Abstract
Insights into structure-function relations of many proteins opens the possibility of engineering peptides to selectively interfere with a protein’s activity. To facilitate the use of peptides as probes of cellular processes, we have developed caged peptides whose influence on specific proteins can be suddenly and uniformly changed by near-UV light. Two peptides are described which, on photolysis of a caging moiety, block the action of calcium-calmodulin or myosin light chain kinase (MLCK). The efficacy of theses peptides is demonstrated in vitro and in vivo by determining their effect before and after photolysis on activities of isolated enzymes and cellular functions known to depend on calcium-calmodulin and MLCK. These caged peptides each were injected into motile, polarized eosinophils, and when exposed to light promptly blocked cell locomotion in a similar manner. The results indicate that the action of calcium-calmodulin and MLCK, and by inference myosin II, are required for the ameboid locomotion of these cells. This methodology provides a powerful means for assessing the role of these and other proteins in a wide range of spatio-temporally complex functions in intact living cells.
Keywords: calmodulin, myosin light chain kinase, smooth muscle, photolysis
Several methods have been used in the past to probe the role of a protein in cell function, each with advantages as well as limitations. Organic compounds are available that can modulate the activity of proteins, but interpretation of effects often is complicated by their relatively slow onset and low selectivity for a specific protein. Impressive progress toward single protein specificity has been made with antisense (1) and homologous recombination (2) methods, which disrupt the expression, and thus the function of a specific protein. Interpretation of effects, or lack thereof, on a cellular function may be complicated, however, by compensatory pathways enhanced by the absence of the targeted protein. Peptides that bind to proteins with high affinity and high selectivity provide a means to rapidly and potently inhibit the activity of selected proteins, but peptides must be microinjected into cells, and microinjection itself can at least transiently alter cell function. It thus would be desirable to have a way to make a peptide that is initially inactive or “caged” because of a strategically placed photolabile moiety (3, 4). Such a peptide could be injected into a cell, and time allowed for it to distribute evenly and for normal cell function to be verified. The peptide’s biological activity then could be unmasked by light-directed removal of the photolabile group. Each cell would be its own control, thereby diminishing effects of cell to cell variability, and active peptides could be produced rapidly (within milliseconds) and with good spatial resolution.
We describe here the preparation and use of photoactivatable caged peptides targeted against calmodulin and myosin light chain kinase (MLCK). Because these two proteins are known to be essential in the control of smooth muscle contractility, the efficacy of the caged peptides was verified in vitro by using smooth muscle MLCK (5) and in smooth muscle cells of toad stomach (6). There is also evidence that calmodulin and MLCK regulate cell motility in fibroblasts (7), but there is little information in other cells such as leukocytes where cell shape changes and intracellular Ca2+ levels fluctuate rapidly, and cells move considerably faster (8). The caged peptides therefore were used to determine the extent to which individual eosinophil cells rely on calmodulin, MLCK, and by inference myosin II to control their rapid ameboid cell motility.
MATERIALS AND METHODS
Synthesis.
Caged peptides were synthesized by automated solid-phase methods using 9-fluorenylmethoxycarbonyl (Fmoc) amino acids. l-Tyrosine protected on its α-amino and α-carboxyl groups was coupled to 2-bromo-2′nitrophenylacetic acid methyl ester through its phenolic group (9). Methyl α-carboxyl caged tyrosine (MW = 374.1, m/z 375.1) then was derivatized with an Fmoc moiety and incorporated into peptides during automated synthesis (Fig. 1A). Cleavage from the solid support and deprotection of side chains (90% trifluoroacetic acid) was followed by HPLC purification of the peptide then by hydrolytic removal of the methyl ester from the cage moiety using 10% aqueous K2CO3. α-Carboxyl 2′-nitrobenzyl tyrosine containing peptides were repurified by HPLC and lyophilized. Peptide compositions were confirmed by amino acid analysis and MS: 5cgY-RS-20: MS, MW = 2434.2, found 2434.5. 5Y-RS-20 (obtained by photolysis of 5cgY-RS-20): MS, MW = 2256.2, found 2256.2. 5,18cgY-RS-20: MS, MW = 2663.2, found 2663.2. 9cgY-LSM1: MS, MW = 1862.3, found 1861.7. LSM1 (obtained by photolysis of 9cgY-LSM1) MS, MW = 1682.3, found 1682.
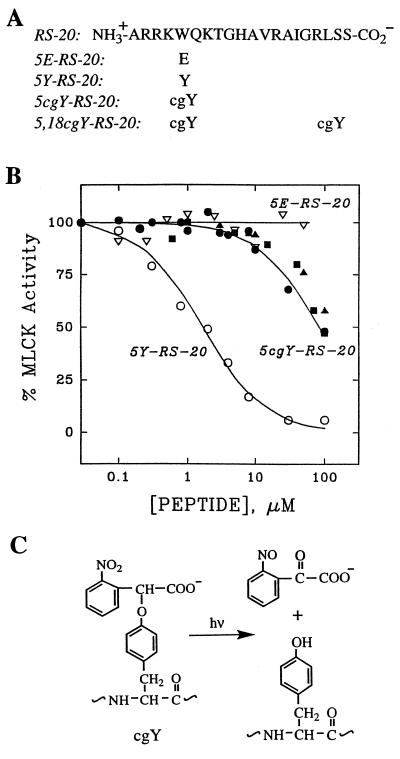
Figure 1.
(A) Amino acid sequences of calmodulin inhibitory peptide RS-20 and variants containing glutamate (E), tyrosine (Y), and caged tyrosine (cgY). Peptide and caged peptide structures were confirmed by amino acid composition and mass spectrometry (see Materials and Methods). (B) Effects of RS-20 peptides on Ca2+/calmodulin-dependent MLCK activity in vitro. 5cgY-RS-20 before (•) and after (○) photoconversion; the photoreleased peptide was indistinguishable from authentic 5Y-RS-20 in inhibitory potency, chromatographic behavior, and mass spectra. There was no detectable inhibition by 5E-RS-20 (▵), whereas 5,18cgY-RS-20 inhibited only weakly both before (▴) and after photolysis (▪). Data points represent means of triplicate measurements with SEM values less than 10% of the mean. (C) Structure and photolysis reaction of cgY.
MLCK Activity and Calmodulin Binding Assays.
MLCK was assayed as described (5) with the following modifications. Various concentrations of peptides were added to the assay mixture containing 0.5 μg/ml smooth muscle MLCK, 0.2 mg/ml smooth muscle regulatory light chain, 100 nM calmodulin, 0.1 mM CaCl2, 100 mM KCl, 1 mM MgCl2 and 30 mM Tris at pH 7.5, 25°C. Phosphorylation was initiated by addition of 50 μM 32P-γ-ATP and continued for 5 min. Constitutively active catalytic domain of MLCK was prepared by trypsinization (5) and assayed in the above mixture containing 1 mM EGTA and no added Ca2+. The selectivity of LSM1 peptide for MLCK over other protein kinases was examined as follows. At a concentration of 100 μM, LSM1 inhibited protein kinase A (PKA), protein kinase C (PKC), and calmodulin-dependent protein kinase II (CaMPKII) activity by 38%, 49%, and 47%, respectively. At a concentration of 37 nM, where LSM1 inhibited MLCK activity by 50%, LSM1 had no significant inhibitory effect on PKA, PKC, or CaMPKII.
LSM1 also inhibited less than 15% of Rho kinase activity (10) at concentrations up to 100 μM. Rho kinase was expressed in COS-7 cells, immunoprecipitated, then assayed for activity by using smooth muscle light chains as a substrate.
Calmodulin binding was assessed by adding 100 nmol aliquots of peptides to a 5-ml calmodulin agarose column (Sigma) equilibrated in 32 μM free Ca2+ at 4°C. Peptides that bound were eluted from the column in 1 mM EGTA. All peptides then were identified by UV spectra and HPLC.
Eosinophils.
Newt eosinophil cells were prepared as described (8, 11). Briefly, cells were settled onto coverslips from 1:5 newt blood in amphibian culture medium, washed and counted in a chamber on an inverted microscope. Cells were injected with 1 mM caged peptide in a vehicle of 20 mM Hepes, pH 7.2, containing 0.1 mM fluorescein dextran (3 kDa) to estimate injection volume and peptide concentration (see below).
Cells were activated with newt serum and motility recorded at 1-sec intervals by a Dage video charge-coupled device camera and stored on video disks. Each cell served as its own control by recording for 5 min before UV exposure. Effects of UV exposure then were determined in the subsequent 5-min period by examining three categories of motility parameters: (i) whole-cell locomotion, shape, and polarity, (ii) lamellipod formation, expansion, ruffling, withdrawal, and distribution around the cell periphery, and (iii) granule motion patterns. Four types of control cells were examined. Cells injected with vehicle only or with 5,18cgY-RS-20 (an inactive variant of RS-20) were subjected to the same UV exposure protocol as the experimental cells. In addition, the behavior of uninjected cells and cells injected with caged peptide was recorded without being exposed to UV light. This method permitted double-blind analysis of responses by two scorers, each positive response to the active peptides and each negative response in control cells requiring consensus. Video records were viewed and scored a total of four times, the final scoring session occurring several weeks after the experiment. Cells remained active on the microscope for up to 3 hr after injection, and the peptides were effective throughout, indicating stability of caged peptides in the cytosol over this period.
Photorelease of peptides was accomplished with the near-UV output of an argon ion laser delivered in a series of 10 pulses each of 5-msec duration spaced 20 msec apart. The extent of photoconversion of caged peptide was estimated to be 45–55% by comparison with caged fluorescein. This estimate is consistent with the finding that enough caged peptide remained after a single exposure to obtain a response to a second exposure (in cells that recovered from the first). Depending on the injection volume, the cyotosolic caged peptide concentrations were estimated to be 20–100 μM, and final free peptide 10–50 μM. The extent and persistence of peptide effects on cell locomotion and granule flow was correlated with the volume of caged peptide introduced into each cell assessed by fluorescein dextran fluorescence.
Exposure of newt eosinophils to high levels of near-UV light resulted in some rundown of motility. This rundown usually could be distinguished from the effects of peptides in that it was nonspecific (e.g., ruffling also was lost) and cells never recovered. To minimize these effects of near-UV light, each series of experiments began with exposure of at least a dozen uninjected cells to determine the maximum dose of laser light that had no detectable effect on any of the motility parameters evaluated. In addition, at least one uninjected cell from each coverslip was exposed to near-UV light and recorded to rule out the possibility that UV sensitivity varied among different batches of cells.
RESULTS
A method was developed to prepare caged peptides for the purpose of inhibiting the activity of the calcium-calmodulin complex, as this complex is believed to play a key role in mediating many of the effects of calcium in cells. Our efforts were focused on RS-20 (Fig. 1A), a 20-aa calcium-calmodulin binding domain of smooth muscle MLCK (12). In an abbreviated structure-activity analysis of RS-20, we found that substitution of hydrophobic residues W-5, L-18, or both with E greatly reduced the affinity of RS-20 for calmodulin as assessed by an in vitro MLCK assay (Fig. 1B). This finding confirmed that hydrophobic side chains are preferred in positions 5 and 18 for high affinity calmodulin binding (13) and showed that negatively charged side chains at these sites interfered with formation of the complex.
Several candidate caged forms of RS-20 were prepared by replacing W-5, L-18, or both with a caged tyrosine that featured a negatively charged photolabile group linked to tyrosine’s phenolic function (Fig. 1C). Analysis of these peptides by a quantitative calmodulin-dependent MLCK assay (5) identified only one that displayed properties appropriate for a caged peptide. The peptide, designated 5cgY-RS-20 (Fig. 1A), was largely inactive before photolysis and then showed smooth and complete photoconversion to 5Y-RS-20, resulting in an increase in apparent calmodulin affinity of about 50-fold (Fig. 1B). The peptide derivative with caged tyrosine at positions 5 and 18, designated 5,18cgY-RS-20, was largely inactive in the MLCK assay both before and after illumination (Fig. 1B), and therefore was used as a control peptide in cellular experiments (see below). Similar conclusions about peptide activities were reached when peptides and caged peptides were tested for binding to calmodulin immobilized on agarose (14).
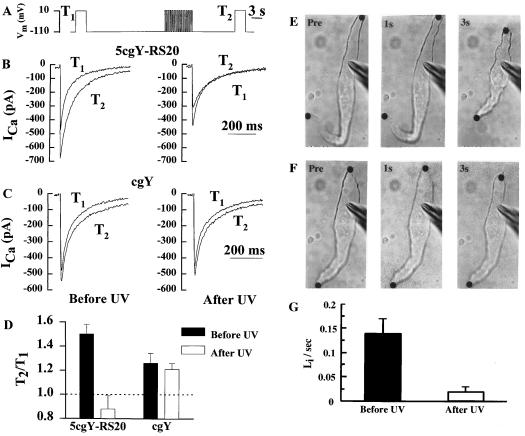
The utility of 5cgY-RS-20 and its compatibility with biological tissues first was tested in freshly isolated smooth muscle cells from the stomach of Bufo marinus. The calcium calmodulin complex regulates an inward calcium current (15) and contractility (6) in these cells. To investigate calcium current regulation, individual cells in the whole-cell patch-clamp mode were dialyzed with 200 μM 5cgY-RS-20 from the recording pipette. Without illumination, the peak calcium current induced by depolarization increased on average by 50 ± 8% (SEM, n = 7) after a conditioning train (Fig. 2B, Left) as expected from previous studies (15). After brief exposure to near-UV light, the enhancement of inward calcium current after the conditioning train was absent (Fig. 2B, Right); on average the current actually decreased by 12 ± 11% (SEM, n = 7). Control experiments demonstrated that inhibition of current enhancement was caused by production of the calmodulin inhibitory peptide and not by nonspecific effects associated with photolysis (Fig. 2C).
Figure 2.
Effects of photolysis of 5cgY-RS-20 on Ca2+ current enhancement and shortening in Bufo marinus smooth muscle cells. (A) Voltage clamp protocol used to demonstrate enhancement of inward Ca2+ currents (ICa) by repetitive depolarization of single smooth muscle cells. (B and C) Currents during initial 3-sec depolarizations before (T1) and 12 sec after the train (T2) are shown for an experiment in which 200 μM 5cgY-RS-20 or 200 μM caged tyrosine (cgY) were included in the pipette. The protocol was carried out on each cell before (Left) and just after (Right) 15-sec exposure to a xenon source UG11 filtered to pass near-UV light. (D) Summary of the effect of 5cgY-RS-20 and cgY on enhancement of inward Ca2+ current. The mean (± SEM) peak inward current after versus before the conditioning train (T2/T1) is shown for seven and five cells with 5cgY-RS-20 and cgY, respectively. (E) Images of a smooth muscle cell attached to a pipette containing 200 μM 5cgY-RS-20 in the whole-cell configuration. Images were observed just before and 1 and 3 sec after depolarization with the train of pulses shown in A. (F) Same cell as in E but after illumination to release 5Y-RS-20. (G) Mean (± SEM) rates of shortening of four cells before and after photorelease of 5Y-RS-20.
To determine the efficacy of the caged peptide to inhibit yet another response known to be calmodulin dependent, we examined contractile responses of individual smooth muscle cells (6). Cells containing 5cgY-RS-20 shortened in response to electrical stimulation at a peak rate of 14 ± 2% of cell length/sec (SEM, n = 4) (Fig. 2E), similar to cells with no peptide. After exposure to near-UV light, the rate of shortening of cells containing the caged peptide slowed to 2 ± 1% of cell length/sec (SEM, n = 4) (Fig. 2F). Normal shortening rates were observed after the same near-UV exposure in the absence or presence of caged tyrosine (not shown), indicating that inhibition of shortening speed was not caused by illumination alone or by photolysis byproducts. For both the contractile and Ca2+ current responses, a smaller number of trials was needed with the caged peptide to achieve better levels of reliability compared with the noncaged RS-20 (6, 15). The impact of cell-to-cell variability was diminished because responses could be compared in the same cell within a few seconds after activation of peptide activity.
This capability to measure control and experimental responses in the same cell allowed us to examine the role of calcium-calmodulin in the rapid locomotion of newt eosinophils. Like many other polarized motile cells, these cells exhibit a tail high/front low calcium gradient that is required for normal cell polarization (8, 11). In migrating fibroblasts, the spatial distribution of cytosolic [Ca2+], calcium-calmodulin, activated MLCK and phosphorylated myosin II provide evidence for the involvement of myosin II in polarization and locomotion (7, 16). Little is known, however, about the role of calmodulin, MLCK, and myosin II in many other cell types, particularly cells like eosinophils that move at least an order of magnitude faster than fibroblasts. We used caged peptides targeted against calmodulin as a way to explore the role of calcium-calmodulin and myosin II in these rapidly moving cells.
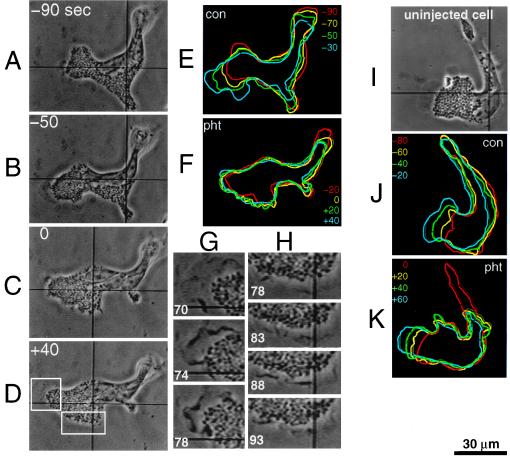
Cells were microinjected with 5cgY-RS-20, allowed to recover, activated with serum, then illuminated briefly to release active peptide. Fig. 3 illustrates a typical response in which locomotion of the cell was arrested soon after illumination. Forward flow of granules also stopped, as did leading lamellipod extensions. Generally similar responses were observed in 55 of 67 cells (82%). The shortest response time was ≈10 sec, with most responsive cells producing a noticeable change in at least one aspect of cell, lamellar, or granule motion within 60 sec. Not all intracellular motion ceased, however, as high magnification images showed phase dark ruffles associated with granule-free membrane extensions continuing to form and move after photorelease of peptide. These membrane extensions were observed over multiple regions of the cell, including areas adjacent to the nucleus where such structures are typically not seen. In some cells, new lamellipods formed transiently in what had been the rear of the cell at the time of peptide release, indicating that the capability to form lamellipods was not impaired. In addition, most cells displayed a sharp reduction in granule motion, but in about 25% of cells granule motion reversed direction or became random. Overall similar results were obtained with longer observation periods. Ten cells injected with the 5cgY-RS-20 showed normal locomotion for 15 min, then after illumination, again lost motility within a 60- to 90-sec observation period. Fourteen of 55 responsive cells recovered somewhat from the effects of the peptide within 5 min, and another 22 recovered within 15 min of release, indicating that the peptide effects were at least partially reversible. Control experiments showed that UV exposure alone altered motility in only four of 31 cells, and those cells never recovered. Also, motile behavior was unaltered in three cells injected with 5,18cgY-RS-20 and irradiated, a protocol that released an inactive peptide and two molar equivalents of the photolysis byproduct. We conclude that acute inhibition of calcium-calmodulin blocked specific aspects of eosinophil motility in the time domain of tens of seconds.
Figure 3.
Acute paralysis of locomotion after photorelease of the calmodulin inhibitory peptide from 5cgY-RS-20. (A–D) Phase contrast images before (−40 and −20) and after (+40, +80, and +120) near-UV exposure (0) with times in sec. (E and F) Color-coded outlines showing cell movements at 20-sec intervals during the 5-min control period (“con”) and immediately after release of the peptide (“pht”). The cell outline 5 min after photolysis was virtually identical to that 40–60 sec after photolysis. (G and H) Enlargements of boxed areas in D showing continued edge ruffling 70–93 sec after peptide release. (I-K) Phase contrast images and outlines of a control cell and its lamellipods at 20-sec intervals before (“con”) and after (“pht”) the same near-UV light exposure used in A-D.
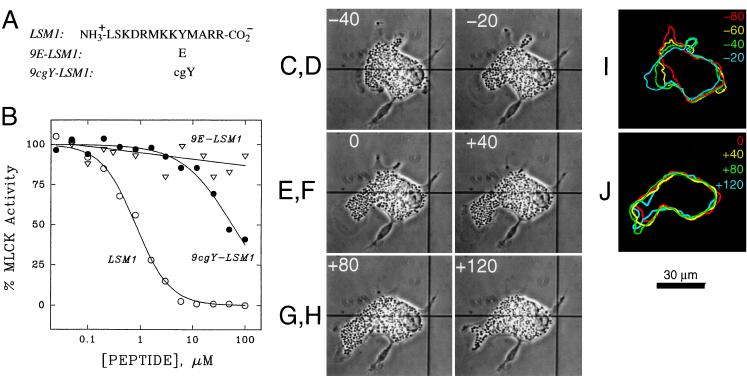
MLCK and myosin II activity are known to depend on calcium-calmodulin (5) and hence the observed effects may reflect interference with myosin II activity. It is possible, however, that the observed effects result from interference with unconventional myosin motors in which calmodulin acts as a functional light chain (17, 18). The observed effects also may result from inhibition of other calmodulin dependent proteins that either directly or indirectly influence the organization or activity of cytoskeletal elements or other motor proteins. To determine which, if any, of the effects of 5cgY-RS-20 were directed at MLCK and consequent inhibition of myosin II, we developed another caged peptide to specifically inhibit this kinase. The autoinhibitory domain peptide from MLCK, designated LSM1 (Fig. 4A), inhibits both calcium calmodulin-dependent and constitutively active forms of MLCK (5) and is ineffective toward calmodulin-dependent protein kinase II, protein kinase A, protein kinase C, and Rho kinase (see Materials and Methods). The tyrosine at position 9 in LSM1 proved critical for inhibitory efficacy, as predicted from mutagenesis studies on MLCK (5). We synthesized a caged form of this peptide, designated 9cgY-LSM1, in which negatively charged tyrosine was inserted in this critical position. 9cgY-LSM1 had 1.6% of the potency of LSM1, but after photolysis, regained its full potency (Fig. 4B).
Figure 4.
Effects on MLCK activity and eosinophil locomotion of variants of MLCK inhibitory peptide LSM1. (A) Amino acid sequences of LSM1 and variants containing glutamate (E) and caged tyrosine (cgY). (B) Effects of LSM1 peptides on constitutively active catalytic domain of MLCK before (•) and after (○) photoconversion; the photoreleased peptide was indistinguishable from authentic LSM1 in inhibitory potency, chromatographic behavior and mass spectra. Loss of inhibitory potency for 9E-LSM1 (▵) illustrates the requirement for Y in position 9 for inhibitory activity. Data represent means of triplicate measurements with SEM values less than 10% of the mean. (C–J) Effects of photorelease of LSM1 on eosinophil locomotion. Phase contrast images of an eosinophil injected with 9cgY-LSM1, before (C and D), during (E), and after (F–H) near-UV exposure with times in sec. Color coded outlines of the same cell at the designated intervals before (I) and after (J) photorelease of LSM1.
Following the same injection and irradiation protocol used for 5cgY-RS-20, we observed polarized eosinophils before and after photorelease of LSM1. LSM1 caused cell locomotion to cease and inhibited forward motion of the leading lamellipod (Fig. 4 C-J), again within tens of seconds. Granule flow in the direction of cell locomotion ceased, and with time, granules moved somewhat toward the rear of the cell. Granule-free lamellar extensions again appeared over a considerable portion of the cell. Cells did not resume normal locomotion or associated behavior during the entire 5-min recording period after photoactivation of 9cgY-LSM1. Some or all of the effects depicted in Fig. 4 C-J were observed in 26 of 30 cells (87%). As the effects of LSM1 were quite similar to those of 5Y-RS-20, the results suggest that calcium-calmodulin regulation of MLCK is a major signaling pathway underlying eosinophil locomotion.
Localized photorelease of caged peptides (5-μm beam diameter at half-maximal intensity) had no effect on more than a dozen cells, regardless of whether the spot was aimed at the cell’s center, front, back, or within the lamellipod. Exposure of half of the cell area was also without effect, unless the cell had a bipolar morphology, in which there was a narrow constriction connecting the exposed and unexposed parts of the cell. In such cells, the unexposed part often rescued the exposed portion of the cell during a few-minute observation period. Thus, the peptides or calmodulin appear to be readily diffusible throughout the cell in the time domain of tens of seconds to minutes.
DISCUSSION
Photorelease within polarized and motile eosinophils of inhibitory peptides directed at calmodulin or MLCK caused rapid inhibition of lamellipod extension, granule flow, and net translation of cells on the substratum. These results show that MLCK is central in the signaling pathways underlying motility because these peptide probes were much less effective toward other kinases that could play a role in regulation of filament assembly and motor protein activity such as calmodulin-dependent protein kinase II, protein kinase C, and Rho kinase. Because MLCK only phosphorylates conventional myosin II, the data strongly implicate myosin II motors in these motile functions. These results extend previous work in which antibodies to MLCK (19) or to myosin II (20) disrupted vertebrate cell motility measured over tens of minutes to hours. Caged peptides allowed observations to be made in the previously inaccessible time domain of seconds. The effects of peptides were observed within a few seconds of photoconversion, suggesting that phosphate turnover in the regulatory light chains of myosin in these cells occurs every few seconds. This turnover would allow dynamic regulation of myosin II in a time frame that matches changes in calcium and cell behavior during chemotactic stimuli (11).
The prompt cessation of locomotion in eosinophils most readily is explained as resulting from inhibition of a forward-directed propulsive force caused by myosin II activity in the cell cortex at the rear and sides of the cell where myosin II has been localized in similar cells (21–23). Such a localization of myosin II is also compatible with its proposed role in tail retraction (24), and a loss of this capability would contribute to inhibition of net cell translation. Experiments with fluorescent biosensors reveal gradients of active calmodulin and active MLCK, which places these regulatory molecules also at the cell cortex near the rear of polarized cells (7). It is of interest to establish the subcellular site of action of the inhibitory peptides to determine whether specific aspects of cell motion can be linked with myosin II activity in specific regions of the cell. Peptides with restricted diffusion currently are under development to make these experiments possible.
Not all cell activity associated with motility was blocked by the peptides as formation of phase dark ruffles at the cell edge, believed to reflect actin polymerization (16) and possibly myosin I activity (25), continued. Inhibition of cortical myosin II based contraction, but not actin polymerization or myosin I activity, also may explain why after activation of the peptides lamellar extensions occurred in regions of the cell where they were normally absent. Similar multilamellar behavior was reported in studies of Dictyostelium in which functional myosin II was deleted by molecular biological techniques (1, 2). These studies led to the proposal that cortical myosin II contraction suppresses global lamellar activity, and the extensive edge ruffling observed in the present experiments is consistent with such a role for myosin II.
It is noteworthy that in studies of Dictyostelium lacking myosin II, cell locomotion was somewhat impaired but still present (1, 2). This apparent contradiction with our observations in eosinophils may be caused by activation of compensatory mechanisms in cells in which myosin II is chronically absent; such compensatory processes would not be evident in cells where myosin II is suddenly inhibited. Alternatively, Dictyostelium may possess redundancy in locomotory mechanisms to permit movement on a variety of surfaces. It has been demonstrated recently that Dictyostelium lacking myosin II cannot move on surfaces to which the cells adhere strongly, consistent with models of cell motility in which myosin II produces much of the force required for tail retraction (24). Thus, studies of myosin II knockouts in Dictyostelium may not be in conflict with the results reported here because newt eosinophils are known to be strongly attached to the substratum under the conditions used (8, 11).
In summary, the present experiments show that calcium-calmodulin-dependent MLCK represents a major molecular control mechanism underlying the processes of cortical contraction to maintain asymmetry, granule flow, and net translation of these rapidly moving cells on the substratum. Myosin II, the likely motor protein involved, appears to be tightly coupled to calmodulin and MLCK activity, and phosphates on the light chains probably turn over rapidly. Edge ruffling and lamellipod formation were not under the control of calmodulin and MLCK, consistent with the idea that these processes are mainly caused by actin polymerization, which is subject to other control mechanisms (7). Rapid eosinophil locomotion likely relies on coordinated actions of both actin polymerization and myosin II motor activity. The results show that regulation of myosin II by the calmodulin-dependent MLCK signaling pathway has the necessary kinetics to dictate the rate and direction of cell movement in response to complex and rapidly changing extracellular signals.
The approach we have used in the design and development of caged peptides is general and can be applied to other peptides by (i) evaluating single amino acid substitutions in an in vitro functional assay to determine the peptide’s structure-activity profile, and (ii) incorporating into synthetic peptides photolabile amino acids designed to interfere with characteristic interactions between the peptide and its target. Large quantities and many varieties of precisely defined caged peptides and, by use of peptide ligation techniques, caged proteins can be prepared by this strategy. Drawbacks of earlier methods used to prepare caged proteins (26–28) include that incorporation of photolabile groups often occurs with varying efficiency into multiple sites, and photolytic conversions are low. Selective incorporation of caged amino acids into proteins by in vitro translation has been achieved (29), but caged proteins were obtained in limited quantities. The capability to prepare quantities of well-defined biologically active caged peptides and to photorelease them within living cells creates opportunities for investigating dynamic cell functions. It now should be possible to generate specific activators or inhibitors of protein–protein interactions in a spatially defined manner and in the time domain in which many cellular processes occur.
Acknowledgments
This work is dedicated to the memory of Fredric S. Fay who passed away on March 18, 1997. Support for this research was provided in part by grants from the National Institutes of Health (to J.W.W., M.I., and F.S.F.) and the National Science Foundation (to J.W.W. and F.S.F.). R.M.D. is a postdoctoral fellow of the Massachusetts Heart Association. J.W.W. is a recipient of a Research Career Development Award from the National Institutes of Health.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviation: MLCK, myosin light chain kinase.
References
- 1.Knecht D A, Loomis W F. Science. 1987;236:1080–1085. doi: 10.1126/science.3576221. [DOI] [PubMed] [Google Scholar]
- 2.DeLozanne A, Spudich J A. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- 3.Adams S R, Tsien R Y. Annu Rev Physiol. 1993;55:755–784. doi: 10.1146/annurev.ph.55.030193.003543. [DOI] [PubMed] [Google Scholar]
- 4.Corrie J E T, Trentham D R. In: Biological Applications of Photochemical Switches. Morrison H, editor. New York: Wiley; 1993. pp. 243–305. [Google Scholar]
- 5.Tanaka M, Ikebe R, Matsuura M, Ikebe M. EMBO J. 1995;14:2839–2846. doi: 10.1002/j.1460-2075.1995.tb07283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itoh T, Ikebe M, Kargacin G J, Hartshorne D J, Kemp B E, Fay F S. Nature (London) 1991;338:164–167. doi: 10.1038/338164a0. [DOI] [PubMed] [Google Scholar]
- 7.Guiliano K A, Taylor L D. Curr Opin Cell Biol. 1995;7:4–12. doi: 10.1016/0955-0674(95)80038-7. [DOI] [PubMed] [Google Scholar]
- 8.Koonce M P, Cloney R A, Berns M W. J Cell Biol. 1984;98:1999–2010. doi: 10.1083/jcb.98.6.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker J W, Martin H, Schmitt F S, Barsotti R J. Biochemistry. 1993;32:1338–1345. doi: 10.1021/bi00056a020. [DOI] [PubMed] [Google Scholar]
- 10.Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. J Biol Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert S H, Perry K, Fay F S. J Cell Biol. 1994;127:489–503. doi: 10.1083/jcb.127.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lukas T J, Burgess R H, Prendergast F G, Lau W, Watterson D M. Biochemistry. 1986;25:1458–1464. doi: 10.1021/bi00354a041. [DOI] [PubMed] [Google Scholar]
- 13.Ikura M, Gore G M, Gronenborn A M, Zhu G, Klee C B, Bax A. Science. 1992;256:632–637. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- 14.Sreekumar, R., Ikebe, M., Fay, F. S. & Walker, J. W. (1998) Methods Enzymol., in press. [DOI] [PubMed]
- 15.McCarron J G, McGown J S, Reardon S, Ikebe M, Fay F S, Walsh J V. Nature (London) 1992;357:74–77. doi: 10.1038/357074a0. [DOI] [PubMed] [Google Scholar]
- 16.Hahn K, DeBiasio R, Taylor D L. Nature (London) 1992;359:736–738. doi: 10.1038/359736a0. [DOI] [PubMed] [Google Scholar]
- 17.Pollard T D, Doberstein S K, Zot H G. Annu Rev Physiol. 1991;53:653–681. doi: 10.1146/annurev.ph.53.030191.003253. [DOI] [PubMed] [Google Scholar]
- 18.Zhu T, Sata M, Ikebe M. Biochemistry. 1996;35:513–522. doi: 10.1021/bi952053c. [DOI] [PubMed] [Google Scholar]
- 19.Wilson A K, Gorgas G, Claypool W D, de Lanerolle P. J Cell Biol. 1991;114:277–283. doi: 10.1083/jcb.114.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honer G, Citi S, Kendrick-Jones J, Jockusch B M. J Cell Biol. 1988;107:2181–2189. doi: 10.1083/jcb.107.6.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conrad P A, Giuliano K A, Fisher G, Collins K, Matsudaira P T, Taylor D L. J Cell Biol. 1993;130:1381–1391. doi: 10.1083/jcb.120.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly C A, Sellers J R, Gard D L, Bui D, Adelstein R S, Baines I C. J Cell Biol. 1996;134:675–687. doi: 10.1083/jcb.134.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukui Y, Lynch T J, Brzeska H, Korn E D. Nature (London) 1989;341:328–331. doi: 10.1038/341328a0. [DOI] [PubMed] [Google Scholar]
- 24.Jay P Y, Pham P A, Wong S A, Elson E L. J Cell Sci. 1995;108:387–393. doi: 10.1242/jcs.108.1.387. [DOI] [PubMed] [Google Scholar]
- 25.Ruppert C, Godel J, Muller R T, Kronschewski R, Reinhard J, Bahler M. J Cell Sci. 1995;108:3775–3786. doi: 10.1242/jcs.108.12.3775. [DOI] [PubMed] [Google Scholar]
- 26.Marriott G. Biochemistry. 1994;33:9092–9097. doi: 10.1021/bi00197a010. [DOI] [PubMed] [Google Scholar]
- 27.Marriott G, Heidecker M. Biochemistry. 1996;35:3170–3174. doi: 10.1021/bi952207o. [DOI] [PubMed] [Google Scholar]
- 28.Chang C, Niblack B, Walker B, Bayley H. Chem Biol. 1995;2:391–400. doi: 10.1016/1074-5521(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 29.Mendel D, Ellman J A, Shultz P G. J Amer Chem Soc. 1991;113:2758–2760. [Google Scholar]