Abstract
BAC libraries generated from restriction-digested genomic DNA display representational bias and lack some sequences. To facilitate completion of genome projects, procedures have been developed to create BACs from DNA physically sheared to create fragments extending up to 200 kb. The DNA fragments were repaired to create blunt ends and ligated to a new BAC vector. This approach has been tested by generating BAC libraries from Drosophila DNA, with average insert lengths between 50 – 150 kb. The libraries lack chimeric clone problems as determined by mapping paired BAC-end sequences to the assembled fly genome sequence. The utility of “sheared” libraries was demonstrated by closure of a previous clone gap and by isolation of clones from telomeric regions, which were notably absent from previous Drosophila BAC libraries.
Keywords: bacterial artificial chromosome, BAC, sheared DNA, cloning, vector, adaptor, telomere, centromere and heterochromatin
Introduction
Bacterial artificial chromosome (BAC) libraries have played a crucial role in many genome mapping and sequencing projects [1–7]. All previous BAC libraries have been constructed from DNA partially-digested with restriction enzymes, such as EcoRI, MboI and HindIII [5, 8–11]. In most mapped and sequenced genomes, a number of regions remain unresolved due to an absence of BAC coverage, in spite of considerable effort. BAC-based physical maps contain persistent clone gaps, and maps created by alignment of BAC-end sequences suffer from gaps and occasional genome assembly errors. Such errors often result from difficulties in assembling regions containing complex repetitive sequence blocks. In addition, genome sequence assemblies derived by the whole genome shotgun (WGS) approach tend to have sequence collapses in regions containing duplicons [6]. Finally, some genomic sequences can not be recovered through conventional BAC cloning presumably due to instability in E. coli, non-uniform distribution of restriction sites in the affected regions, or potential toxicity of the cloned DNA. It has been impossible to clone high AT (>70%) content DNA as large inserts in E. coli, as was observed for Plasmodium falciparum [12] and Dictyostelium discoideum [13]. Telomeric and/or subtelomeric regions are underrepresented in most BAC libraries most likely due to problematic patterns of restriction sites [14, 15]. Obviously the ends of chromosomes are inaccessible to cloning by partial restriction strategies due to the absence of restriction sites in telomeric repeat sequences. To circumvent this problem, half-YAC clones have been created from the human genome to isolate telomeric regions, but at considerable expense [14, 16]. To reduce the cloning bias generated by restriction digestion, we developed procedures to clone large fragments from physically sheared genomic DNA. Four BAC libraries were constructed, two for Drosophila melanogaster, one for D. pseudoobscura and one for human. One of the D. melanogaster BAC libraries has been characterized extensively by mapping end-sequences to the assembled genome sequence. The usefulness of these D. melanogaster libraries has been demonstrated by preparing physical maps of the telomeric regions [17, 18] and by closing a gap in the genome assembly of the X chromosome.
Results
Development of cloning procedures
Our overall goal was to construct BAC libraries with a more random representation of the genome than those generated through partial genomic restriction digestion and hopefully suitable for closing gaps in existing physical maps. To this end, a new cloning vector was developed which enables cloning of blunt-ended DNA fragments. High-molecular-weight (HMW) DNA embedded in agarose blocks was sheared through repeated freezing and thawing. To increase the fraction of blunt-ended DNA fragments, the agarose blocks were incubated consecutively with Mung Bean nuclease and T4 DNA polymerase, or only with T4 DNA polymerase. The “polished” DNA was then isolated from the agarose blocks by electro-elution. Initial attempts to clone these fragments directly into a blunt-ended vector resulted in very low cloning efficiencies, at least 50-fold lower as compared with results from partial restriction-digest fragments. Moreover, the recombinant clones were a minor fraction, with most clones lacking inserts. The low efficiency combined with high levels of non-insert clones made it impractical to create BAC libraries in this way (data not shown). To suppress the non-insert background problem, the blunt ended DNA was ligated to an adaptor to create 5′ protruding, non-complementary (CACA) ends (Figure 1). The ligation products were then size-fractionated by pulsed-field gel electrophoresis permitting the simultaneous removal of excess free adaptor and isolation of the sized genomic fragments. The size-fractionated DNA retains 5′extending ends which can be ligated to the complementary 5′(TGTG) ends of the BstXI-digested pTARBAC6 cloning vector (Figure 1).
Figure 1.
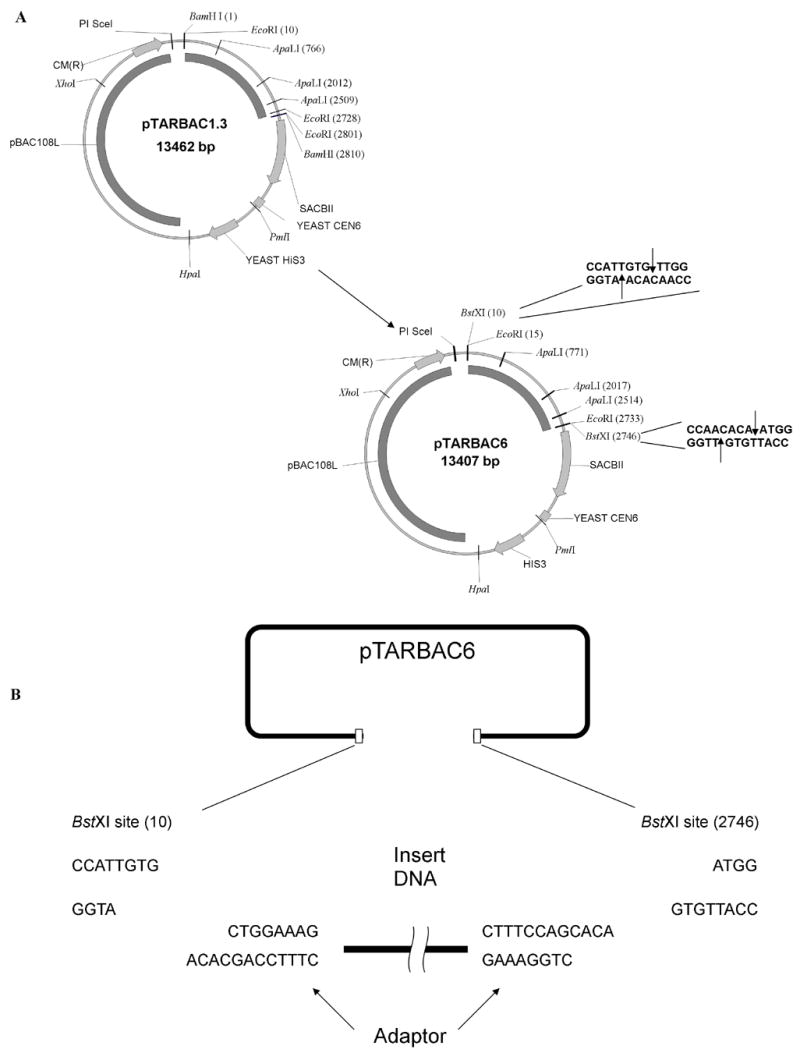
A: Construction of pTARBAC6 vector: The pTARBAC6 vector was generated from pTARBAC1.3 by replacing the two BamHI-EcoRI fragments with synthetic adapters to regenerate the BstXI sites while eliminating the BamHI sites. The restriction enzyme names followed by the numbers in the parenthesis indicate the base position of each restriction enzyme cutting site. The sequences of two BstXI sites are shown and their cutting sites are indicated by the arrows. B: Cloning blunt-ended sheared DNA into vector: Digestion of pTARBAC6 vector generates two cohesive ends (TGTG) which are not complementary strand each other, thus avoiding self-ligation of the vector. Blunt-ended sheared DNA is ligated with an adaptor which has blunt end and protruding end (CACA). The DNA with the adaptor is compatible with ligation with the cohesive ends of the vector.
Construction of a new cloning vector
BstXI restriction sites were removed from the pTARBAC1 vector resulting in the intermediate pTARBAC1.3 vector [11]. Two new BstXI sites were inserted at positions flanking a stuffer fragment to be removed when creating a BAC library. A small EcoRI (2,728)–EcoRI (2801) fragment in the pTARBAC1.3 vector was removed when the two BstXI sites were inserted (Figure 1). BstXI recognizes a 6 base-pair palindrome interrupted by an arbitrary six base-pair sequence (CCANNNNNNTGG). The newly inserted BstXI sites contain the same arbitrary sequence (TTGTGT) but are placed within the pTARBAC6 vector in inverted orientation (Figure 1). Digestion of the vector with BstXI results in two fragments: a 10.6 kb vector replicon and a 2.7 kb removable stuffer fragment. The two 5′ ends (TGTG) of the cloning vector are not complementary to each other, which suppresses vector self-ligation [19, 20], but are complementary to the adapter-ends ligated to the sheared DNA fragments.
Construction of BAC libraries
Two Drosophila melanogaster BAC libraries designated as CHORI-221 and CHORI-223 were constructed from sheared DNA using the pTARBAC6 vector. Each library is composed of 18,000 clones arrayed into 384-well microtiter dishes (Table 1). We hypothesize that some cloning gaps in previous BAC libraries [RPCI-98: [5]] might result from the combined instability of multiple elements, which perhaps can be propagated separately using smaller insert clones. In addition, stable sequences adjacent to unclonable elements will be under-represented in large insert libraries. The CHORI-223 library was therefore constructed with a ~50 kb average insert size as compared to 115 kb for the CHORI-221 library. Two BAC libraries designated as CHORI-222 and CHORI-16 were constructed from sheared D. pseudoobscura and sheared human DNA (Table 1) and are composed of 8,200 and 151,000 arrayed clones, respectively.
Table 1. Summary of fly and human BAC libraries.
| Libraries | RPCI-11 | CHORI-16 | RPCI-98 | CHORI-221 | CHORI-222 | CHORI-223 |
|---|---|---|---|---|---|---|
| Cloning vector | pBACe3.6 | pTARBAC6 | pBACe3.6 | pTARBAC6 | pTARBAC6 | pTARBAC6 |
| DNA source | White blood cells | White blood cells | Adult fly | Adult fly | Adult fly | Adult fly |
| Methods | EcoRI | sheared | EcoRI | sheared | sheared | sheared |
| Empty wells (%) | 5,334 (1.2%) | 3,340 (2.1%) | 243 (1.3%) | 360 (1.9%) | 443 (4.8%) | 258 (1.4%) |
| Non-insert clones (%) | 9/1,028 (0.9%) | 20/415 (4.8%) | 9/272 (3.3%) | 16/157 (10%) | 6/96 (6.3%) | 2/192 (1.0%) |
| Recombinant clones | ~437,415 | ~ 151,058 | ~17,588 | ~16,265 | ~8,218 | ~17,992 |
| Insert size | 175 kb | 84 kb | 165 kb | 115 kb | 152 kb | 48 kb |
| Standard deviation | 34 kb | 36 kb | 33 kb | 33 kb | 52 kb | 17 kb |
| Library redundancy | 23.1-fold | 4.1-fold | 22-fold | 15.6-fold | 10.4-fold | 7.2-fold |
| cfu per μg | 131,200 | 30,700 | 66,900 | 18,800 | 147,300 | 217,100 |
Fly and human BAC libraries have been constructed using EcoRI partially digested DNA 5,9 and sheared DNA. The “Methods” row indicates whether the libraries were constructed by using EcoRI partially digested DNA or sheared DNA. Empty wells were manually scored for each plate. The “Libraries” row indicates the official library nomenclature for clone registry in public databases. The “Recombinant clones” row represents total wells after empty wells and estimated total non-insert clones were subtracted in each segment. The “Insert size” row shows the average insert size of each library. The “cfu per μg” indicates colony forming unit per μg of size-fractionated insert DNA corresponding to each library. The cloning efficiencies for the RPCI-98 and RPCI-11 libraries were not described previously, but calculated for comparison in this study. The library redundancies were estimated using 120 Mb and 3,200 Mb as the Drosophila and human genome sizes, respectively.
Average insert sizes
BAC insert sizes were sampled for the four libraries by pulsed-field gel electrophoresis. The average insert sizes were 115 kb, 152 kb, 48 kb and 84 kb with standard deviations of 33 kb, 52 kb, 17 kb and 36 kb for the CHORI-221, CHORI-222, CHORI-223 and CHORI-16 libraries, respectively. The insert size distributions for the libraries are shown in Figure 2. The fractions of non-insert clones are estimated to be 10% (16/157), 6.3% (6/96), 1.0% (2/192) and 4.8% (20/415) for the CHORI-221, CHORI-222 segment 2, CHORI-223 and CHORI-16 libraries, respectively (Table 1).
Figure 2.
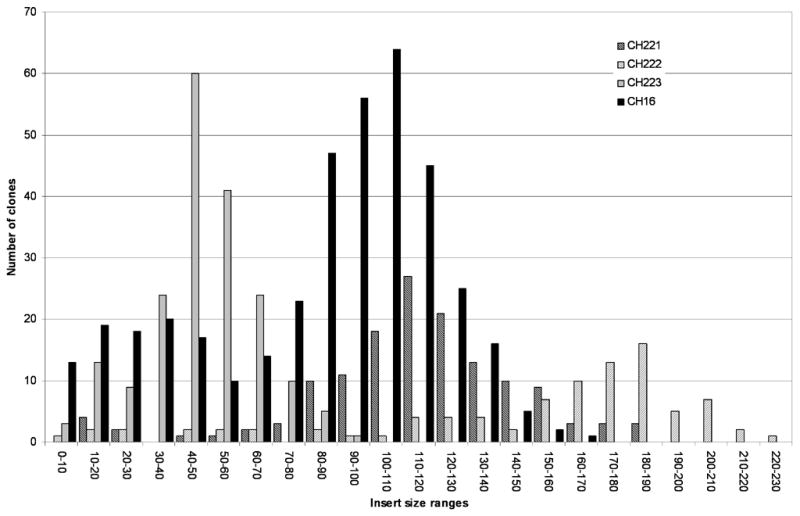
Insert size distribution of the CHORI-221, CHORI-222, CHORI-223 and CHORI-16 libraries. The horizontal axis is divided into 23 10-kb resolution bins ranging from 0–230 kb. The vertical axis presents the number of clones in each insert size range. Gray background with black diagonal stripe, white background with gray diagonal stripe, gray and black bars represent insert sizes of BAC clones derived from CHORI-221, CHORI-222, CHORI-223 and CHORI-16 libraries, respectively.
Efficiency of constructing sheared DNA BAC libraries
BAC cloning efficiency is defined by the number of independently-transformed cells, also known as colony-forming units (cfu), generated per microgram of size-fractionated DNA. Using conventional BAC cloning from partially restriction-digested genomic DNA, a sharp decrease in the cloning efficiency is always observed with increasing insert size. For instance, cloning from the 130 kb DNA size-fraction is at least 5-fold higher than from the 170-kb fraction (unpublished observations). We observed a similar decrease in cloning efficiency, inversely proportional to DNA size, when we cloned sheared genomic DNA. It is important to consider the relative cloning efficiency from sheared DNA versus partially-digested DNA. The results of comparison of cloning efficiencies by the different methods are presented in Table 1, for five BAC libraries. We have constructed fly (CHORI-221 and CHORI-223) and human (CHORI-16) BAC libraries from sheared DNA. We have also constructed many more BAC libraries from EcoRI-partially digested DNA, for instance the fly (RPCI-98) and human (RPCI-11) libraries [5, 9]. The efficiency for constructing the CHORI-221 library was 3.6-fold lower than that of the RPCI-98 library even though the average insert size of CHORI-221 is 50 kb less than that of RPCI-98. Similarly the efficiency for creating CHORI-16 was 4.2-fold lower than that of RPCI-11, while the average insert size of CHORI-16 is 95-kb less than that of RPCI-11. The high cloning efficiency for CHORI-223 was mainly due to the small average size of sheared fragments (48 kb). We were able to generate the CHORI-222 library with 152 kb average insert size from sheared DNA at very high cloning efficiency, but were never able to reproduce this for the other libraries.
Mapping BAC ends
Cloning artifacts which combine unrelated genomic fragments in a single derivative clone can be a major impediment for genome mapping and sequencing if they occur at significant levels. Chimeric clone levels in Yeast Artificial Chromosome (YAC) libraries were always high with chimeric clones representing at least 10% of the total. By contrast, BAC libraries prepared by partial restriction-digest cloning have a relatively low numbers of chimeras: levels of 1% or lower have been reported for the human, mouse and rat BAC libraries used in support of genome projects [9–11]. To ascertain the levels of such cloning artifacts resulting from the blunt-end cloning strategy, paired “CHORI-221” BAC end sequences were mapped onto Release 3 of the D. melanogaster genome sequence assembly [7]. A small number of BAC end sequences containing highly repetitive sequences were aligned onto Release 5 D. melanogaster genome sequence assembly (http://www.fruitfly.org/sequence/release5genomic.shtml). High quality sequences were obtained from raw sequence trace files for 288 BACs by trimming vector-, adapter- and low quality 3′-sequences. A total of 239 BACs had high quality sequences at both ends. These BAC-end sequences were mapped by BLASTN to the fly genome sequence. A total of 227 clones (95%) map to unique locations and display a virtual insert size compatible with the properties of the BAC library. Nine BACs could not be used to test chimera levels because at least one end lacked unique sequence or lacked a corresponding BLASTN hit in the Release 3 and 5 fly genome sequence. These BAC ends are probably representing heterochromatic regions. The remaining three clones had unmapped heterochromatic sequences at both ends. No chimeric BACs were identified in this analysis. To exhibit the distribution of BACs from CHORI-221 and RPCI-98 libraries, the number of mapped BACs to all chromosome arms is summarized in Table 2.
Table 2. Summary of clone distribution on each chromosome arm.
| CHORI-221 library | RPCI-98 library | ||||
|---|---|---|---|---|---|
| Chromosomes | % (converted size) of each chromosome | Number of clones observed | Number of clones expected | Number of clones observed | Number of clones expected |
| X (21.9 Mb) | 14.7% (16.4 Mb) | 43 | 33 | 1,136 | 1,229 |
| 3L (23.4 Mb) | 21.0% (23.4 Mb) | 53 | 48 | 1,825 | 1,750 |
| 3R (27.9 Mb) | 25% (27.9 Mb) | 67 | 57 | 2,095 | 2,087 |
| 2L (22.2 Mb) | 19.9% (22.2 Mb) | 25 | 45 | 1,699 | 1,661 |
| 2R (20.3 Mb) | 18.2% (20.3 Mb) | 36 | 41 | 1,562 | 1,519 |
| 4 (1.2 Mb) | 1.1% (1.2 Mb) | 3 | 2 | 18 | 90 |
| Heterochromatin | 3 | ||||
| No unique match | 9 | ||||
This table summarizes the number of clones mapped and expected on each chromosome arm from CHORI-221 and RPCI-98 libraries, respectively. The RPCI-98 clone mapping information was obtained from a publicly available database. A total of two hundred twenty-seven and 8,335 clones from CHORI-221 and RPCI-98 libraries, respectively, were uniquely mapped within the expected distance on the same chromosome arms. The percentage of X chromosome was calculated based on the fact that the X chromosome should be represented at 75% with respect to the autosomes, because the genomic DNA was prepared from a 50/50 mixed population of adult males and females. The expected numbers of clones on each chromosome were calculated by the following formula: total number of uniquely mapped clones (227 for CH221 library and 8,335 for RP98) × percentage of each chromosome. The row “Heterochromatin” represents the number of clones for which both end sequences were aligned to heterochromatic sequences. Both end sequences of 9 clones contained repetitive sequences, thus it was not possible to localize them to a specific chromosome region. These clones are summarized in the row “No unique match”.
Screening D. melanogaster BAC libraries
Most eukaryotic genomes contain regions of highly repetitive sequences, designated as heterochromatin, that are mainly clustered in centromeric and telomeric regions. Most genome analyses have been focused on the euchromatic portion of the genomes [5] where most of the genes are found. The repetitive nature of the heterochromatic regions has made mapping and sequencing difficult. Heterochromatic regions are often under-represented in BAC libraries constructed by the partial restriction-digest approach. For instance, we previously reported under-representation of α-satellite sequences with the 340 bp EcoRI repeat in the human BAC library, presumably due to the abundance of EcoRI sites and the use of EcoRI for the cloning strategy [9]. To examine if sheared libraries affect the efficiency of cloning heterochromatic regions, three fly libraries were compared: the RPCI-98 BAC library generated by cloning EcoRI restriction fragments and the CHORI-221 and CHORI-223 BAC libraries generated from sheared DNA. To this end, the libraries were initially screened with the 359 satellite, a major variant of the D. melanogaster 1.688 satellite DNA family. This DNA sequence consists of a tandemly-repeated 359 bp monomer lacking EcoRI sites. The 359 satellite expands across the centromere of the X chromosome and has been estimated to account for about 11 Mb (6.1%) of the D. melanogaster genome [21, 22]. The three libraries each contain 18,000 clones with the genomic redundancies estimated to be 22-, 15.6- and 7.2-fold, respectively, by taking the average insert size of the clones into account. Only six hybridizing clones were found in the RPCI-98 library as compared to 26 and 218 hybridizing clones in the CHORI-221 and CHORI-223 libraries, respectively (Table 3). This indicates improved cloning efficiency of genomic regions containing this satellite sequence following the new strategy. Nevertheless, the fraction of positive clones in the “sheared” libraries was much less than the 6.1% fraction of the genome corresponding to the 1.688 satellite, indicating that additional factors creating cloning bias against these heterochromatic regions exist. Further hybridizations were performed using seven additional heterochromatin repeat probes (Table 3). In principal, larger insert clones would be more useful for construction of physical maps. It is remarkable that a larger fraction of positive clones was detected in CHORI-223 (48 kb average insert size) as compared to CHORI-221 (115 kb average insert size) for all probes except Mst77F. The aberrant results with the Mst77F probe may relate to its distribution in the genome. It is not exclusively derived from heterochromatic regions, and hybridizes to a euchromatic gene on chromosome 3 and to heterochromatic pseudogenes on the Y chromosome [23]. Three major dodeca satellite blocks, two of which are larger than 200 kb in size, have been found in the centromeric region (h53) of the D. melanogaster chromosome 3 [24]. Mapping BACs in the h53 region revealed that none of the 110 hybridization positive clones from the RPCI-98 library (Table 3) was localized onto the two large dodeca satellite blocks, indicating all the positive clones are derived from regions containing small dodeca satellite blocks. By contrast, several sheared DNA clones from CHORI-221 and CHORI-223 are mapping to the large dodeca satellite blocks (Villasante et at, unpublished results). It is therefore important to note that the number of hybridization positive clones using repeat probes may not reflect the true representation of the library.
Table 3. Comparison of number of hybridization positive clones.
| Probes | RPCI-98 | CHORI-221 | CHORI-223 |
|---|---|---|---|
| 359 satellite | 6 | 26 | 218 |
| 356 satellite | 21 | 11 | 79 |
| 353 satellite | 25 | 6 | 148 |
| Dodecasatellite | 110 | 21 | 70 |
| 18HT satellite | 4 | 1 | 5 |
| Circe | 296 | 68 | 219 |
| Doc2 | 324 | 57 | 160 |
| Mst77F | 28 | 23 | 14 |
The satellite sequences derive from different heterochromatic regions (chromosomes X, Y and 3). Circe and Doc2 are retrotransposons found only in heterochromatin. All probes derive from the heterochromatin. The Mst77F probe however hybridizes with the a euchromatic gene on chromosome 3 and with heterochromatic pseudogenes on the Y chromosome 23.
Extension of contigs toward telomere and closing gaps
The most distal regions of the telomeres of D. melanogaster chromosomes had not previously been cloned and mapped, reflecting deficiencies of the earlier BAC libraries [5, 25]. Physical maps of telomeric regions of each chromosome arm have now been prepared using the new BAC libraries (CHORI-221 and CHORI-223). The physical maps of chromosome arms XL, 2L, 2R, 3L, 3R and 4R were extended by ~270 kb, ~45 kb, ~90 kb, ~35 kb, ~20 kb and ~35 kb, respectively, from the most distal clones obtained from earlier libraries and have permitted the detailed structural analysis of the telomeric regions of D. melanogaster [18]. A persistent clone gap (~45 kb) near the tip of X chromosome has been reported between the genomic sequences with GeneBank accession numbers of AABU01002701 and AE003417 [25]. No clones spanning this gap could be found in three libraries generated from partial genomic digest fragments created with EcoRI (RPCI-98), NdeII and HindIII (the European Drosophila Genome Project “DrosBAC” BACN and BACH libraries), respectively. The end of the most distal clone (RP98-37P7) contained several copies of the 1.688 satellite repeat (Figure 3). A more distal sequence contig derived from small-insert clones was available but was separated from the most distal BAC through a gap in clone coverage. When the CHORI-221 library was screened with a probe isolated from RP98-37P7, nine clones were found that span the gap (Figure 3). The results described above demonstrate the utility of these libraries for construction of physical maps from highly repetitive regions.
Figure 3.
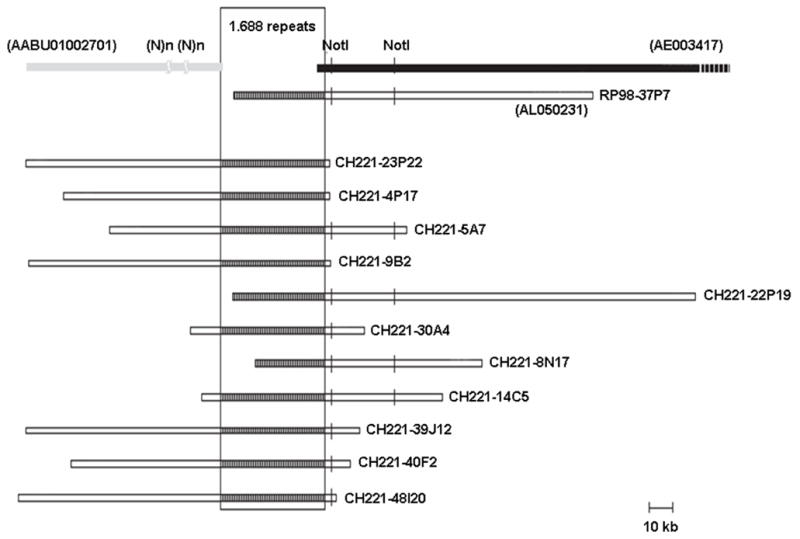
A schematic representation of redundant BAC clones derived from CHORI-221 and covering a gap between sequence accession numbers AABU01002701 and AE003417at the left end of the X chromosome (cytological band 1A1): The region containing 1.688 repeats is shown with a rectangle. A total of 11 clones were identified using a probe isolated from the RP98-37P7 BAC.
Discussion
We present here procedures enabling the construction of BAC libraries derived from sheared DNA. A new BAC vector, pTARBAC6, was constructed to clone sheared DNA following adaptor ligation. Three Drosophila BAC libraries have been constructed, and characterized by BAC-end sequence mapping, screening with heterochromatic probes and probes flanking a prior cloning gap. No chimeric clones were found in a sample of 227 BAC clones with unique end-sequence pairs (Table 2). An increased fraction of clones containing highly repetitive sequences could be identified in the libraries generated from sheared DNA (Table 3). Interestingly, the smaller insert library presented better representation in terms of number of hybridization positive clones using various probes derived from heterochromatic regions. It is not clear why the smaller insert library had more positive clones for these probes, but we speculate that it may be due to cloning of relatively short regions of heterochromatic DNA situated between highly repetitive or toxic regions. The sheared DNA libraries have provided clones for regions with highly repetitive sequences and for sub-telomeric regions [7, 17, 18, 26]. Although we have not shown that sheared DNA BACs are more randomly distributed than BACs generated by restriction-digest (Table 2), we demonstrated that the sheared DNA libraries have a different and complementary distribution with better representation of problematic and repeat-rich regions at telomeres (Figure 3) and in centromeric heterochromatin. The D. melanogaster “sheared DNA” libraries have permitted the characterization of previously unmapped regions of the fly genome. Construction of BAC libraries from sheared DNA is now feasible but may need careful consideration for highly complex genomes similar to the human genome. This deliberation relates to the logistics of making a BAC library from a typical mammalian genome of 3 billion base pairs. The current cloning efficiency of size-fractionated sheared DNA was about 30,000 colony-forming units per microgram for the human CHORI-16 BAC library. At the observed 80 kb insert size, we were able to generate a BAC library representing about 4.1-fold coverage of the human genome at great effort and expense. One would need to create about 375,000 BAC clones from 12 micrograms of size purified DNA. In practice, this corresponds to 24 successful preparative pulsed-field gels to purify the DNA size fraction and 2,000 transformations. Moreover, with one notable exception for the CHORI-222 D. pseudoobscura BAC library, we have never been able to generate BACs with insert sizes in the range of 150 kb or larger from sheared DNA. Nevertheless, the high costs of creating BACs from blunt-ended fragments may be justified for genomes which will be mapped and sequenced at a high-quality “finished” level including the centromeric and telomeric heterochromatin regions. For instance, nearly all human telomeric sequences were previously isolated from a dedicated half-YAC library at high cost [14, 16]. To demonstrate the usefulness of cloning sheared DNA, we have recently created a BAC library from mouse genomic DNA and successfully isolated most of telomeric and sub-telomeric sequences (unpublished results).
Materials and Methods
Construction and preparation of BAC vectors
The pTARBAC1.3 vector was constructed from the pTARBAC1 vector [27] by eliminating two ApaLI and one BstXI sites by cross-over PCR [11]. The pTARBAC1.3 vector was digested with BamHI and EcoRI, and separated in a 0.7 % agarose gel. The BamHI-BamHI (10.6 kb) and EcoRI-EcoRI (2.8 kb) fragments were recovered from the gel and ligated to a partial-duplex linker (BglII-BstXI-EcoRI) composed of overlapping oligonucleotides (GATCTCCATTGTGTTGGG; AATTCCCAACACAATGGA). This resulted in the pTARBAC6 vector (Figure 1). Approximately 12.5 μg pTARBAC6 vector DNA was digested with 10 units of ApaLI (New England Biolabs) in 500 μl reaction mixture [50 mM potassium acetate, 20 mM Tris-acetate (pH7.9), 10 mM magnesium acetate, 1 mM dithiothreitol, 100 μg/ml BSA] at 37°C for 15 min. Subsequently 3 units of calf intestinal alkaline phosphatase (Roche) was added into the reaction mixture, and the incubation was continued at 37°C for another 1 hr. The DNA was extracted with 500 μl phenol:chloroform:isoamyl alcohol (25:24:1), then with 500 μl chloroform and precipitated with 500 μl isopropanol after mixing of 50 μl 3 M sodium acetate (pH 5.2) and 1 μl 20 mg/ml glycogen (Roche). The pellet was rinsed with 500 μl 70% ethanol twice and dried in a laminar hood for 15 minutes. To prepare for cloning, all recovered vector DNA (10–12 μg) was digested with 10 units of BstXI (New England Biolabs) at 55°C for 1 hr. The BstXI-digested DNA was loaded in a large preparative well in 0.7% agarose gel (25-cm long and 15-cm wide) and run at 3V/cm for 16 hr. The vector fragment (10.6 kb) was sliced from the gel and recovered by electrodialysis [28].
Preparation of high-molecular-weight DNA
High-molecular-weight DNA from mixed-sex adult flies was prepared in agarose blocks (InCert agarose: FMC) as previously described [5]. The D. melanogaster BAC libraries CHORI-221 and CHORI-223 were prepared from an isogenic y1; cn1 bw1 sp1 strain [29]. The D. pseudoobscura BAC library CHORI-222 was prepared from the strain Tucson 14011-0121.4 which was obtained from the Drosophila Species Stock Center, Tuscon, Arizona (http://stockcenter.arl.arizona.edu/). Preparation of agarose embedded DNA from anonymous human blood samples has been described [9].
Construction of a sheared DNA library
Four agarose blocks (~100 μg DNA) are each cut into 4 small pieces. The agarose blocks are transferred into a 15-ml conical screw-cap polypropylene tube (Corning 430790) and frozen & thawed four times by cycling between on dry ice for 3 minutes and room temperature for 10 minutes. The agarose-embedded DNA is treated with Mung Bean nuclease (40 units, New England Biolabs) in the commercially supplied buffer [50 mM sodium acetate (pH5.0), 30 mM NaCl, 1 mM ZnSO4] in 1.5 ml reaction mixture at 30°C for 2 hrs. The enzyme is inactivated by adding 3 μl of 20% SDS in the reaction mixture and maintaining the mixture at room temperature for 30 min. The agarose blocks are transferred into dialysis tubing (3/4 in. diameter, molecular weight exclusion limit of 12,000–14,000 daltons; Invitrogen). DNA is electro-eluted into 300 μl sterile 0.5 × TBE buffer at 3 V/cm for 3 hr [28, 30]. The DNA is dialyzed in TE [10 mM Tris-HCl (pH8.0), 1 mM EDTA] buffer at 4°C overnight. The DNA solution and agarose pieces are recovered and treated with 6 units of T4 DNA polymerase (New England Biolabs) in a 500 μl reaction mixture at 16°C for 1 hr. The reaction is stopped by adding 10 μl 0.5 M EDTA and 10 μl 10 mg/ml Proteinase K and incubating at 37°C for 1 hr. Proteinase K is inactivated by adding 10 μl 100 mM PMSF and incubating at room temperature for 1 hr. The solution is dialyzed against water on a floating dialysis membrane (47-mm diameter, 0.025-μm pore-size microdialysis filters: Millipore) for 3 hr. Alternatively, after freeze and thaw cycles the agarose embedded DNA is kept on ice with 105 units of T4 DNA polymerase (New England Biolabs) in 500 μl reaction mixture [50 mM NaCl, 10 mM Tris-HCl (pH7.9), 10 mM MgCl2, 1 mM dithithreitol, 50 μg/ml BSA, 100 μM of each dNTP] and incubated at 16°C for 20 min. The reaction buffer is removed and then the T4 DNA polymerase is inactivated by 300 μg proteinase K (Roche) in 1.5 ml TE50 [10 mM Tris-HCl (pH8.0), 50 mM EDTA] buffer at 37°C for 1 hr. Proteinase K is inactivated with 1.5 ml 2 mM PMSF in TE50 buffer at room temperature for 1 hr. DNA is electroeluted in the same manner. Lyophilized BstXI adaptor (18 μg) consisting of 5′-phosphorylated CTGGAAAG and CTTTCCAGCACA (Invitrogen Cat. No.N-408-18) is dissolved in 100 μl water at a concentration of 28 μM, stored at −20°C and thawed on ice prior to use. The dialysed genomic DNA (~750 μl) is transferred into a microcentrifuge tube and the DNA is ligated to 280 pmole of BstXI adaptor with 10 units T4 DNA ligase (Invitrogen) in a final volume of 1 ml [400 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 1 mM ATP, 1 mM DTT, 5% (w/v) polyethylene glycol 8000]. The ligation mixture (1-ml) is loaded into a preparative well of a 1% agarose gel and separated in a contour-clamped homogeneous electrical field [CHEF [31]] unit as described [30]. Size-fractionated DNA electro-eluted from a gel slice is ligated to the BstXI sites of the pTARBAC6 vector and then used to transform E. coli DH10B (Invitrogen) by electroporation. Approximately 0.5 μg of sheared, adaptor-ligated and size-fractionated DNA is obtained from a single size fractionation on a single gel using 100 μg of agarose embedded DNA. The transformed cells were spread on 22 × 22 cm LB agar plates containing 5% sucrose and 20 μg/ml chloramphenicol and incubated at 37°C for 20 hr. Colonies were picked and arrayed into 48 384-well microtiter dishes containing LB media with 7.5% glycerol and 20 μg/ml chloramphenicol. The BAC vector and libraries are available from BACPAC Resources (http://bacpac.chori.org) at Children’s Hospital and Research Center at Oakland.
Insert size analysis
BACs from the CHORI-221 (192 clones), CHORI-222 (96 clones), CHORI-223 (192 clones) and CHORI-16 (422 clones) libraries were inoculated in 96-deep well blocks containing LB medium with 20 μg/ml chloramphenicol. BAC DNA was isolated using an automated plasmid isolation robot (AutoGen960, Autogen) and digested with PI-SceI (New England Biolabs) for the CHORI-221, -222, -223 and NotI (New England Biolabs) for CHORI-16. The DNA was loaded in 1% agarose gel in 0.5 x TBE buffer, run in CHEF units and analyzed as described [30]. The size (10.6 kb) of the vector was subtracted from the size of the linearized DNA fragments for determining insert sizes for the CHORI-221, CHORI-222 and CHORI-223 libraries.
Mapping of paired BAC end sequences
The BAC-end sequencing procedure originally developed at The Institute for Genome Research (TIGR) was modified as described [10]. A total of 288 BACs were sequenced on an ABI-377 DNA machine using the following primers for the T7 and SP6 ends respectively: GCCGCTAATACGACTCACTATAGGGAGAG and GTTTTTTGCGATCTGCCGTTTC. Low quality sequences were trimmed from the 3′ raw sequence data when more than three Ns appeared in a set of 20 bases after 400th base read. Vector and adaptor (GGAAAG) sequences were trimmed from the 5′end. The trimming process was automated through use of a PERL script. The end sequences were matched against the Release 3 sequence assembly of the D. melanogaster genome [7] using BLASTN [32]. The alignment results provide accurate chromosomal map locations and a virtual insert size. A simple graphic interface program designated as “chrmapV11.pl” was designed using PERL/Gtk to visualize the mapping results for all BAC clones.
Hybridization
RPCI-98 [5], CHORI-221 and CHORI-223 libraries containing ~18,000 clones each were gridded in duplicate with a specific pattern on nylon membranes using an automated gridding robot (BioRobotics). The high-density replica filters were processed with alkaline solution, neutralization solution and Protease solution as described [10]. A total of 8 probes were labeled using random priming, and hybridized to the filters. The 18HT satellite, Doc2 and Mst77F DNA was isolated from the centromeric region of chromosome Y [33, 34]. The other 5 probes have been described elsewhere: 359, 356, 353 satellites [21, 35], Dodecasatellite [36] and Circe [37].
Acknowledgments
The authors thank Dr. John Elliot for useful technical advice for developing the cloning procedure and Joseph W. Carlson for the assistance to align BAC end sequences to D. melanogaster genome sequences. The authors are grateful to Joseph Catanese, Susan Rhodes, Jeff Froula, and Barbara Swiatkiewicz for their technical advice and assistance. K.O. has been supported by a Postdoctoral Fellowship for Research Abroad from the Japan Society for the Promotion of Science (JSPS). The work was funded by grants from the US Department of Energy (DE-FG02-94ER61883) to Pieter de Jong and the Ministerio de Educación y Ciencia (BFU2005-07690-C02-01) to Alfredo Villasante. Funding for construction of the CHORI-222 and CHORI-223 BAC libraries was provided by Gerald M. Rubin from NIH grant HG00750.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gregory SG, et al. A physical map of the mouse genome. Nature. 2002;418:743–750. doi: 10.1038/nature00957. [DOI] [PubMed] [Google Scholar]
- 2.McPherson JD, et al. A physical map of the human genome. Nature. 2001;409:934–941. doi: 10.1038/35057157. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Krzywinski M, et al. Integrated and sequence-ordered BAC- and YAC-based physical maps for the rat genome. Genome Res. 2004;14:766–779. doi: 10.1101/gr.2336604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoskins RA, et al. A BAC-based physical map of the major autosomes of Drosophila melanogaster. Science. 2000;287:2271–2274. doi: 10.1126/science.287.5461.2271. [DOI] [PubMed] [Google Scholar]
- 6.Gibbs RA, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 7.Celniker SE, et al. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 2002;3:RESEARCH0079. doi: 10.1186/gb-2002-3-12-research0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim UJ, et al. Construction and characterization of a human bacterial artificial chromosome library. Genomics. 1996;34:213–218. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- 9.Osoegawa K, et al. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 2001;11:483–496. doi: 10.1101/gr.169601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osoegawa K, et al. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 2000;10:116–128. [PMC free article] [PubMed] [Google Scholar]
- 11.Osoegawa K, et al. BAC resources for the rat genome project. Genome Res. 2004;14:780–785. doi: 10.1101/gr.2033904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eichinger L, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riethman HC, et al. Integration of telomere sequences with the draft human genome sequence. Nature. 2001;409:948–51. doi: 10.1038/35057180. [DOI] [PubMed] [Google Scholar]
- 15.Mefford HC, Trask BJ. The complex structure and dynamic evolution of human subtelomeres. Nat Rev Genet. 2002;3:91–102. doi: 10.1038/nrg727. [DOI] [PubMed] [Google Scholar]
- 16.Riethman HC, Moyzis RK, Meyne J, Burke DT, Olson MV. Cloning human telomeric DNA fragments into Saccharomyces cerevisiae using a yeast-artificial-chromosome vector. Proc Natl Acad Sci USA. 1989;86:6240–6244. doi: 10.1073/pnas.86.16.6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abad JP, et al. TAHRE, a novel telomeric retrotransposon from Drosophila melanogaster, reveals the origin of Drosophila telomeres. Mol Biol Evol. 2004;21:1620–1624. doi: 10.1093/molbev/msh180. [DOI] [PubMed] [Google Scholar]
- 18.Abad JP, et al. Genomic analysis of Drosophila melanogaster telomeres: full-length copies of HeT-A and TART elements at telomeres. Mol Biol Evol. 2004;21:1613–1619. doi: 10.1093/molbev/msh174. [DOI] [PubMed] [Google Scholar]
- 19.Klickstein LB. In: Ausubel FM, et al., editors. John Wiley & Sons, Inc; New York: 1991. [Google Scholar]
- 20.Saitoh T, et al. TWEAK induces NF-kappaB2 p100 processing and long lasting NF-kappaB activation. J Biol Chem. 2003;278:36005–36012. doi: 10.1074/jbc.M304266200. [DOI] [PubMed] [Google Scholar]
- 21.Losada A, Villasante A. Autosomal location of a new subtype of 1.688 satellite DNA of Drosophila melanogaster. Chromosome Res. 1996;4:372–383. doi: 10.1007/BF02257273. [DOI] [PubMed] [Google Scholar]
- 22.Abad JP, et al. Pericentromeric regions containing 1.688 satellite DNA sequences show anti-kinetochore antibody staining in prometaphase chromosomes of Drosophila melanogaster. Mol Gen Genet. 2000;264:371–377. doi: 10.1007/s004380000331. [DOI] [PubMed] [Google Scholar]
- 23.Russell SR, Kaiser K. Drosophila melanogaster male germ line-specific transcripts with autosomal and Y-linked genes. Genetics. 1993;134:293–308. doi: 10.1093/genetics/134.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Losada A, Abad JP, Agudo M, Villasante A. Long-range analysis of the centromeric region of Drosophila melanogaster chromosome 3. Chromosome Res. 1996;8:651–653. doi: 10.1023/a:1009250411192. [DOI] [PubMed] [Google Scholar]
- 25.Benos PV, et al. From first base: the sequence of the tip of the X chromosome of Drosophila melanogaster, a comparison of two sequencing strategies. Genome Res. 2001;11:710–730. doi: 10.1101/gr.173801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoskins RA, et al. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 2002;3:RESEARCH0085. doi: 10.1186/gb-2002-3-12-research0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng C, et al. Large-insert BAC/YAC libraries for selective re-isolation of genomic regions by homologous recombination in yeast. Genomics. 2001;77:27–34. doi: 10.1006/geno.2001.6616. [DOI] [PubMed] [Google Scholar]
- 28.Strong SJ, Ohta Y, Litman GW, Amemiya CT. Marked improvement of PAC and BAC cloning is achieved using electroelution of pulsed-field gel-separated partial digests of genomic DNA. Nucleic Acids Res. 1997;25:3959–3961. doi: 10.1093/nar/25.19.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brizuela BJ, Elfring L, Ballard J, Tamkun JW, Kennison JA. Genetic analysis of the brahma gene of Drosophila melanogaster and polytene chromosome subdivisions 72AB. Genetics. 1994;137:803–813. doi: 10.1093/genetics/137.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osoegawa K, et al. An improved approach for construction of bacterial artificial chromosome libraries. Genomics. 1998;52:1–8. doi: 10.1006/geno.1998.5423. [DOI] [PubMed] [Google Scholar]
- 31.Chu G, Vollrath D, Davis RW. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986;234:1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Abad JP, et al. Genomic and cytological analysis of the Y chromosome of Drosophila melanogaster: telomere-derived sequences at internal regions. Chromosoma. 2004;113:295–304. doi: 10.1007/s00412-004-0318-0. [DOI] [PubMed] [Google Scholar]
- 34.Agudo M, et al. Centromeres from telomeres? The centromeric region of the Y chromosome of Drosophila melanogaster contains a tandem array of telomeric HeT-A- and TART-related sequences. Nucleic Acids Res. 1999;27:3318–24. doi: 10.1093/nar/27.16.3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lohe AR, Brutlag DL. Multiplicity of satellite DNA sequences in Drosophila melanogaster. Proc Natl Acad Sci USA. 1986;83:696–700. doi: 10.1073/pnas.83.3.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abad JP, et al. Dodeca satellite: a conserved G+C-rich satellite from the centromeric heterochromatin of Drosophila melanogaster. Proc Natl Acad Sci USA. 1992;89:4663–4667. doi: 10.1073/pnas.89.10.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Losada A, Abad JP, Agudo M, Villasante A. The analysis of Circe, an LTR retrotransposon of Drosophila melanogaster, suggests that an insertion of non-LTR retrotransposons into LTR elements can create chimeric retroelements. Mol Biol Evol. 1999;16:1341–1346. doi: 10.1093/oxfordjournals.molbev.a026044. [DOI] [PubMed] [Google Scholar]


