Abstract
The human osteosarcoma-derived cell line SAOS-2, exhibits many of the phenotypic characteristics of osteoblasts including the deposition of types I and V collagens in an extracellular matrix. Lesser amounts of collagen XI chains were also detected. The cell layer collagen contains hydroxylysyl pyridinoline cross-links but without the accompanying lysyl pyridinoline typical of human bone collagen. This indicates that the lysine residues at the two helical cross-linking loci are fully hydroxylated. The isoform of lysyl hydroxylase, LH1, known to be required for full hydroxylation at these sites, was shown to be highly expressed by SAOS-2 cells. Our findings provide insight on the mechanism of post-translational overmodification of lysine residues in collagen made by osteosarcoma tumors, and may be relevant for understanding a similar overmodification observed in osteoporotic bone.
Keywords: Bone, type I collagen, type V collagen, osteosarcoma, pyridinoline, lysyl hydroxylase, protein mass spectrometry
Introduction
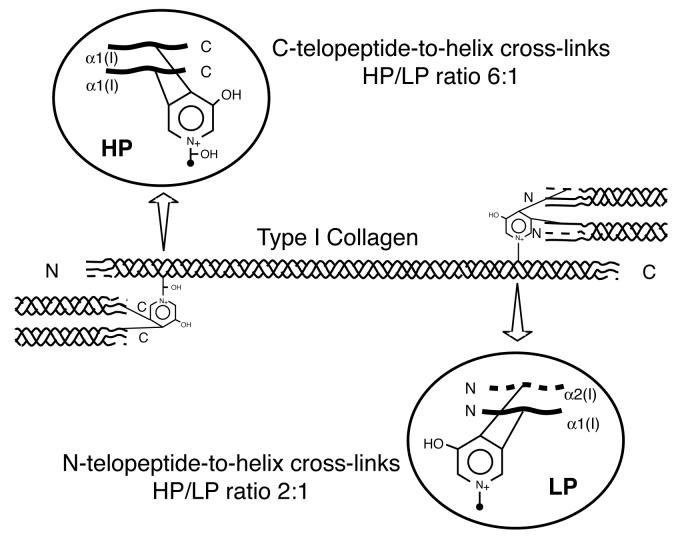
Bone type I collagen can be distinguished from type I collagen of other tissues by its characteristic post-translational chemistry [1]. In particular, it has a distinctive profile of cross-linking amino acids that reflects the degree of hydroxylation of specific lysines in the telopeptide and helical cross-linking domains of the α1(I) and α2(I) chains. Hydroxylysyl pyridinoline (HP) cross-links are formed from two telopeptide and one helical hydroxylysine (Figure 1). Lysyl pyridinoline (LP) cross-links are a post-translational variant seen most prominently in bone collagen that are formed from two telopeptide hydroxylysines and a helical lysine [2,3].
Figure 1.
Intermolecular cross-links in type I collagen
Diagram showing the sites, type and ratio of pyridinoline cross-links in bone type I collagen. Pyridinolines stabilize type I collagen fibrils of human bone at both ends of the molecule. HP is more abundant at the C-telopeptide-helix cross-link site (HP/LP ratio, 6:1) and LP is more abundant at the N-telopeptide-helix site (HP/LP ratio, 2:1) [19].
Lysines are hydroxylated in collagen by the lysyl hydroxylase (PLOD, LH) family of enzymes [4]. The human genome has three genes PLOD1, PLOD2 and PLOD3 that encode three different isoforms, lysyl hydroxlyase 1, 2 and 3 (LH1, LH2, LH3) [5-8]. Early studies showed that LH1 could hydroxylate lysine residues in the triple helical domains of α1(I) and α2(I) chains of type I collagen [4]. There is evidence that LH2 (which has 2 splice forms, LH2a and LH2b, [9]) hydroxylates cross-linking lysines in collagen telopeptide domains [10]. LH3, the most recently identified isoform [7], is essential for lysine hydroxylation in basement membrane type IV collagen [11]. LH3 also has glucosyl and galactosyl transferase activity, and so could add sugars to specific hydroxylysine side-chains [12]. Expression of lysyl hydroxylase isoforms is thought to be a factor in regulating tissue-dependent differences in cross-linking of collagen.
Lysyl hydroxylases are essential for normal development. Null mutations in PLOD 1 cause Ehlers Danlos syndrome type VIA (EDS-VIA) in which hydroxylysine is absent from skin collagen and deficient in other tissues [13-15]. In normal bone collagen the ratio of LP (lysyl pyridinoline) to HP (hydroxylysyl pyridinoline) is much higher than in other collagens (cartilage, tendon, ligament, etc.) that use the pyridinoline cross-linking pathway (0.25 to 0.5 versus <0.1). This is because the helical lysines, which donate the ring-nitrogen arm of the pyridinoline structure, are only partially hydroxylated in bone collagen [16,17]. Mutations in PLOD2 cause Bruck Syndrome, a bone disorder resembling osteogenesis imperfecta [18], in which the bone collagen lacks pyridinolines and the other cross-links based on hydroxylysine aldehydes. Since the telopeptide lysines are not hydroxylated, this implies that LH2 is the telopeptide lysine hydroxylase [10]. Mice in which plod3 is null have defective basement membranes due to underhydroxylation of collagen type IV [11]. These genetic and molecular studies indicate that the three isoforms of lysyl hydroxylase act in a tissue-specific and collagen site-specific manner in the post-translational processing of collagens.
The cross-linking lysine residues of bone collagen appear to be partially hydroxylated in a distinctive pattern, resulting in the characteristic HP:LP ratio and the prominence of pyrrole cross-links in bone collagen [2,3,19]. How this is controlled is not fully clear but differential expression of LH1, 2 and 3 by osteoblasts seems to be involved. The overall content of hydroxylysine in human bone collagen has been shown to vary greatly with stage of bone maturity [20], being higher, for example, in osteosarcoma bone [21,22], osteoporotic cancellous bone [23] and woven repair bone [24] than in normal mature bone. Elevated levels of hydroxylysyl pyridinoline (a marker of bone degradation) and a high HP:LP ratio in the urine of osteosarcoma patients [25,26] is likely to be due to the degradation of bone collagen with a high degree of lysine hydroxylation.
Here we report on the post-translational quality of collagen deposited extracellularly by SAOS-2 osteosarcoma cells [27]. These cells express an osteoblastic phenotype [27-31], mineralize their matrix in the presence of β-glycerophosphate [32-35] and have been used extensively in bone biology research [36-40]. The SAOS-2 cell line synthesizes primarily types I and V collagens and deposits them in a copious extracellular matrix [35,41]. We show here, the collagen contains pyridinoline cross-links at a level typical of human bone collagen but the ratio of HP:LP cross-links is high. The demonstrated high expression level of the LH1 enzyme can explain the high HP:LP ratio.
Materials and methods
Cell culture
The SAOS-2 cell line (ATCC # HTB 85) was maintained as monolayer cultures in a humidified 37°C incubator, 5% CO2 in air. McCoys media containing 10% FBS and 50 μg/ml ascorbate was exchanged every other day and the cells were cultured for 4 weeks. The harvested medium from the entire culture period was pooled and frozen for collagen analysis.
Isolation of collagen from the cultured cell extracellular matrix
After a month in culture, cell layers were washed with PBS and then extracted with 1M NaCl, 50 mM Tris, pH 7.5, containing 1 mM phenyl methyl sulfonyl fluoride (PMSF), 1 mM n-ethyl maleimide (NEM), 5 mM ethylenediaminetetraacetic acid (EDTA) for 2 days at 4°C. The NaCl extract (N) and the insoluble residue were separated by centrifugation at 18000 RPM (25000g). The residue was further digested with 0.1 mg/ml pepsin in 0.5M acetic acid for 24 hours at 4°C to solubilize the cross-linked collagen. Type I collagen was purified from the pepsin digest as a precipitate by centrifugation after adding NaCl to 0.8M. The insoluble residue was not analyzed further.
Collagen in the NaCl extract was precipitated by adding NaCl to 4.5 M. A portion was the treated with 0.1 mg/ml pepsin in 0.5M acetic acid and run on SDS-PAGE. The medium was acidified, digested with 0.1mg/ml pepsin and collagen was precipitated at 0.8M NaCl.
Gel electrophoresis
Collagen types in the medium and cell layer were identified by electrophoresis of the pepsin-digested protein. All samples were dried, dissolved in Laemmli sample buffer without reducing agent and heated at 100°C for 3 min. Collagen chains were electrophoresed on 6% gels [42] and visualized by Coomassie blue staining. Human bone type I collagen was used as standard. Gels were scanned densitometrically and the NIH Image software program with the gel electrophoresis macro was used to determine approximate ratios of collagen types in the extracts [43].
Mass spectrometry
Collagen in the 1M NaCl extract was separated by SDS-PAGE under reducing conditions and stained with Coomassie Blue. Individual bands were cut from the gel and subjected to in-gel trypsin digestion. Electrospray MS was performed on the tryptic peptides using an LCQ Deca XP ion-trap mass spectrometer equipped with in-line liquid chromatography (LC) (ThermoFinnigan) using a C8 capillary column (300μm × 150mm; Grace Vydac 208MS5.315). For protein identification, MS/MS spectra were searched again the NCBI nrfasta database using the SEQUEST database search algorithm.
Collagen cross-link analysis
Aliquots of pepsin-extracted collagen were hydrolyzed in 6M HCl, 110°C for 24 hours. Hydroxylysyl pyridinoline (HP) and lysyl pyridinoline (LP) cross-linking residues were resolved and quantified by RP-HPLC and fluorometry [44, 45]. An aliquot of the hydrolysate was colorimetrically assayed for hydroxyproline as a measure of collagen content [46]. Pyridinoline content was expressed as moles per mole of collagen. Human bone type I collagen (fetal and adult) was run in comparison.
Preparation of polyclonal antibody
Recombinant LH1 antigen was generated in E. coli as a maltose-binding protein (MBP)-LH1 fusion product. The coding sequence of LH1 was amplified from SAOS-2 cells by RT-PCR using primer CAT ACC GAA TTC ATG CGG CCC CTG CTG CTA CT (92-111 of PLOD1 cDNA, containing an EcoRI site) and (AAG AAA GGT CCA AGA GGGTC 2307-2288 of PLOD1 cDNA, adjacent to a StuI site), then digested with EcoRI and StuI, and cloned into the EcoRI-EcoRV sites of Blue Script (Stratagene, La Jolla, CA) as clone “SAOS-LH1”. The LH insert was transferred from SAOS-LH1 into the pMAL-cRI vector (New England BioLabs) as an EcoRI-HindIII fragment, placing LH in frame with MBP, expressed in E. coli strain XL1-blue (Stratagene), and gel-purified from the inclusion bodies by SDS-PAGE. In-frame fusion was confirmed by N-terminal sequencing [47] of the LH peptide after release from MBP by factor Xa digestion.
Antibodies were generated in rabbits (R and R Rabbitry; Stanwood, WA) by repeated subcutaneous injections of the MBP-LH suspension (100 μg initial injection, and 5 boosts of 50 μg each at two week intervals). LH1-directed antibodies were enriched from crude serum by adsorption to a poly-histidine-LH1 (His-Tag-LH1) fusion protein, bound to a nickel column made with His-Bind Resin (Novagen). The His-TagLH1 fusion construct was made in pET-15b (Novagen).
Immunoblot analysis
Total lysates from plates of cultured cells were prepared by draining the plate for 2-3 min, shaking out all excess liquid, adding 0.5ml SDS sample buffer (2% w/v SDS, 5% v/v b-mercaptoethanol, 10 mM EDTA, 50 mM Tris, 0.1% w/v bromophenol blue, 10% v/v glycerol, pH 7.0), scraping the plate contents into a microfuge tube, and incubating at 95-100°C for 5 min with occasional vortexing. Lysates were cleared of insoluble material by centrifugation at 10,000 g. Total tissue lysates were prepared by powdering frozen tissue under liquid nitrogen in a SPEX 6700 Freezer/Mill (SPEX Industries), suspending it in about 10 volumes of SDS sample buffer, and heating as above. LH1 protein was enriched from placental homogenates by differential precipitation with ammonium sulfate and concanavalin-A affinity chromatography (4). Equal aliquots of protein in samples were separated by SDS-PAGE on 8% polyacrylamide gels, and transferred to PVDF membranes using a semi-dry electrotransfer apparatus (Millipore). Blots were blocked, and probed according to standard procedures (48), using 500X diluted purified LH1 antibody (see above), and 5000X diluted goat anti-rabbit IgG-horse radish peroxidase (BioRad) as secondary antibody. Bound horseradish peroxidase was detected by chemiluminescence using enhanced Luminol reagent (New England Nuclear) and Hyperfilm-MP (Amersham).
RNAase protection assays
A riboprobe template was generated by cloning the 208 bp PstI-NspI fragment of LH1 cDNA (nucleotides 947-1154) [5] between the PstI and EcoRV sites of the plasmid, pBC KS (Stratagene). The cDNA was generated by RT-PCR from the human osteosarcoma cell line, SAOS-2. An antisense RNA probe, including 46 bases of vector sequence and 208 bases of cDNA sequence, was synthesized from the T7 promoter, using the Riboprobe T7/T3 Combination kit from Promega. Probe was hybridized to cellular RNA and unbound probe was digested, using the RPII RNAase Protection kit from Ambion. Protected probe was resolved by acrylamide gel electrophoresis and detected by autoradiography and quantified by densitometry.
Northern blot analysis
RNA from SAOS-2 cells, normal fetal human skin and Swarm Rat chondrosarcoma (RCS-LTC) chondrocytes [47] was electrophoresed (5 μg/lane) and probed following the protocol described in [48]. Blots were prehybridized and the membrane bound RNA was hybridized to 32P-labelled LH1 cDNA. Detection was by autoradiography.
RT-PCR analysis of lysyl hydroxylase gene expression
RNA was extracted using TriZol (Invitrogen) and purified according to the manufacturer's protocol. cDNA was synthesized using the Thermoscript RT-PCR system (Invitrogen). A co-RT-PCR assay was developed that yielded 748, 879, 434 and 378 bp products specific for LH1, LH2, G3PDH and COL1A1, respectively, in a single reaction. The primers used were as follows: LH1; Fwd: GTTTCCAGCCCCTGCGAGCGCCGC; Rev: CAGGTTCCTCGCTCTCACATGGC LH2; Fwd: ATGGGGGGATGCACGGTGAAGCCTC Rev: ATCTACTGCAGACAAGTCGACTGTATCG G3PDH; Fwd; AGCTTGCTCAAGAAGACAGACC Rev: GAGGACATCCCCTCTTCTCAC COL1A1; Fwd GTGGACCTCCGGCTCCTGCTCC Rev GAAGTCCAGGCTGTCCAGGGATGC. The primers designed for LH2 do not discriminate between LH2a and LH2b splice forms. During each mRNA amplification reaction, each cycle of PCR included 1 min of denaturation at 94°C, 1 min of annealing at 60°C and 3 min of extension at 72°C for 35 cycles.
The assay was applied to mRNA extracted from SAOS-2 cells, human foreskin fibroblasts, CH1 human chondrosarcoma cells (a gift from Dr. Linda Sandell), mature human osteoblasts and mRNA from fetal human bone, cartilage, skin and various other fetal human tissues. To keep the amplification within the linear range for all the four gene products in the same reaction, the cycle number was reduced to 25, and the sensitivity increased by 32P-labelling. To assess relative levels of LH1 message, dCT32P labelled RT-PCR products were separated on 6% polyacrylamide gels and semi-quantified by densitometry using NIH Image software [49-51]. G3PDH, COL1A1 or LH2 was used as reference.
Comparative genome analysis of LH1
Regions of the PLOD1 (LH1) gene and COL10A1 gene (reference) were amplified by multiplex-PCR using 300 ng genomic DNA from human fetal liver or SAOS-2 cells as described in [51]. Using primers for LH1: Fwd AGGACTGGAAGGAGAAGTACATCC, Rev GGTCCCACCTTGTTGTTGCCCAG and COL10A1: Fwd GGCCCAGCTGGCATAGCAACTAAGGG Rev CTCCCTGAAGCCTGATCCAGGTAGCC yielded products of 800bp and 530 bp respectively. PCR was carried out at 1 min of denaturation at 94°C, 1 min of annealing at 60°C and 3 min of extension at 72°C for 35 cycles. After electrophoresis, products were quantified by densitometry.
Results
Characterization of collagen
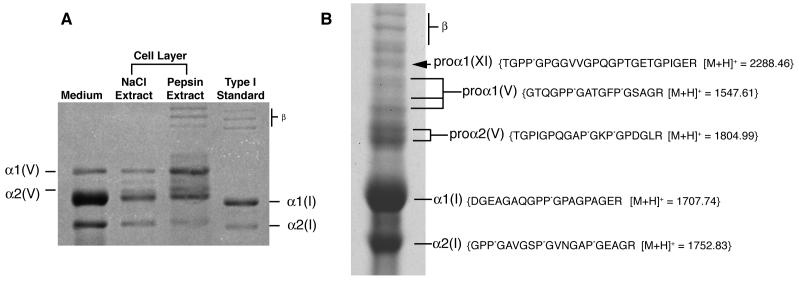
On pepsin digestion the medium and cell layer gave chains of types I and V collagens on SDS-PAGE by Coomassie staining (Figure 2A). The α1(I) and α2(I) chains ran slightly slower than pepsin-extracted control preparations from human bone, suggesting a higher level of post-translational modification.
Figure 2.
Biochemical analysis of collagen
A) Collagen types synthesized by the SAOS-2 cell line in culture
The medium and cell layer on pepsin digestion showed types I and V collagen α-chains on SDS-PAGE. The α1(I) and α2(I) chains ran slightly slower than pepsin extracted control preparations from human bone, suggesting post-translational over-modification. Mass spectrometry identified the band between the α1(V) and α2(V) to be a degradation product of the α1(V) chain. Bands marked “β” are dimers of collagen chains.
B) Identification of α1(XI) collagen chains by mass spectrometry
SDS-PAGE (6%) of collagen extracted by 1M NaCl from the extracellular matrix of SAOS-2 cells (+DTT). Arrow indicates a band identified as a pro-form of the α1(XI) collagen chain. The sequence of a tryptic peptide from this band unequivocally identified by mass spectrometry is shown. Mass spectrometry also identified pro-forms of α1(V) and α2(V), as well as processed α1(I) and α2(I) from this gel. Bands marked “β” are dimers of collagen chains.
Of the total collagen synthesized, 65% was present in the medium, 2% in the 1M NaCl extract and 33% in the pepsin extract of the cell layer. The insolubility of the cell-layer collagen without pepsin digestion indicated extensive covalent cross-linking.
Type I collagen was the major collagen synthesized by the cell line accounting for 80% of the total medium plus cell layer collagen. Most of the collagen found in the medium was type I (Table 1). About equal amounts of types I and V collagen were recovered in the pepsin extract of the cell layer (Table 1), indicating a preferential deposition of type V collagen in cross-linked extracellular collagen fibrils. Type III collagen was not detected.
Table 1.
Distribution of collagens type I and type V in the SAOS-2 cell culture extractsa.
| Type I collagen | Type V collagen | |
|---|---|---|
| Medium | 90.2 | 9.8 |
| NaCl extract | 75 | 25 |
| Pepsin extract | 47.7 | 52.3 |
Values expressed as percentages of total collagen in each extract
In-gel trypsin digestion and mass spectroscopic analysis of individual bands resolved by SDS-PAGE from the non-pepsin-treated1M NaCl fraction identified higher molecular weight forms of α1(V) and α2(V) shown in Fig. 2B. The band indicated by the arrow in Figure 2B was a pro-form of the α1(XI) chain. Presumably these components of the cell-layer matrix are partially processed pro-forms that are not yet cross-linked. This cell line is known to express the COL11A2 gene and export α2(XI) chains of type XI collagen [41].
Cross-link analysis
RP-HPLC analysis of pyridinoline cross-links in the cell layer collagen showed HP alone in contrast with HP:LP ratios in the range of 2 to 4:1 typical of human adult bone collagen and human fetal bone collagen (Fig. 3, middle and lower panels) [3]. Cross-linking lysines at the triple-helical sites in the SAOS-2 collagen appear therefore to be fully hydroxylated, whereas in bone collagen they are partially hydroxylated. The total pyridinoline (HP+LP) concentration in the SAOS-2 collagen (0.28 mol/mol of collagen) was similar to that of adult human bone (0.24 mol/mol collagen) [1]. On further analysis, the pyridinolines were shown to be present primarily in type I collagen (data not shown).
Figure 3.
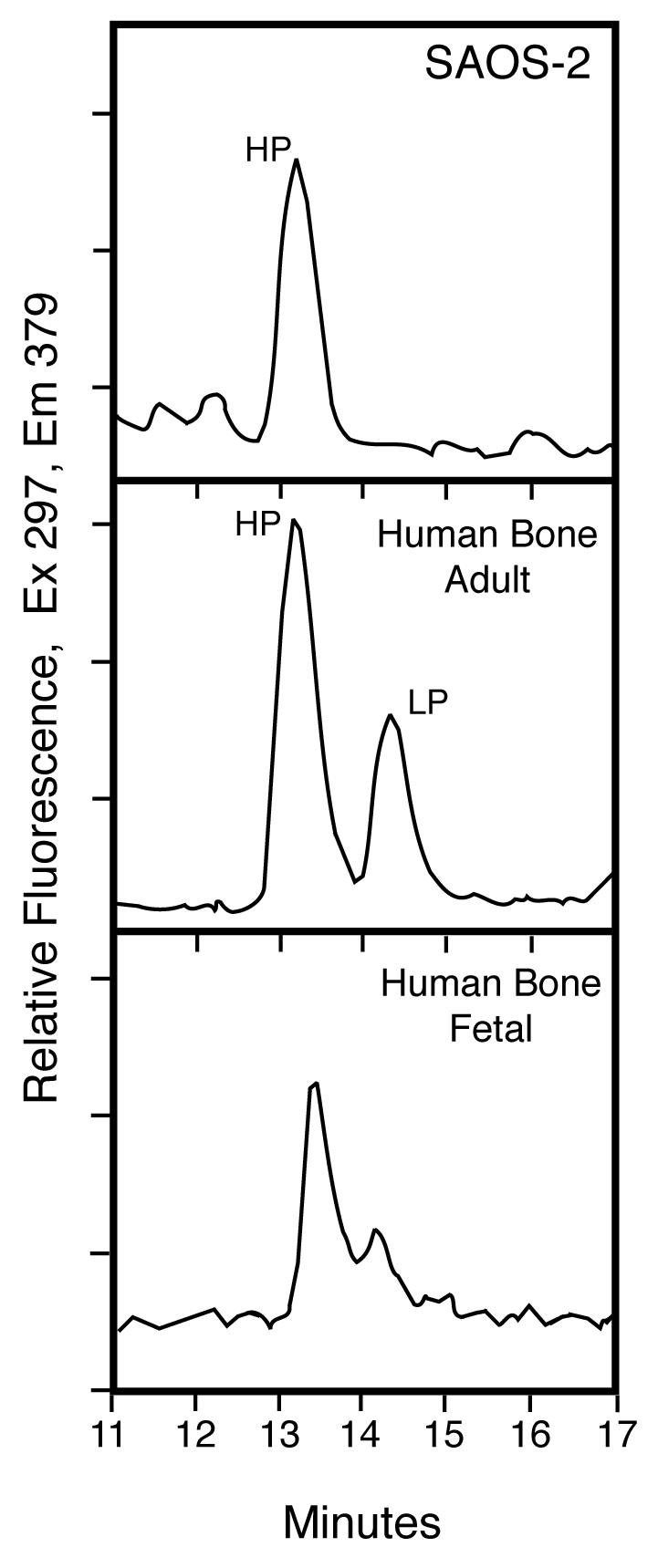
Pyridinoline cross-links in SAOS-2 cell layer collagen compared with human bone
RP-HPLC analysis of pyridinoline cross-links in the cell layer collagen showed HP alone (upper panel) in contrast with the ratio of about 2:1 for HP:LP typical of adult human bone (middle panel). This result indicates that the triple-helical domain cross-linking lysines were fully hydroxylated in the SAOS-2 collagen, whereas in bone collagen they are only partially hydroxylated.
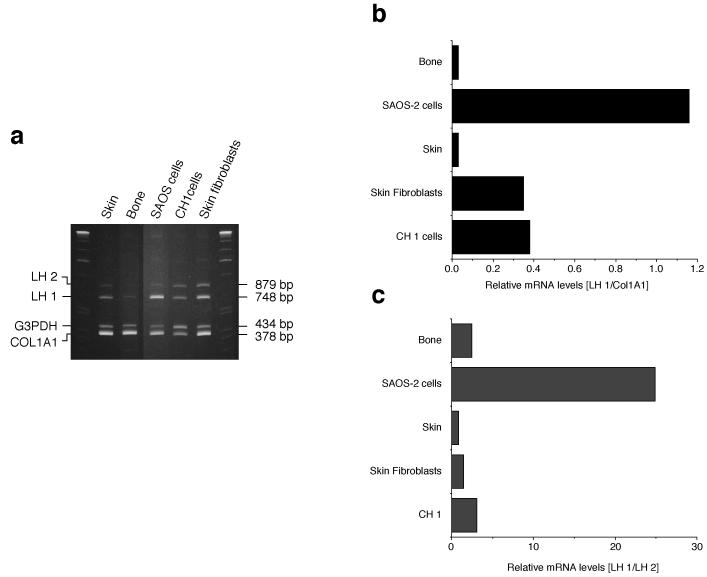
Expression of lysyl hydroxylase
The high HP:LP cross-link ratio in SAOS-2 cell collagen implied a high level of LH1 activity. In order to determine if LH1 expression was unusual in SAOS-2 cells, a co-RTPCR assay was developed (Fig. 4a). The abundance of LH1 RNA was compared to those of three other RNAs encoding COL1A1, G3PDH, and LH2 in human bone osteoblasts, skin fibroblasts, CH1 cells and skin. Semi-quantitative comparison of LH1 mRNA to each of the other RNAs (Fig. 4b, LH1/COL1A1and 4c, LH1/LH2) showed that LH1 expression in SAOS-2 cells was unusually high. Regardless of the RNA chosen as standard, LH1 RNA was 10-30 fold higher in SAOS-2 cells than in bone or skin. Since cultured cells derived from other connective tissues (fibroblasts from skin, and a human cartilage chondrosarcoma cell line [CH1]) did not exhibit this elevated LH1 mRNA the effect is unlikely to be a culture artifact. LH2 mRNA levels in SAOS-2 cells were not decreased when compared to skin fibroblasts and bone tissue, suggesting that the high LH1 mRNA levels are not due to a compensatory mechanism in these cells (data not shown).
Figure 4.
Expression of lysyl hydroxylase mRNA
a) 6% polyacrylamide gels showing the ethidium bromide-stained RT-PCR (35 cycles) products of LH 1, LH2, G3PDH and COL1A1 from human skin, mature human osteoblasts, SAOS-2 cells, cultured human chondrosarcoma cells (CH1) and cultured skin fibroblasts.
b, c) A similar RT-PCR (25 cycles) experiment was performed after labelling with dCT32P to assess relative levels of LH1 message. For each tissue or cell culture, RNA was amplified and resolved on a 6% gel. Autoradiograghs were scanned to compare bands quantitatively. As shown in the bar graphs, regardless of the RNA chosen as reference (COL1A1 or LH2), LH1 mRNA was much higher (10 to 30 fold) in SAOS-2 cells than in osteoblasts, skin and cultured cells.
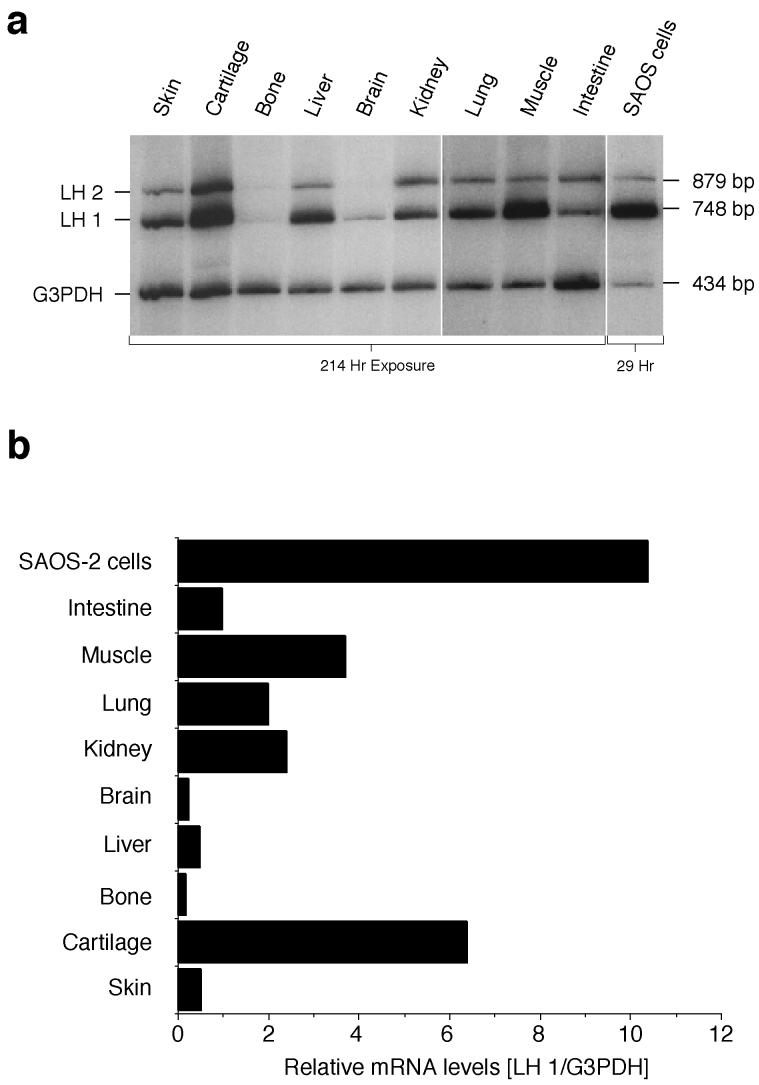
A wider survey of LH1 mRNA levels in human fetal tissues including bone and skin (Fig 5a), standardized to G3PDH, showed considerable variation, with particularly high levels in cartilage and in muscle (Fig. 5b). However, none of the tissues exhibited levels approaching that of SAOS-2 cells.
Figure 5.
Co-RT-PCR survey of LH1 mRNA levels in human tissues
LH1 and G3PDH RNAs were co-amplified from fetal human tissues, using dCT32P labelling conditions.
a) Products were separated by electrophoresis on 6% polyacrylamide gels, and detected by autoradiography.
b) Values represent the LH1 mRNA levels normalized to G3PDH as internal standard, as determined by quantitative scans of each lane. Large differences of LH1 mRNA levels were seen between the tissues, with particularly high levels in cartilage and muscle. None, however, exhibited levels approaching that of the SAOS-2 cells.
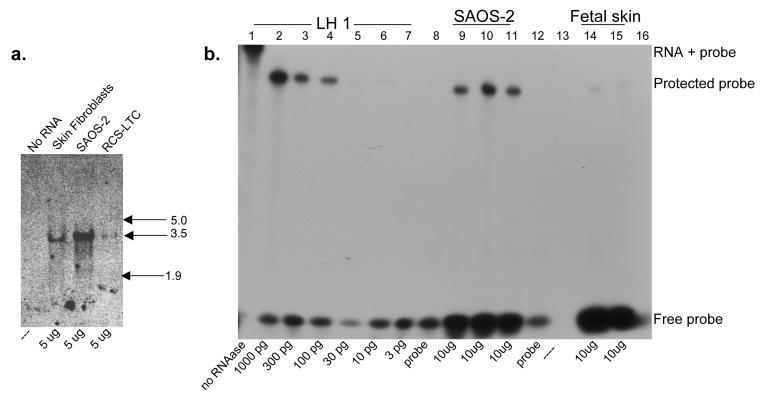
The high level of LH1 mRNA in SAOS-2 cells relative to skin fibroblasts was confirmed by Northern blot analysis (Fig. 6a), and quantified by RNAase protection assay (Fig. 6b), which indicated a 30-fold higher level of LH1 in the SAOS-2 cells than in human fetal skin. Since the relative levels of LH1 in bone and skin were comparable and because LH1 is a crucial enzyme for lysyl hydroxylation in skin [13-15], human skin fibroblasts and human fetal skin were used for this comparison rather than osteoblasts. Mutations in PLOD1 result in loss of lysine hydroxylation in skin type I collagen [14]. The RCS-LTC chondrocytes showed a reduced level of LH1 when compared to skin fibroblasts. The collagen from this cell line is known to have a lower HP:LP ratio when compared to normal cartilage [45].
Figure 6.
a: Northern Blot analysis of LH1 mRNA in SAOS-2 cells and skin
5 μg of SAOS-2, human fetal skin and RCS-LTC chondrocyte cell line total RNA was resolved on a 1.2% agarose-formaldehyde gel, blotted to nitrocellulose membrane, probed with a 32P-labelled LH1 cDNA, and detected by autoradiography. RNA kilobase markers are shown at right.
b: Quantitation of LH1 mRNA in SAOS-2 cells and skin by RNAase protection.
RNA was hybridized to a 32P-labelled LH1 antisense probe, digested with nuclease to remove the non-homologous portion of the probe, resolved on a denaturing 6% acrylamide gel, and detected by autoradiography. Lane 1, Full-length probe prior to nuclease digestion. Lanes 2-7, Control LH1 RNA reactions containing 1000, 300, 100, 30, 10, and 3 pg LH1 RNA respectively. Lanes 9-11, Replicate reactions containing 10 μg SAOS-2 RNA. Lanes 14 and 15, Replicate reactions containing 10 μg human fetal skin RNA. Lane 8 and12, Probe only. Lanes 13 and 16 are empty.
Immunoblot analysis
Western blot analysis was used to determine if LH1 protein is expressed in SAOS-2 cells. As shown in Fig. 7, an antibody raised against recombinant LH1 binds the predicted 82-85 kDa LH1 band in SAOS-2 cell lysates. SAOS-2 cells gave several fold higher levels than skin fibroblasts of this LH1 band. The high mRNA levels in SAOS-2 cells were therefore reflected in higher protein levels.
Figure 7.
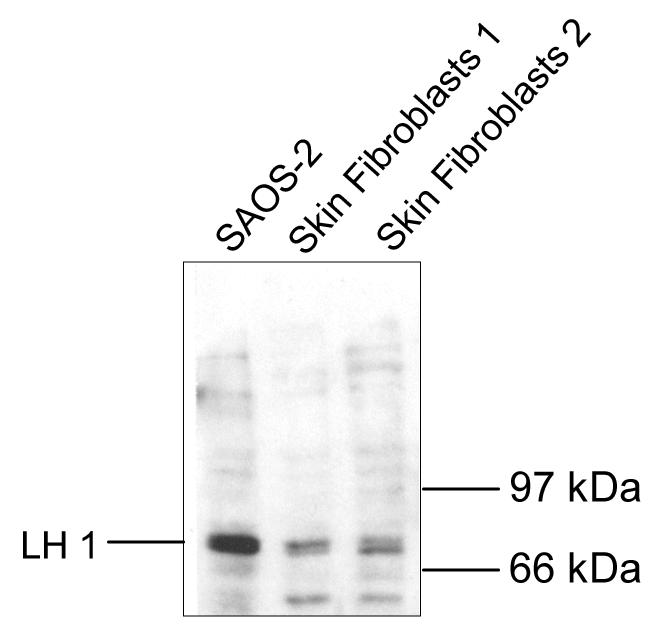
Expression of LH1 protein in cultured SAOS-2 cells
SDS-soluble protein from SAOS-2 cells (lanes 1) and cultured human skin fibroblasts (lanes 2, 3) were separated by SDS-PAGE, blotted to PVDF membrane, probed with an antibody to LH1, and detected with a chemiluminescence based secondary antibody assay. The background bands below 66 kDa are unidentified.
Comparative genome analysis
One possible mechanism for the elevated LH1 expression could be amplification of the PLOD1 gene in SAOS-2 cells, due either to karyotype abnormalities or to tandem duplication of the gene. To address this possibility, a multiplex-PCR assay was developed, which compared the abundance of the PLOD1 sequence in genomic DNA to that of a reference gene, COL10A1. The relative abundance of PLOD1 and collagen X PCR products was the same in SAOS-2 DNA as in normal human fetal liver DNA (Fig. 8), indicating a normal PLOD1 copy number in SAOS-2 cells. The high levels of LH1 RNA and protein are due to elevated expression and/or stability of these products in SAOS-2 cells.
Figure 8.
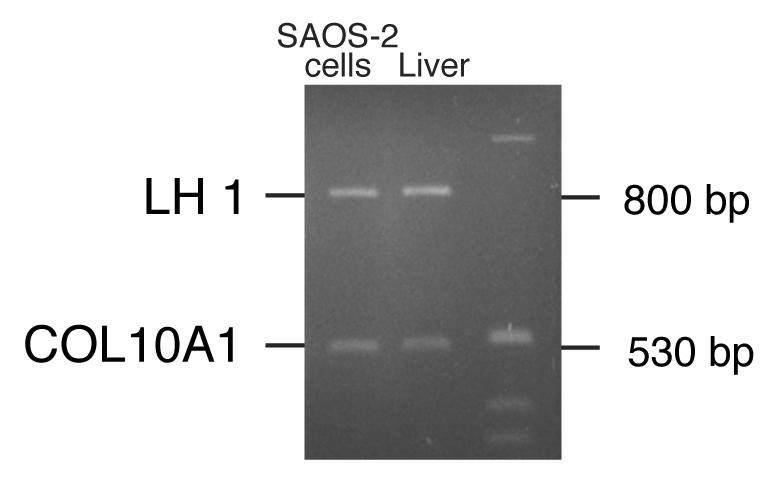
LH1 gene copy number in SAOS-2 cells and human liver cells
A 6% polyacrylamide gel showing multiplex-PCR products of the LH1 gene (PLOD1) and type X collagen gene. The relative abundance of PLOD1 and type X collagen PCR products was similar in SAOS-2 DNA and in normal fetal liver DNA, indicating a normal LH1 copy number in SAOS-2 cells. Thus, the high levels of LH1 mRNA are due to elevated expression and/or stability of these products in SAOS-2 cells.
Discussion
The results of protein analysis show that human SAOS-2 osteosarcoma cells in monolayer culture deposit and cross-link type I and type V collagen as the principal components of the extracellular matrix (Figs. 2, 3), as in human bone [52, 53] and osteosarcoma tumors [21]. No type III collagen was detected. Type III collagen can be a minor component of bone [52], where it is concentrated at soft tissue insertion sites [54]. Shapiro and Eyre found type III collagen in the soft tissue regions of human osteosarcoma [21].
About four-fifths of the synthesized collagen was type I and one-fifth type V. Mass spectrometry after SDS-PAGE and in-gel trypsin digestion of extracted matrix collagen confirmed that the dominant chains were α1(I), α2(I), α1(V) and α2(V). However, the α1(XI) chain of type XI collagen was also identified (Fig. 2B). By sensitive Western blot analysis, α2(XI) chains were previously detected in extracts of SAOS-2 cell layers, but the level was much lower than Coomassie Blue-stainable collagen I and V chains [41]. The α1(XI) and α2(XI) chains are presumably incorporated into trimers, perhaps in the form of hybrid V/XI molecules as suggested by findings on bovine bone matrix by Niyibizi and Eyre (1989) [52]. The lack of any α3(XI) chain (i.e., α1(II)) rules out a typical type XI collagen molecule of composition [α1(XI)][ α2(XI)][ α3(XI)]). Interestingly the COL11A2 gene has been found to be expressed in low amounts by normal mouse osteoblasts [55] and by osteochondrogenic tumors [56]. Together these findings indicate that the collagen phenotype of SAOS-2 cells is similar to that of human bone, but with a higher level of lysyl hydroxylation. Indeed the SAOS-2 cells express abundant osteoblast markers [27] and appear to have a more osteoblast-like phenotype than other human osteosarcoma cell lines [28].
The presence of hydroxylysyl pyridinoline (HP) cross-links and essential absence of the lysyl pyridinoline (LP; Fig. 3) that features in normal bone collagen [3] implies an essentially complete hydroxylation of Lys 87 and Lys 930 in α1(I) and Lys 87 and Lys 933 in α2(I) chains. RNAase protection assays (fig. 6B) and Western blot analysis (Fig. 7) showed higher LH1 expression in SAOS-2 cells compared with normal human skin fibroblasts. This may explain the complete hydroxylation of the cross-linking lysines in SAOS-2 type I collagen. In Ehlers Danlos Syndrome Type VI, the lack of hydroxylation of these lysines is due to absent LH1 activity [14], which in bone collagen results in a low HP:LP ratio [13,17].
Of the tissues surveyed by semi-quantitative RT-PCR (Figs. 4, 5), cartilage had the highest level of LH1 expression relative to G3PDH, but still less than that of SAOS-2 cells (Fig. 5). Both cartilage and SAOS-2 matrix collagens had a similarly high ratio of HP:LP indicating that the helical-domain cross-linking lysines were fully hydroxylated [3].
The findings support a role for LH1 in efficiently hydroxylating cross-linking lysine residues in the triple-helical region of collagen. Although elevated levels of LH1 have not yet been associated with any form of human disease, high levels of hydroxylysine and of glycated-hydroxylysine residues have been found in osteoporotic bone collagen, diabetic bone and skin collagen (reviewed in [57]), and osteosarcoma bone collagen [21,22]. Resorption of bone collagen with a high degree of lysine hydroxylation can explain the high HP:LP ratio in urine of osteosarcoma patients (25,26). Upregulation of LH1 activity in osteosarcoma cells can explain this. SAOS-2 cells may be useful for studying the mechanism of regulation of LH1 gene expression, which is unclear.
Finally, it is worth noting that lysyl oxidase (LOX) was recently implicated in tumor metastasis [58]. This enzyme initiates collagen cross-linking and it has been shown that the mechanical properties of a collagen substratum can modulate the behavior of osteoblasts and osteosarcoma cells in vitro [59]. A stiff matrix directs human mesenchymal stem cells to an osteoblast lineage [60]. It is possible, therefore, that variations in the post-translational chemistry of collagen that alter cross-linking and matrix stiffness can affect the attachment and migration of bone forming cells.
ACKNOWLEDGEMENTS
This work was supported in part by NIH grants AR 37318, AR 36794 and AR052896. We thank Desiree Engel for technical assistance and Kae Ellingsen for help in manuscript preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Eyre DR. Biochemical basis of collagen metabolites as bone turnover markers. In: Raisz L, Rodin G, Bilezikian JP, editors. Principles of Bone Biology. Academic Press; London: 1996. pp. 43–152. [Google Scholar]
- 2.Eyre DR, Wu JJ. Collagen cross-links. Top Curr Chem. 2005;247:207–29. [Google Scholar]
- 3.Eyre DR, Dickson IR, Van Ness K. Collagen cross-linking in human bone and articular cartilage. Age-related changes in the content of mature hydroxypyridinium residues. Biochem J. 1988;252:495–500. doi: 10.1042/bj2520495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kivirikko KI, Myllyla R. Posttranslational enzymes in the biosynthesis of collagen: intracellular enzymes. Methods Enzymol. 1982;82(Pt A):245–304. doi: 10.1016/0076-6879(82)82067-3. [DOI] [PubMed] [Google Scholar]
- 5.Hautala T, Byers MG, Eddy RL, Shows TB, Kivirikko KI, Myllyla R. Cloning of human lysyl hydroxylase: complete cDNA-derived amino acid sequence and assignment of the gene (PLOD) to chromosome 1p36.3----p36.2. Genomics. 1992;13:62–9. doi: 10.1016/0888-7543(92)90202-4. [DOI] [PubMed] [Google Scholar]
- 6.Valtavaara M, Papponen H, Pirttila AM, Hiltunen K, Helander H, Myllyla R. Cloning and characterization of a novel human lysyl hydroxylase isoform highly expressed in pancreas and muscle. J Biol Chem. 1997;272:6831–4. doi: 10.1074/jbc.272.11.6831. [DOI] [PubMed] [Google Scholar]
- 7.Valtavaara M, Szpirer C, Szpirer J, Myllyla R. Primary structure, tissue distribution, and chromosomal localization of a novel isoform of lysyl hydroxylase (lysyl hydroxylase 3) J Biol Chem. 1998;273:12881–6. doi: 10.1074/jbc.273.21.12881. [DOI] [PubMed] [Google Scholar]
- 8.Passoja K, Rautavuoma K, Ala-Kokko L, Kosonen T, Kivirikko KI. Cloning and characterization of a third human lysyl hydroxylase isoform. Proc Natl Acad Sci U S A. 1998;95:10482–6. doi: 10.1073/pnas.95.18.10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeowell HN, Walker LC. Tissue specificity of a new splice form of the human lysyl hydroxylase 2 gene. Matrix Biol. 1999;18:179–87. doi: 10.1016/s0945-053x(99)00013-x. [DOI] [PubMed] [Google Scholar]
- 10.van der Slot AJ, Zuurmond AM, Bardoel AF, Wijmenga C, Pruijs HE, Sillence DO, Brinckmann J, Abraham DJ, Black CM, Verzijl N, DeGroot J, Hanemaaijer R, TeKoppele JM, Huizinga TW, Bank RA. Identification of PLOD2 as telopeptide lysyl hydroxylase, an important enzyme in fibrosis. J Biol Chem. 2003;278:40967–72. doi: 10.1074/jbc.M307380200. [DOI] [PubMed] [Google Scholar]
- 11.Ruotsalainen H, Sipila L, Vapola M, Sormunen R, Salo AM, Uitto L, Mercer DK, Robins SP, Risteli M, Aszodi A, Fassler R, Myllyla R. Glycosylation catalyzed by lysyl hydroxylase 3 is essential for basement membranes. J Cell Sci. 2006;119:625–35. doi: 10.1242/jcs.02780. [DOI] [PubMed] [Google Scholar]
- 12.Heikkinen J, Risteli M, Wang C, Latvala J, Rossi M, Valtavaara M, Myllyla R. Lysyl hydroxylase 3 is a multifunctional protein possessing collagen glucosyltransferase activity. J Biol Chem. 2000;275:36158–63. doi: 10.1074/jbc.M006203200. [DOI] [PubMed] [Google Scholar]
- 13.Walker LC, Overstreet MA, Siddiqui A, De Paepe A, Ceylaner G, Malfait F, Symoens S, Atsawasuwan P, Yamauchi M, Ceylaner S, Bank RA, Yeowell HN. A novel mutation in the lysyl hydroxylase 1 gene causes decreased lysyl hydroxylase activity in an Ehlers-Danlos VIA patient. J Invest Dermatol. 2005;124:914–8. doi: 10.1111/j.0022-202X.2005.23727.x. [DOI] [PubMed] [Google Scholar]
- 14.Walker LC, Marini JC, Grange DK, Filie J, Yeowell HN. A patient with Ehlers-Danlos syndrome type VI is homozygous for a premature termination codon in exon 14 of the lysyl hydroxylase 1 gene. Mol Genet Metab. 1999;67:74–82. doi: 10.1006/mgme.1999.2824. [DOI] [PubMed] [Google Scholar]
- 15.Krane SM, Pinnell SR, Erbe RW. Lysyl-protocollagen hydroxylase deficiency in fibroblasts from siblings with hydroxylysine-deficient collagen. Proc Natl Acad Sci U S A. 1972;69:2899–903. doi: 10.1073/pnas.69.10.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinmann B, Eyre DR, Shao P. Urinary pyridinoline cross-links in Ehlers-Danlos syndrome type VI. Am J Hum Genet. 1995;57:1505–8. [PMC free article] [PubMed] [Google Scholar]
- 17.Eyre D, Shao P, Weis MA, Steinmann B. The kyphoscoliotic type of Ehlers-Danlos syndrome (type VI): differential effects on the hydroxylation of lysine in collagens I and II revealed by analysis of cross-linked telopeptides from urine. Mol Genet Metab. 2002;76:211–6. doi: 10.1016/s1096-7192(02)00036-7. [DOI] [PubMed] [Google Scholar]
- 18.Bank RA, Robins SP, Wijmenga C, Breslau-Siderius LJ, Bardoel AF, van der Sluijs HA, Pruijs HE, TeKoppele JM. Defective collagen crosslinking in bone, but not in ligament or cartilage, in Bruck syndrome: indications for a bone-specific telopeptide lysyl hydroxylase on chromosome 17. Proc Natl Acad Sci U S A. 1999;96:1054–8. doi: 10.1073/pnas.96.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanson DA, Eyre DR. Molecular site specificity of pyridinoline and pyrrole cross-links in type I collagen of human bone. J Biol Chem. 1996;271:26508–16. doi: 10.1074/jbc.271.43.26508. [DOI] [PubMed] [Google Scholar]
- 20.Lehmann HW, Bodo M, Frohn C, Nerlich A, Rimek D, Notbohm H, Muller PK. Lysyl hydroxylation in collagens from hyperplastic callus and embryonic bones. Biochem J. 1992;282(Pt 2):313–8. doi: 10.1042/bj2820313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shapiro FD, Eyre DR. Collagen polymorphism in extracellular matrix of human osteosarcoma. J Natl Cancer Inst. 1982;69:1009–16. [PubMed] [Google Scholar]
- 22.Lehmann HW, Wolf E, Roser K, Bodo M, Delling G, Muller PK. Composition and posttranslational modification of individual collagen chains from osteosarcomas and osteofibrous dysplasias. J Cancer Res Clin Oncol. 1995;121:413–8. doi: 10.1007/BF01212948. [DOI] [PubMed] [Google Scholar]
- 23.Bailey AJ, Wotton SF, Sims TJ, Thompson PW. Post-translational modifications in the collagen of human osteoporotic femoral head. Biochem Biophys Res Commun. 1992;185:801–5. doi: 10.1016/0006-291x(92)91697-o. [DOI] [PubMed] [Google Scholar]
- 24.Glimcher MJ, Shapiro F, Ellis RD, Eyre DR. Changes in tissue morphology and collagen composition during the repair of cortical bone in the adult chicken. J Bone Joint Surg Am. 1980;62:964–73. [PubMed] [Google Scholar]
- 25.Acil Y, Springer I, Behrens P, Ullrich KP, Hedderich J, Bruns J. Differential enhancement of collagen crosslink excretion in cases of osteosarcoma and chondrosarcoma. J Cancer Res Clin Oncol. 2003;129:583–8. doi: 10.1007/s00432-003-0470-6. [DOI] [PubMed] [Google Scholar]
- 26.Behrens P, Bruns J, Ullrich KP, Acil Y, Gille J. Pyridinoline cross-links as markers for primary and secondary bone tumors. Scand J Clin Lab Invest. 2003;63:37–44. [PubMed] [Google Scholar]
- 27.Rodan SB, Imai Y, Thiede MA, Wesolowski G, Thompson D, Bar-Shavit Z, Shull S, Mann K, Rodan GA. Characterization of a human osteosarcoma cell line (Saos-2) with osteoblastic properties. Cancer Res. 1987;47:4961–6. [PubMed] [Google Scholar]
- 28.Pautke C, Schieker M, Tischer T, Kolk A, Neth P, Mutschler W, Milz S. Characterization of osteosarcoma cell lines MG-63, Saos-2 and U-2 OS in comparison to human osteoblasts. Anticancer Res. 2004;24:3743–8. [PubMed] [Google Scholar]
- 29.Murray E, Provvedini D, Curran D, Catherwood B, Sussman H, Manolagas S. Characterization of a human osteoblastic osteosarcoma cell line (SAOS-2) with high bone alkaline phosphatase activity. J Bone Miner Res. 1987;2:231–8. doi: 10.1002/jbmr.5650020310. [DOI] [PubMed] [Google Scholar]
- 30.Barrett MG, Belinsky GS, Tashjian AH., Jr. A new action of parathyroid hormone. receptor-mediated stimulation of extracellular acidification in human osteoblast-like SaOS-2 cells. J Biol Chem. 1997;272:26346–53. doi: 10.1074/jbc.272.42.26346. [DOI] [PubMed] [Google Scholar]
- 31.Farley JR, Hall SL, Herring S, Tarbaux NM, Matsuyama T, Wergedal JE. Skeletal alkaline phosphatase specific activity is an index of the osteoblastic phenotype in subpopulations of the human osteosarcoma cell line SAOS-2. Metabolism. 1991;40:664–71. doi: 10.1016/0026-0495(91)90081-7. [DOI] [PubMed] [Google Scholar]
- 32.Schroder HC, Boreiko O, Krasko A, Reiber A, Schwertner H, Muller WE. Mineralization of SaOS-2 cells on enzymatically (silicatein) modified bioactive osteoblast-stimulating surfaces. J Biomed Mater Res B Appl Biomater. 2005;75:387–92. doi: 10.1002/jbm.b.30322. [DOI] [PubMed] [Google Scholar]
- 33.Fassina L, Visai L, Asti L, Benazzo F, Speziale P, Tanzi MC, Magenes G. Calcified matrix production by SAOS-2 cells inside a polyurethane porous scaffold, using a perfusion bioreactor. Tissue Eng. 2005;11:685–700. doi: 10.1089/ten.2005.11.685. [DOI] [PubMed] [Google Scholar]
- 34.Fedde KN. Human osteosarcoma cells spontaneously release matrix-vesicle-like structures with the capacity to mineralize. Bone Miner. 1992;17:145–51. doi: 10.1016/0169-6009(92)90726-t. [DOI] [PubMed] [Google Scholar]
- 35.McQuillan DJ, Richardson MD, Bateman JF. Matrix deposition by a calcifying human osteogenic sarcoma cell line (SAOS-2) Bone. 1995;16:415–26. doi: 10.1016/8756-3282(95)90186-8. [DOI] [PubMed] [Google Scholar]
- 36.Sevetson B, Taylor S, Pan Y. Cbfa1/RUNX2 directs specific expression of the sclerosteosis gene (SOST) J Biol Chem. 2004;279:13849–58. doi: 10.1074/jbc.M306249200. [DOI] [PubMed] [Google Scholar]
- 37.Bertaux K, Broux O, Chauveau C, Jeanfils J, Devedjian JC. Identification of CBFA1-regulated genes on SaOs-2 cells. J Bone Miner Metab. 2005;23:114–22. doi: 10.1007/s00774-004-0549-4. [DOI] [PubMed] [Google Scholar]
- 38.Cheng YY, Huang L, Lee KM, Li K, Kumta SM. Alendronate regulates cell invasion and MMP-2 secretion in human osteosarcoma cell lines. Pediatr Blood Cancer. 2004;42:410–5. doi: 10.1002/pbc.20019. [DOI] [PubMed] [Google Scholar]
- 39.Raval P, Hsu HH, Anderson HC. Osteoinductive ability of confluent Saos-2 cell correlates with enhanced expression of bone morphogenetic proteins. J Orthop Res. 1996;14:605–10. doi: 10.1002/jor.1100140415. [DOI] [PubMed] [Google Scholar]
- 40.Hunt TR, Schwappach JR, Anderson HC. Healing of a segmental defect in the rat femur with use of an extract from a cultured human osteosarcoma cell-line (Saos-2). A preliminary report. J Bone Joint Surg Am. 1996;78:41–8. doi: 10.2106/00004623-199601000-00006. [DOI] [PubMed] [Google Scholar]
- 41.Goto T, Matsui Y, Fernandes RJ, Hanson DA, Kubo T, Yukata K, Michigami T, Komori T, Fujita T, Yang L, Eyre DR, Yasui N. Sp1 Family of Transcription Factors Regulates the Human alpha2 (XI) Collagen Gene (COL11A2) in Saos-2 Osteoblastic Cells. J Bone Miner Res. 2006;21:661–73. doi: 10.1359/jbmr.020605. [DOI] [PubMed] [Google Scholar]
- 42.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 43.Hauselmann HJ, Fernandes RJ, Mok SS, Schmid TM, Block JA, Aydelotte MB, Kuettner KE, Thonar EJ. Phenotypic stability of bovine articular chondrocytes after long-term culture in alginate beads. J Cell Sci. 1994;107(Pt 1):17–27. doi: 10.1242/jcs.107.1.17. [DOI] [PubMed] [Google Scholar]
- 44.Eyre DR, Koob TJ, Van Ness KP. Quantitation of hydroxypyridinium crosslinks in collagen by high-performance liquid chromatography. Anal Biochem. 1984;137:380–8. doi: 10.1016/0003-2697(84)90101-5. [DOI] [PubMed] [Google Scholar]
- 45.Fernandes RJ, Schmid TM, Eyre DR. Assembly of collagen types II, IX and XI into nascent hetero-fibrils by a rat chondrocyte cell line. Eur. J. Biochem. 2003;270:3243–3250. doi: 10.1046/j.1432-1033.2003.03711.x. [DOI] [PubMed] [Google Scholar]
- 46.Stegemann H. Microdetermination of hydroxyproline with chloramine-T and p-dimethylaminobenzaldehyde. Hoppe Seylers Z Physiol Chem. 1958;311:41–5. [PubMed] [Google Scholar]
- 47.Fernandes RJ, Schmid TM, Harkey MA, Eyre DR. Incomplete processing of type II procollagen by a rat chondrosarcoma cell line. Eur J Biochem. 1997;247:620–4. doi: 10.1111/j.1432-1033.1997.00620.x. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 49.Robbins JR, Thomas B, Tan L, Choy B, Arbiser JL, Berenbaum F, Goldring MB. Immortalized human adult articular chondrocytes maintain cartilage-specific phenotype and responses to interleukin-1beta. Arthritis Rheum. 2000;43:2189–201. doi: 10.1002/1529-0131(200010)43:10<2189::AID-ANR6>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 50.Bolton MC, Dudhia J, Bayliss MT. Quantification of aggrecan and link-protein mRNA in human articular cartilage of different ages by competitive reverse transcriptase-PCR. Biochem J. 1996;319(Pt 2):489–98. doi: 10.1042/bj3190489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grandinetti KB, Spengler BA, Priedler JL, Ross RA. Loss of one HuD allele on chromosome #1p selects for amplification of the N-myc protooncogene in human neuroblastoma cells. Oncogene. 2006;25:706–712. doi: 10.1038/sj.onc.1209095. [DOI] [PubMed] [Google Scholar]
- 52.Niyibizi C, Eyre DR. Identification of the cartilage alpha 1(XI) chain in type V collagen from bovine bone. FEBS Lett. 1989;242:314–8. doi: 10.1016/0014-5793(89)80492-2. [DOI] [PubMed] [Google Scholar]
- 53.Niyibizi C, Eyre DR. Bone type V collagen: chain composition and location of a trypsin cleavage site. Connect Tissue Res. 1989;20:247–50. doi: 10.3109/03008208909023894. [DOI] [PubMed] [Google Scholar]
- 54.Keene DR, Sakai LY, Burgeson RE. Human bone contains type III collagen, type VI collagen, and fibrillin: type III collagen is present on specific fibers that may mediate attachment of tendons, ligaments, and periosteum to calcified bone cortex. J Histochem Cytochem. 1991;39:59–69. doi: 10.1177/39.1.1983874. [DOI] [PubMed] [Google Scholar]
- 55.Sugimoto M, Kimura T, Tsumaki N, Matsui Y, Nakata K, Kawahata H, Yasui N, Kitamura Y, Nomura S, Ochi T. Differential in situ expression of alpha2(XI) collagen mRNA isoforms in the developing mouse. Cell Tissue Res. 1998;292:325–32. doi: 10.1007/s004410051063. [DOI] [PubMed] [Google Scholar]
- 56.Matsui Y, Kimura T, Tsumaki N, Nakata K, Yasui N, Araki N, Hashimoto N, Uchida A, Ochi T. Splicing patterns of type XI collagen transcripts act as molecular markers for osteochondrogenic tumors. Cancer Lett. 1998;124:143–8. doi: 10.1016/s0304-3835(97)00468-0. [DOI] [PubMed] [Google Scholar]
- 57.Dominguez LJ, Barbagallo M, Moro L. Collagen overglycosylation: a biochemical feature that may contribute to bone quality. Biochem Biophys Res Commun. 2005;330:1–4. doi: 10.1016/j.bbrc.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 58.Erler JT, Bennewith KL, Nicolau M, Dornhofer N, Kong C, Le QT, Chi JT, Jeffrey SS, Giaccia AJ. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–6. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 59.Masi L, Franchi A, Santucci M, Danielli D, Arganini L, Giannone V, Formigli L, Benvenuti S, Tanini A, Beghe F, et al. Adhesion, growth, and matrix production by osteoblasts on collagen substrata. Calcif Tissue Int. 1992;51:202–12. doi: 10.1007/BF00334548. [DOI] [PubMed] [Google Scholar]
- 60.Engler AJ, Sen S, Sweeny HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]