Summary
Histone tail modifications play a fundamental role in the processes that establish chromatin structure and determine gene expression [1–4]. One such modification, histone methylation was considered irreversible until the recent discovery of histone demethylases. Lsd1 was the first demethylase to be identified [5]. Lsd1 is highly conserved from yeast to humans but its function has primarily been studied using biochemical approaches. The mammalian ortholog has been shown to demethylate mono and dimethyl K4 and K9 residues of histone H3 [5, 6]. Here we describe the effects of Lsd1 mutation in Drosophila. The inactivation of dLsd1 strongly affects the global level of mono and dimethyl-H3-K4 methylation and results in elevated expression of a subset of genes. dLsd1 is not an essential gene, but animal viability is strongly reduced in mutant animals in a gender specific manner. Interestingly, dLsd1 mutants are sterile and possess defects in ovary development, indicating that dLsd1 has tissue specific functions. Mutant alleles of dLsd1 suppress positional effect variegation, suggesting a disruption of the balance between euchromatin and heterochromatin. Taken together, these results show that dLsd1-mediated H3-K4 demethylation has a significant and specific role during Drosophila development.
Results and Discussion
Originally Lsd1 was found as a component of co-repressor complexes [7–11]. Lsd1 demethylase activity was only discovered recently [5] and was found to be modulated by its associated proteins, such as CoREST [12, 13]. Lsd1 depletion in mammalian cells correlates with increased gene expression and elevated levels of H3-K4 methylation at target promoters [5]. However Lsd1 can also act as a co-activator and demethylates H3-K9, a repressive mark [6].
Lsd1 is evolutionary conserved [5] but little is known about its biological function. To address this question we have generated flies carrying a mutation in the sole Drosophila gene, CG17149/dLsd1, that exhibits high homology to Lsd1 (Fig. S1A). dLsd1 contains both a putative amine oxidase domain and a SWIRM domain (Fig. S1B). In the Exelixis collection of mutants [14] we found two piggyBac insertions in the vicinity of CG17149/dLsd1: f03544 (designated as dLsd11) and f00678 (dLsd12). Using FRT sites in the piggyBac transposon [15] to promote trans recombination between dLsd11 and dLsd12 we generated a deletion allele of dLsd1, dLsd1ΔN (Fig. S1B,C).
Southern blot analysis confirmed the authenticity of the dLsd1 alleles (Fig. S2A, S2B). dLsd1ΔN lacks the presumptive promoter region and the N-terminal portion of the gene, including the SWIRM domain (Fig. S1B). Quantitative PCR analysis using primers specific for the 5′ end of dLsd1 confirmed the absence of these sequences in dLsd1ΔN homozygous flies (Fig. S2C). Low levels (<20%) of 3′ transcripts persist in the mutant animals (data not shown) but any potential products would lack the putative nuclear localization signal and the SWIRM domain and are unlikely to be functional. The SWIRM domain is thought to function in protein-protein interactions, DNA protein interaction and enzyme catalysis [16–19]. Inactivation of this domain greatly reduces the stability and demethylase activity of Lsd1 [16, 17]. Western blot analysis showed that dLsd1 is expressed at high levels in wild-type (wt) flies but no dLsd1 protein was detected in dLsd1ΔN homozygous flies (Fig. S2D). Hence dLsd1ΔN is, most likely, a null allele.
This collection of mutant alleles provided us with the opportunity to study the biological function of dLsd1 in an animal model system.
First, we assessed the effects of dLsd1 mutation on viability. Crosses of dLsd1ΔN heterozygous animals gave only one-third of the expected number of homozygous dLsd1ΔN progeny (Table S1). Interestingly, this reduction in viability is more dramatic in the male progeny (approximately 90% of the viable dLsd1ΔN homozygotes were females) (Table S2).
dLsd1ΔN mutants are sterile. In these animals, ovary development is severely impaired (Fig. 1C,D). The Drosophila ovary consists of approximately 16 ovarioles, chains of developing egg-chambers with a germarium at the anterior tip. The germarium contains germline stem cells (GSC) and somatic stem cells (SSC) which give rise respectively to the germline cysts and to follicle cells (Fig. 1C,E,G,I,M) [20]. Interestingly, DNA staining shows that dLsdΔN mutant ovaries lack proper ovariole structures (Fig. 1D,F) and the formation of egg chambers is abnormal at very early stages. Both the germline and follicle cells appear abnormal (Fig. 1H, L, N) and, strikingly, the 16 cells cysts fail to be properly encapsulated by follicle cells (Fig. 1H). In males, the testes are morphologically intact, but DNA staining suggests defects during spermatogenesis (data not shown). Interestingly, dLsd1ΔN homozygotes also have a held-out wing phenotype (Fig. 1B) that renders them unable to fly.
Figure 1. dLsd1ΔN mutant animals have developmental defects.
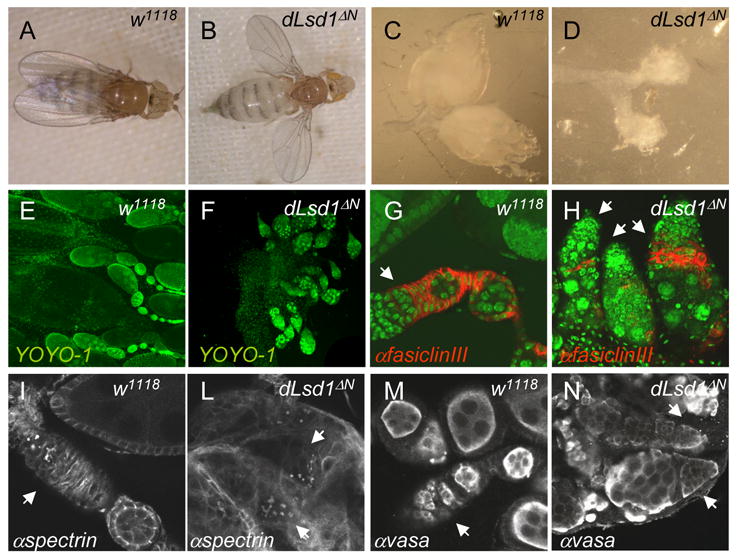
A, B: dLsd1ΔN mutant flies have a held-out wing phenotype. Images of wild-type (w1118) and dLsd1ΔN flies. C, D: dLsd1ΔN mutant ovaries are smaller than wild-type ovaries. Images of the ovaries of wild-type and dLsd1ΔN flies. E, F: dLsd1ΔN mutant ovaries lack proper ovariole structures. DNA staining (YOYO-1) of a wild-type ovary versus the ovary of dLsd1ΔN flies. G, H: In dLsd1ΔN mutant ovaries follicle cells fail to encapsulate the cyst. Wild-type and dLsd1 mutant ovarioles were stained with YOYO-1 (green) and anti-fasiclin III (red) to outline somatic cells. I, L, M, N: Egg chambers of dLsd1ΔN mutant ovaries are abnormal at very early stages. Wild-type and dLsd1ΔN mutant ovarioles were stained with anti-αspectrin (white), to show the fusomes (I,L) and anti-vasa (white) to label the germ cells (M,N). The germarium is indicated by arrows.
To confirm that these defects are due specifically to dLsd1 loss and are not the result of secondary mutations, we performed complementation tests with a deficiency (Df(3L)ED4858) that uncovers the dLsd1 gene (Table 3). Trans-heterozygotes carrying dLsd1ΔN and Df(3L)ED4858 recapitulated the phenotypes observed in dLsd1ΔN homozygous flies (Table S3).
We conclude that dLsd1 mutation reduces viability in a gender-dependent manner, causes abnormal ovary development, and results in animal sterility. Collectively, these results point to important roles for dLsd1 in the late stages of development. dLsd1 levels are highest in the embryonic stages (Fig. S2E,F), suggesting that dLsd1 might also have functions during early stages of development that may be masked in the homozygous dLsd1 mutants by maternally supplied products.
Biochemical studies with human Lsd1 have led to reports that Lsd1 can act both as a co-repressor of transcription by demethylating H3-K4 [5], and as a co-activator by demethylating H3-K9 [6]. To ask which of these potential activities is predominant in Drosophila, we examined the levels of histone modification in dLsd1 mutant flies. The global level of mono and dimethyl-H3-K4 was considerably higher in dLsd1 mutants than in wild-type flies (Fig. 2A,B); this effect was particularly striking in adult males (Fig. 2B). In contrast, we found no increase in the global levels of methyl-H3-K9, indeed the level of dimethyl and trimethyl-H3-K9 decreased slightly in dLsd1 mutants (Fig. 2A,B). The levels of monomethyl-H4-K20, dimethyl-H3-K27, dimethyl-H4-K20, trimethyl-H3-K36 and acetyl-H3-K9 were unaltered in dLsd1ΔN mutant flies (Fig. 2A). Interestingly, ovaries contain a higher level of dLsd1 and a lower level of monomethyl-H3-K4 than the rest of the adult female (Fig. 2C), potentially explaining why dLsd1 mutation selectively perturbs ovary development.
Figure 2. dLsd1 specifically affects global levels of H3-K4 mono and dimethylation.
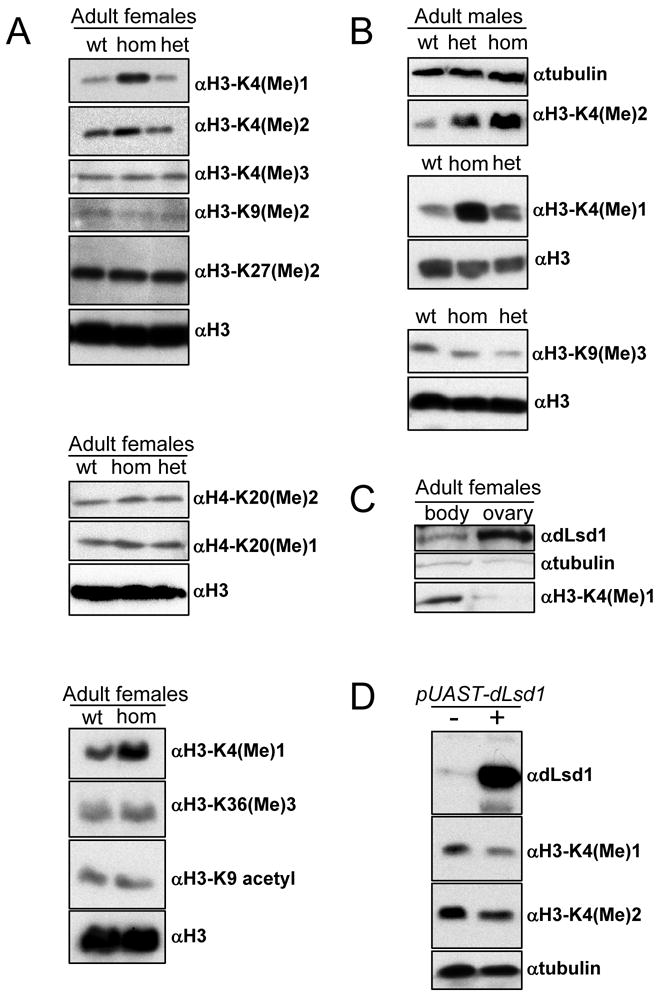
A: Western blot analysis of protein extracts from w1118(wt), dLsd1ΔN/dLsd1ΔN (hom) and dLsd1ΔN/TM3 (het) females using antibodies specific for different modified histone H3 and H4 residues as indicated on the right. Equal loading was assessed by blotting with either a histone H3 specific antibody or a tubulin specific antibody. B: Western blot analysis of protein extracts from w1118, dLsd1ΔN/dLsd1ΔN and dLsd1ΔN/TM3 males. C: dLsd1 is highly expressed in the ovary. Western blot analysis of dLsd1 expression and of the level of H3-K4 monomethylation in the ovary. dLsd1 protein level are higher in the ovary than in the rest of the body and this correlates with lower levels of H3-K4 monomethylation. D: Ectopic expression of dLsd1 affects H3-K4 methylation levels and gene expression. Ectopic expression of the pUAST-dLsd1 transgene was driven by the Actin5C-GAL4 driver. dLsd1 expression was verified by Western blot analysis. The level of mono and dimethyl H3-K4 in w1118 versus dLsd1 overexpressing flies are shown. Equal loading was assessed using a tubulin specific antibody.
As a further test, we generated UAST-dLsd1 transgenic flies and found that increased levels of dLsd1 reduced di and monomethyl-H3-K4 (Fig. 2D), confirming that dLsd1 is a critical determinant of the global level of H3-K4 methylation. Surprisingly, these animals lacked any clear developmental defects.
H3-K4 methyl residues are highly enriched in euchromatin. To test whether the elevated levels of mono and dimethyl-H3-K4 in dLsd1 mutants alter the balance between euchromatin and heterochromatin, we used three variegating systems (T(2;3)Stubblevariegated (Sbv), In(1)y3Pyellow and In(1)whitem4h) and asked whether dLsd1 alleles modify positional effect variegation (PEV). PEV is the mosaic pattern of gene silencing observed when genes that are normally located in euchromatin regions are placed into a heterochromatic environment [21]. Suppressors of PEV include mutants of heterochromatin-associated proteins, such as HP1 and the histone H3-K9 methyltransferase Su(var)3–9 [22].
T(2;3)Sbv translocation juxtaposes the Sb mutation and the centric heterochromatin of the second chromosome, resulting in mosaic flies with both Sb and normal bristles [23, 24]. Activation of dominant Sb results in stubble bristles. When T(2;3)Sbv was crossed to dLsd1ΔN, we observed a significant increase in the frequency of Sb bristles (Fig. 3A). Similar results were found using the yellow locus [25, 26]. Analysis of bristles in the wing of In(1)y3P/+; +; dLsd1ΔN/+ flies (Fig. 5B) showed that a single allele of dLsd1ΔN or dLsd12 suppressed yellow variegation, giving a 25% reduction of yellow bristles (Fig. 3C). Suppression of variegation was also observed using wm4h (Fig. S3). These results indicate that dLsd1 alleles are potent suppressors of PEV and suggest that the absence of dLsd1 alters chromatin structure.
Figure 3. dLsd1 is a suppressor of PEV.

Heterozygotes for the dLsd1ΔN allele were crossed to either T(2;3)Sbv or In(1)y3P. The effects of dLsd1ΔN on Stubble variegation of T(2;3)Sbv are shown in A. The effects of dLsd1ΔN on Yellow variegation are shown in B. Red asterisks indicate the yellow bristles in In(1)y3P/+;+; Me1/dLsd1ΔN flies. C: Quantification of the percentage of yellow bristles in the indicated genotypes.
Mono and dimethyl forms of H3-K4 are linked to transcriptional activation; the increased level of these modifications in dLsd1ΔN mutants suggests that dLsd1 normally represses transcription. Previous studies with human cells have shown that Lsd1 regulates the expression of neuron-specific genes [12]. To ask whether this function is conserved, we depleted dLsd1 from SL2 cells using RNAi and examined the expression of neuron-specific genes, such as Nicotinic Acetylcholine Receptor beta (nAcrβ) and Na channel (Nach). dLsd1 depletion increased the level of monomethyl-H3-K4 and increased the expression of both nAcrβ and Nach (Fig. 4A), indicating that Lsd1’s role in the repression of neuron-specific genes is conserved between Drosophila and humans.
Figure 4. Neuron specific genes and Hox genes are misexpressed in SL2 cells depleted for dLsd1 and in dLsd1ΔN mutant animals.

A: Western blot analysis of SL2 cells treated for 8 days with white and dLsd1 dsRNA. Three different dsRNAs directed against the N-terminal (Nt), Central (Ce) or C-terminal (Ct) region target dLsd1 protein with the same efficacy while dsRNA against the white gene (wh), used as negative control, does not affect dLsd1 protein expression. qPCR analysis of the expression of the indicated genes 8 days after dsRNA treatment of SL2 cells. The expression level is normalized against the white dsRNA treated control. Experiments were performed in triplicate and standard deviation of the mean is indicated. B: Western blot analysis of wild-type (wt), dLsd1ΔN/dLsd1ΔN (hom) and dLsd1ΔN/TM3 (het) flies with an antibody specific for dLsd1 and with anti-tubulin (loading control). qPCR analysis of the expression of the indicated genes in wild-type (+) and dLsd1 mutant (−) third instar larvae (L3) and adult flies respectively 1, 2 and 3 days after eclosion (A1, A2, A3). Rp49 was used as a control (data not shown). Experiments were performed in triplicate and standard deviation of the mean is indicated.
The homeobox (Hox) gene locus is subject to extensive H3-K4 methylation by Trithorax group proteins [27]. We therefore asked whether the expression level of the Hox genes Ultrabithorax (Ubx) and abdominal-A (abdA), is affected by dLsd1 depletion. Ubx and abdA mRNA levels increased two-fold in SL2 cells treated with dLsd1 double stranded RNA (dsRNA) (Fig. 4A). These changes were specific and were not seen with other control genes (dDP and Hid). To verify the relevance of these observations in vivo we compared the expression of these genes in wild-type and dLsd1ΔN mutant flies. A significant upregulation of each of these targets was found in dLsd1ΔN mutant flies (Fig. 4B and data not shown), confirming the importance of dLsd1-mediated repression in vivo. Intriguingly we observe that this upregulation is age dependent: the difference in gene expression is minimal in larval stages, consistent with this the Hox gene expression pattern in imaginal discs from dLsd1ΔN mutant larvae and in embryos is largely unaltered (Fig. S4). However the level of nAcrβ, Ubx and Abd-B gradually and significantly increases with age after eclosion (Fig. 4B), suggesting that dLsd1 function is especially important for the regulation of gene expression in adult tissues.
Using Drosophila melanogaster, we describe the consequences of eliminating Lsd1 function in an animal model. The results help to place the previously described biochemical activities of Lsd1 into a biological context and show that dLsd1 is a key determinant of the global levels of mono and dimethyl-H3-K4 in vivo. dLsd1 mutation impacts heterochromatin homeostasis and leads to ectopic expression of a subset of genes in flies. Curiously we observe that the consequences of the global upregulation of H3-K4 methylation on animal development are restricted to a few specific organs. One of the clearest defects is in the ovary where dLsd1ΔN mutant egg chambers are abnormal early in development and follicle cells fail to encapsulate the cyst. This defect is consistent with the observation that dLsd1 is highly expressed in the ovary, and indicates that dLsd1 mediated demethylation of H3-K4 plays a crucial role in this organ. We suggest that, in dLsd1ΔN mutants, the elevated expression level of dLsd1 target genes causes tissue specific defects. We note that both Hox genes and neuron-specific genes are among the target upregulated in dLsd1ΔN mutants. The upregulation occurs late in development and this may explain the lack of homeotic phenotypes or other early developmental defects in dLsd1ΔN mutants. The stronger changes in histone modification seen in dLsd1ΔN mutant males compared to females and the sex dependent lethality caused by dLsd1 mutation raise the possibility that dLsd1 may have a sex-specific distribution on chromatin alternatively its mutation may alter the chromatin distribution of the male specific lethal complex.
Whereas mammalian Lsd1 has been shown to be able to demethylate H3-K9 [6], we saw no increase in the global level of dimethylated H3-K9. This indicates that this activity is either not conserved in Drosophila, or that is redundant with other demethylases, or that is restricted to specific loci and/or specific tissues.
This study opens the road to further studies aimed at delineating the specific functions of dLsd1 in the control of gene expression. Genome-wide studies will be necessary to identify all the dLsd1 regulated genes and to identify the target genes responsible for each of the developmental defects.
One important implication of the tissue specific defects seen in the dLsd1ΔN mutant animals is the possibility that dLsd1 may be manipulated in vivo to modulate specific biological processes. The results support the idea that global changes in the levels of histone methylation can impact specific developmental processes. These results also highlight the need for additional studies to understand how histone methylation is dynamically regulated in vivo and how these changes contribute to normal development and disease.
Supplementary Material
Acknowledgments
We would like to thank Spyros Artanavis-Tsakonas and Douglas Dimlich for providing fly stocks and members of the Dyson laboratory for technical assistance and helpful discussion. We also thank Doug Renny for technical assistance in the generation of dLsd1 transgenic lines. We thank Laurel Raftery and Xiaodong Wu for insightful discussion and Anders Näär, Bradley Bernstein and Martha Betson for critical comments on the manuscript. This study was supported by NIH grants PO1CA095281, GM53203 and CA64402 (to N.D). N.S.M was supported by Fellowships from the Canadian Institute of Health Research and the MGH Fund for Medical Discovery. J.Y.J. was supported by a Tosteson Postdoctoral Fellowship from the Massachusetts Biomedical Research Corporation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wysocka J, Milne TA, Allis CD. Taking LSD 1 to a new high. Cell. 2005;122:654–658. doi: 10.1016/j.cell.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 2.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 3.Trewick SC, McLaughlin PJ, Allshire RC. Methylation: lost in hydroxylation? EMBO Rep. 2005;6:315–320. doi: 10.1038/sj.embor.7400379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister AJ, Kouzarides T. Reversing histone methylation. Nature. 2005;436:1103–1106. doi: 10.1038/nature04048. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 6.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 7.Hakimi MA, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci U S A. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Humphrey GW, Wang Y, Russanova VR, Hirai T, Qin J, Nakatani Y, Howard BH. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J Biol Chem. 2001;276:6817–6824. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- 9.Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 10.You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc Natl Acad Sci U S A. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, Federoff HJ, Rose DW, Rosenfeld MG, Brehm P, Mandel G. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- 12.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 Histone Demethylase Activity by Its Associated Factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 13.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 14.Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D, Killpack K, Laufer A, Mazzotta J, Smith RD, Stevens LM, Stuber C, Tan LR, Ventura R, Woo A, Zakrajsek I, Zhao L, Chen F, Swimmer C, Kopczynski C, Duyk G, Winberg ML, Margolis J. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 15.Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Yang Y, Wang F, Wan K, Yamane K, Zhang Y, Lei M. Crystal structure of human histone lysine-specific demethylase 1 (LSD1) Proc Natl Acad Sci U S A. 2006;103:13956–13961. doi: 10.1073/pnas.0606381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stavropoulos P, Blobel G, Hoelz A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nat Struct Mol Biol. 2006;13:626–632. doi: 10.1038/nsmb1113. [DOI] [PubMed] [Google Scholar]
- 18.Tochio N, Umehara T, Koshiba S, Inoue M, Yabuki T, Aoki M, Seki E, Watanabe S, Tomo Y, Hanada M, Ikari M, Sato M, Terada T, Nagase T, Ohara O, Shirouzu M, Tanaka A, Kigawa T, Yokoyama S. Solution structure of the SWIRM domain of human histone demethylase LSD1. Structure. 2006;14:457–468. doi: 10.1016/j.str.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Da G, Lenkart J, Zhao K, Shiekhattar R, Cairns BR, Marmorstein R. Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes. Proc Natl Acad Sci U S A. 2006;103:2057–2062. doi: 10.1073/pnas.0510949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spradling AC. Germline cysts: communes that work. Cell. 1993;72:649–651. doi: 10.1016/0092-8674(93)90393-5. [DOI] [PubMed] [Google Scholar]
- 21.Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14:377–392. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- 22.Schotta G, Ebert A, Dorn R, Reuter G. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin Cell Dev Biol. 2003;14:67–75. doi: 10.1016/s1084-9521(02)00138-6. [DOI] [PubMed] [Google Scholar]
- 23.Sinclair DAR, Mottus RC, Grigliatti TA. Genes which suppress position-effect variegation in Drosophila melanogaster are clustered. Mol Gen Genet. 1983;191:326–333. [Google Scholar]
- 24.Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhadra U, Bhadra MP, Birchler JA. Interactions among dosage-dependent trans-acting modifiers of gene expression and position-effect variegation in Drosophila. Genetics. 1998;150:251–263. doi: 10.1093/genetics/150.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frolov MV, Birchler JA. Mutation in P0, a dual function ribosomal protein/apurinic/apyrimidinic endonuclease, modifies gene expression and position effect variegation in Drosophila. Genetics. 1998;150:1487–1495. doi: 10.1093/genetics/150.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


