin R, Bina M. Nucleic Acids Res. 1987;15:3703–3721. doi: 10.1093/nar/15.9.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smirnov I V, Krylov D, Makarov V L. J Biomol Struct Dyn. 1991;8:1251–1266. doi: 10.1080/07391102.1991.10507881. [DOI] [PubMed] [Google Scholar]
- 61.Saavedra R A, Huberman J A. Cell. 1986;45:65–70. doi: 10.1016/0092-8674(86)90538-6. [DOI] [PubMed] [Google Scholar]
- 62.Morse R H, Pederson D S, Dean A, Simpson R T. Nucleic Acids Res. 1987;15:10311–10330. doi: 10.1093/nar/15.24.10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krajewski A, Razin S V. Mol Biol Rep. 1993;18:167–175. doi: 10.1007/BF01674427. [DOI] [PubMed] [Google Scholar]
- 64.Bode J, Henco K, Wingender E. Eur J Biochem. 1980;110:143–152. doi: 10.1111/j.1432-1033.1980.tb04849.x. [DOI] [PubMed] [Google Scholar]
- 65.Weischet W O, Tatchell K, van Holde K, Klump H. Nucleic Acids Res. 1978;5:139–160. doi: 10.1093/nar/5.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simpson R T. J Biol Chem. 1979;254:10123–10127. [PubMed] [Google Scholar]
- 67.Morse R H, Cantor C R. Nucleic Acids Res. 1986;14:3293–3310. doi: 10.1093/nar/14.8.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McGhee J D, Nickol J M, Felsenfeld G, Rau D C. Nucleic Acids Res. 1983;11:4065–4075. doi: 10.1093/nar/11.12.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sinden R R, Carlson J O, Pettijohn D E. Cell. 1980;21:773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]