Abstract
Not much is known about the features that determine the biological stability of a molecule retained in the endoplasmic reticulum (ER). Ig light (L) chains that are not secreted in the absence of Ig heavy (H) chain expression bind to the ER chaperone BiP as partially folded molecules until they are degraded. Although all Ig L chains have the same three-dimensional structure when part of an antibody molecule, the degradation rate of unassembled Ig L chains is not identical. For instance, the two nonsecreted murine Ig L chains, κNS1 and λFS62, are degraded with half-lives of approximately 1 and 4 hr, respectively, in the same NS1 myeloma cells. Furthermore, the BiP/λFS62 Ig L chain complex appears to be more stable than the BiP/κNS1 complex. Here, we used the ability of single Ig domains to form an internal disulfide bond after folding as a measure of the folding state of κNS1 and λFS62 Ig L chains. Both of these nonsecreted L chains lack the internal disulfide bond in the variable (V) domain, whereas the constant (C) domain was folded in that respect. In both cases the unfolded V domain provided the BiP binding site. The stability of BiP binding to these two nonsecreted proteins was quite different, and both the stability of the BiP:Ig L chain complex and the half-life of the Ig L chain could be transferred from one Ig L chain isotype to the other by swapping the V domains. Our data suggest that the physical stability of BiP association with an unfolded region of a given light chain determines the half-life of that light chain, indicating a direct link between chaperone interaction and delivery of partially folded substrates to the mammalian degradation machinery.
Many cellular and extracellular proteins are translocated into the endoplasmic reticulum (ER) of eukaryotic cells to reach their final destination. During or immediately after translocation, covalent modifications such as N-linked glycosylation, peptidyl-proline isomerization, and/or disulfide bond formation occur as the polypeptide folds (1). The newly synthesized polypeptides usually assemble, fold, and acquire their three-dimensional conformation with the aid of ER-resident molecular chaperones. One of the best-characterized chaperones is BiP, a lumenal hsp70 protein that transiently interacts with a number of different polypeptides (reviewed in ref. 2). Retention in the ER of polypeptides that have not yet reached their mature state is described as quality control (3, 4) or architectural editing (5). Malfolded structures that cannot be rescued will eventually undergo rapid nonlysosomal degradation, a process typically described as occurring in a pre-Golgi compartment. Lately, evidence has accumulated indicating that degradation of both soluble and integral ER-membrane proteins does not occur within the secretory pathway but requires retro-translocation to the cytosol for proteasome-mediated proteolysis (reviewed in ref. 6). Very recent genetic studies in the yeast S. cerevisiae have identified components that appear to be directly involved in the retro-translocation event such as Sec61p, Sec63p, and Kar2p, the yeast homolog of BiP (7, 8).
Typical ER-degradation substrates are nonsecreted mutant proteins such as the PiZ variant of human alpha-1-antitrypsin (9), mutant insulin (10), or mutant procollagen subunits (11). Similarly, subunits of oligomeric proteins that are produced in stoichiometric excess or whose assembly is impaired are usually not transported but undergo degradation [e.g., unassembled T cell receptor chains (12, 13), isolated subunits of the Na, K-ATPase (14), or unassembled Ig chains (15–17)]. It is reasonable to postulate that unassembled molecules are degraded because they do not attain a mature fold. In the case of Ig light (L) chains that are not able to homodimerize, a disulfide bond is formed in only one of the two domains if the Ig H chain is absent (18). Interestingly, various incompletely assembled and folded Ig L chains were not degraded at the same rate. For instance, the κ Ig L chain (κNS1) synthesized by NS1 cells (19) has a half-life of approximately 50 min (15), whereas half of newly synthesized λ1 FS62 Ig L chains (λFS62) (20) expressed in these same cells are still remaining after 3–4 hr (16).
At present, not much is known about the criteria used by a cell to determine the half-life of polypeptides retained in the ER. Because BiP binds to unassembled and partially folded Ig L chains until they are degraded but BiP is not degraded with its ligand (15, 16, 21), it seemed very likely that BiP participates in the delivery of the substrate to the degradation machinery. This is supported by a similar correlation between release from BiP and degradation of other BiP ligands such as unassembled Ig heavy chains (ref. 22; K. Schröder and I.G.H., unpublished data), subunits of the Na, K-ATPase (14), and an assembly-defective form of the vacuolar storage glycoprotein phaseolin (23).
If BiP release from a polypeptide is indeed a rate-limiting step in the delivery of the ligand to the degradation machinery, it should be possible to transfer the half-life of a given substrate onto another molecule by exchanging the BiP-binding portion, provided that the overall folding capacity of the molecule remains undisturbed. In the present study, we used κNS1 and λFS62 Ig L chains to test this hypothesis. We found that both of these partially folded BiP-bound molecules lacked the internal disulfide bond in the variable (V) domain but have formed the bond in the C domain, suggesting that only the C domain can assume the mature fold in these nonsecreted Ig L chains. Furthermore, our data support the conclusion that the V domain determines both the physical stability of a BiP/Ig L chain complex as well as the biological stability of the nonassembled Ig L chain.
MATERIALS AND METHODS
Cell Culture and Transfections.
For studies on Ig L chain expression and turnover, we used the NS1 murine myeloma expressing nonsecreted κNS1 (19) or stable transfectants of a light chain loss variant of NS1 that were expressing the mutant λFS62 (NSFS62) (16). We also made additional stable Ig L chain transfectants from Ig-negative X63Ag8.653 myelomas (24) or H62 hybridomas (25). Electroporation and selection of stable transfectants of the X63Ag8.653 and H62 cells were performed as described by Allen et al. (26), and all resulting clones were cultured as described (15). Transient expression of Ig L chains was obtained by transfecting COS-1 monkey fibroblasts (27) with the various pSVL constructs by using the DEAE-dextran procedure as described (28). Ig L chain synthesis was analyzed 40 hr after transfection.
cDNA Cloning, Site-Directed Mutagenesis, and Construction of Expression Vectors.
The cDNAs encoding κNS1 and λFS62 were obtained by reverse transcription of κNS1-mRNA isolated from NS1 cells and of λFS62-mRNA isolated from NSFS62 cells. In both cases, PstI restriction sites were engineered upstream of the Ig L chain coding regions by the primers used for PCR, and the amplified material was ligated into the pGEM-T vector (Promega). PCR-mediated site directed mutagenesis (28) of the cloned Ig L chain cDNAs resulted in a pairwise replacement of the cysteine codons with serine codons in the V or C domains of both the κNS1 and λFS62 Ig L chains. The C-terminal fifth cysteine codon remained in both Ig L chains. The fidelity of all PCR products was determined by sequencing. The PstI-excised cDNA fragments were ligated into pUC19DL vector (a kind gift of W. Müller, Genetics Institute, Cologne), and the cDNA fragment was reexcised by using XbaI and SalI and ligated into an XhoI/XbaI-opened pSVL expression vector (Pharmacia).
The pENS1.neo expression vector contains the Ig H chain enhancer, the genomic sequence encoding the wild type κNS1 expressed in NS1 cells, and the neomycin resistance gene. It was created by ligation of the V domain-encoding gene segment isolated as a 2-kb EcoRI/XbaI fragment from pBS+Vκ (18) and the 8.8-kb EcoRI/XbaI fragment of pEVHCκ.neo containing the C domain-encoding gene segment of the mouse κ Ig L chain (29). Construction of the pSV2.neoλ1FS62 expression vector was described earlier (16). To construct the pSV2.neoVλ1FS62/Cκ expression vector that encodes the chimeric Ig L chain Vλ1FS62/Cκ (abbreviated herein as VλCκ), the V domain encoding 0.9-kb BamHI fragment of pA8–6.λ1FS62 (16) was subcloned in the correct orientation into BamHI-linearized pBluescript(+) to reexcise it as an EcoRI/XbaI fragment that was then ligated to the 8.8-kb EcoRI/XbaI fragment of pEVHCκ.neo (29). The pSV2.neoVκ/Cλ expression vector, encoding the chimeric Ig L chain VκNS1/Cλ (abbreviated herein as VκCλ), was constructed as follows: the ends of a 7.6-kb XbaI fragment of pSV2.neoλ1FS62 containing the Ig H enhancer and the resistance genes was religated. The resulting plasmid was linearized with SacI to insert the 2-kb SacI fragment containing the V domain-encoding gene segment isolated from pBS+Vκ (18). After XbaI linearization, the plasmid was ligated to the λ1 C domain-encoding gene segment isolated as a 3.5-kb XbaI fragment from pA8–6λ1 (kind gift of S. Weiss, Gesellschaft für Biologische Forschung, Braunschweig, Germany). After ensuring that the orientation of the insert was correct, the expression vectors were used for stable transfections.
Metabolical Labeling, Pulse–Chase Experiments, Immunoprecipitations, and Western Blot Analysis.
Transfected COS-1 cells were incubated in methionine- and cysteine-free culture medium for 1 hr and then metabolically labeled with 100 μCi (1 Ci = 37 GBq) of 35S-Translabel (ICN) for 3 hr. The cells were washed once with PBS containing 20 mM N-ethylmaleimide (NEM) and solubilized in a lysis buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 0.5% Nonidet P-40, and 0.5% deoxycholate to which 20 mM NEM and 10 units of apyrase (Sigma) were added.
To determine the folding status of wild-type (wt) and mutagenized κNS1 and λFS62, labeled Ig L chains were immunoprecipitated from cell lysates by using specific goat anti-mouse λ or κ antisera (Southern Biotechnology Associates) followed by protein A-Sepharose beads (Pharmacia). Immune complexes were electrophoresed (12% SDS/PAGE) under nonreducing conditions, and the labeled proteins were visualized by autoradiography. To determine the half-lives of wt and chimeric Ig L chains, cells were starved in methionine- and cysteine-free medium, metabolically labeled with 100 μCi of 35S-Translabel (ICN), and chased in the presence of excess unlabeled methionine and cysteine as indicated in the figure legend of individual experiments. Cells (106) were taken at each time point and solubilized in 0.5 ml of 2× lysis buffer (without NEM and apyrase), and Ig L chains were immunoprecipitated from lysates containing equivalent amounts of label protein as determined by TCA-precipitable radioactive counts. It should be noted that the anti-Ig L chain antisera react specifically with the C domains of the various Ig L chain proteins. The protein A-Sepharose-bound immune complexes were washed as described (15) and separated under reducing conditions on a 12% SDS/PAGE, and the labeled proteins were visualized by autoradiography. Quantitation of the labeled Ig L chains for half-life determinations was done by phosphorimaging (Fujix BAS 1000 using the program macbas v 1.0).
Western blot analysis was performed on immunoprecipitated proteins as described (16). BiP was detected with a mouse monoclonal anti-KDEL antibody (StressGen Biotechnologies, Victoria, Canada) in combination with horseradish peroxidase (HRP)-conjugated goat anti-mouse Ig G (H+L) (Bio-Rad), which also stained the Cκ-containing Ig L chains. HRP-conjugated anti-mouse λ antibodies were used to detect Cλ-bearing molecules. The HRP conjugates were visualized by using ECL and the BM chemiluminescence blotting substrate (Boehringer Mannheim).
RESULTS
The κNS1 and λFS62 Ig L chains represent ideal model proteins to study criteria that determine the half-life of proteins. First, all Ig L chains have only two distinct domains (a V and a C domain), each of which folds independently and is stabilized by a single internal disulfide bond after it folds. This makes it relatively easy to monitor the folding state of the protein. By assessing disulfide bond formation on nonreducing gels, four different species can be resolved upon gradual in vitro reduction of a completely folded Ig L chain including two distinct partially oxidized forms (containing a disulfide bond in either of the domains) in addition to the completely reduced and the completely oxidized forms (16, 30). Second, both κNS1 and λFS62 bind to BiP and are only partially folded (i.e., a disulfide bond is formed in only one domain). Third, in spite of these similarities, the half-lives of the κNS1 and λFS62 are quite different as are the stabilities of the respective BiP/Ig L chain complexes (16). Because these two L chains differ in both the V and the C domains, domain-swapping experiments are feasible, which should allow us to determine whether BiP binds to one of the domains and how the stability of BiP binding affects the half-life of a given protein.
Determination of the Folding State of BiP-Bound Ig L Chains.
In the absence of Ig H chain expression, κNS1 as well as λFS62 occur as partially folded BiP-bound molecules in the cell. It seems that BiP binding to the Ig L chain molecule prevents disulfide bond formation in one of the two Ig L chain domains because the sulfhydryl groups in the second Ig L chain domain can oxidize in vitro under conditions that lead to dissociation of Ig L chains from BiP. This is deduced from previous data showing that, upon size-fractionation of total cellular proteins, essentially all Ig L chains are recovered in fractions corresponding to the molecular mass of BiP:Ig L chain complexes. In these fractions the Ig L chains contain only one disulfide bond. In contrast, the majority of the Ig L chains were recovered as monomers containing two disulfide bonds when the lysate was treated with MgATP to dissociate BiP:Ig L chain complexes before size fractionation analysis (ref. 16; M. R. Knittler and I.G.H., unpublished data).
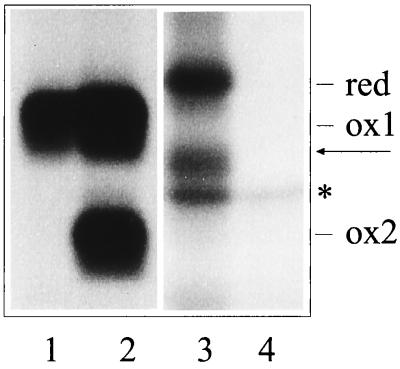
As a first step to further characterizing these two Ig L chains, we determined which domain remained unfolded. This was achieved by replacing of the paired cysteines with serine residues to prevent disulfide bond formation in either the V (V domain mutant) or the C domain (C domain mutant). Wild-type or mutant Ig L chains were transiently expressed in COS-1 cells and investigated for their migration behavior on SDS gels run under nonreducing conditions (Fig. 1). In previous studies, we found that if the NS1 cells were lysed in the presence of N-ethyl maleimide (NEM) to prevent postlysis oxidation, most of the κNS1 Ig L chains migrated as a single species containing only a single disulfide bond, whereas if NEM was not included, half or more of the molecules would undergo postlysis oxidation in the remaining domain (16). Despite the presence of NEM, the wild-type κNS1 expressed by COS-1 cells resolves into two bands corresponding to a partially (ox1) and a completely (ox2) oxidized Ig L chain form (Fig. 1, lane 2). The V domain mutant (Fig. 1, lane 1) resolves as a single species that migrates at the same position as the partially oxidized wild-type molecules (indicated as ox1 in Fig. 1). In contrast, the C domain mutant continues to resolve into two bands, neither of which comigrates with either of the wild-type forms (lane 3). Because the V domain mutant is composed of a single form that migrates at the same position as the partially folded κNS1 species, we conclude that in the wild-type NS1 Ig L chain the C domain is oxidized but the V domain is not unless it is allowed to oxidize after cell lysis. Mutation of the V domain cysteines prevents this from happening. Indeed, the C domain mutant consists of one species that migrates as a completely reduced molecule (red in lane 3) and a second species that occurs when the V domain is artificially oxidized. In summary, these data show that in vivo the partially folded wild-type κNS1 Ig L chain has formed the disulfide bond in the C domain whereas the disulfide bond in V domain has not formed. The same result was found with the analysis of the λFS62 Ig L chains (data not shown) confirming that, in both BiP-bound unassembled Ig L chains, it is the V domain that is not completely folded.
Figure 1.
Identification of the disulfide bond-containing domain in unassembled κNS1 Ig L chains. COS-1 cells were transfected with the various κNS1 cDNAs in a transient expression system. Biosynthetically labeled material was immunoprecipitated by anti-κ-antibodies from the lysate prepared in the presence of NEM from cells that expressed the κNS1V domain mutant (lane 1), the κNS1 wt-Ig L chain (lane 2), the κNS1 C domain mutant (lane 3), or no Ig L chains (lane 4). The migration of the completely reduced (no S—S bond, red), partially oxidized (S—S bond only in the C domain, ox 1), and completely oxidized (S—S bond in both domains, ox2) forms of the Ig L chains is indicated. An additional migration form (arrow) was observed with the C domain mutant and most likely represents a molecule containing a disulfide bond in the V domain. Isolation of the polypeptide marked by an asterisk (∗) is most likely due to nonspecific interactions, as it is also seen in the material precipitated from nontransfected cells (lane 4). Various amounts of this protein coisolated with the different Ig L molecules (i.e., compare lanes 2 and 3). Note that lanes 3 and 4 are from a longer exposure of the autoradiograph, because smaller amounts of labeled material was reproducibly recovered with the C domain mutant.
Analysis of the Physical Stability of BiP/Ig L Chain Complexes.
Although nonsecreted Ig L chains are bound to BiP in vivo, not all BiP/Ig L chain complexes are preserved to the same extent during immunoprecipitation experiments. Whereas λFS62 can be coimmunoprecipitated with BiP quantitatively, a substantial portion of the κNS1 Ig L chains do not remain bound to BiP and achieve disulfide bond formation in the second (V) domain during cell lysis and immunoprecipitation (ref. 16; see also Fig. 1, lane 2). It is noteworthy that λFS62, which remains bound to BiP in vitro, is not readily oxidized whereas the κNS1 Ig L chain, which is more easily dissociated from BiP, is also more easily oxidized upon cell lysis. We interpreted these findings to mean the two BiP/Ig L chain complexes exhibit different physical stabilities under immunoprecipitation conditions and that postlysis oxidation is a reflection of the loss of BiP association.
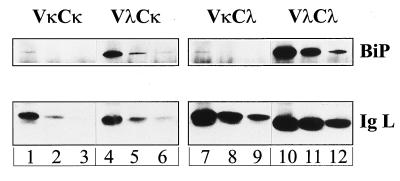
The observation that the two Ig L chains exhibit different stabilities with respect to BiP binding prompted us to investigate whether this property can be attributed to one of the Ig L chain’s domains. Chimeric Ig L chains in which the V domain of the κNS1 chain is combined with the C domain of the λFS62 chain (VκCλ) or vice versa (VλCκ) were made and used to generate stable transfectants of X63Ag8.653 myeloma cells, which do not express any Ig chains. In addition, cells that stably express the prototype κNS1 (VκCκ) or λFS62 (VλCλ) Ig L chains were obtained. Cell lysates were immunoprecipitated with antisera specific for the constant portion of the Ig L chains. Because essentially all Ig L chains are bound to BiP in the cells (16), we reasoned that the amount of BiP that survived coprecipitation with the Ig L chain construct should reflect the stability of the respective BiP:Ig L chain complex. Indeed, when comparing the same amount of immunoprecipitated Ig L chains, it appears that an unstable BiP:κNS1 Ig L chain complex (which is stoichiometric in vivo, ref.16) is stabilized upon replacing the Vκ by the Vλ domain (Fig. 2). The data clearly show that the amount of coprecipitated BiP is low whenever the precipitated Ig L chains bear the Vκ domain, whereas a greater amount of BiP is coprecipitated with molecules that bear the Vλ domain. These results imply that the V domain of unassembled Ig L chains is directly involved in BiP interaction and strongly suggest that Vλ has a higher affinity for BiP than Vκ.
Figure 2.
The amount of BiP coimmunoprecipitated with Ig L chains depends on the V domains of the Ig L chains. The amount of immunoprecipitated Ig L chains corresponds to the equivalent of 2 × 105 (lanes 1, 4, 7, and 10), 105 (lanes 2, 5, 8, and 11), or 0.5 × 105 (lanes 3, 6, 9, and 12) X63Ag8.653 cells. Immunoprecipitation as well as immunoblotting of the Ig L chains were performed by using antisera that recognize the C domain but not the V domain of the Ig L chains. BiP was detected with a mouse monoclonal IgG antibody against the C-terminal KDEL sequence. This antibody and the Ig L chains that bear the Cκ domain (VκCκ and VλCκ, respectively, lanes 1–6) were detected with an anti-mouse IgG (H+L) antiserum. The Cλ domain-bearing Ig L chains (VκCλ and VλCλ, respectively, lanes 7–12) were stained by anti-λ antibodies.
Analysis of the Biological Stability of Ig L Chains.
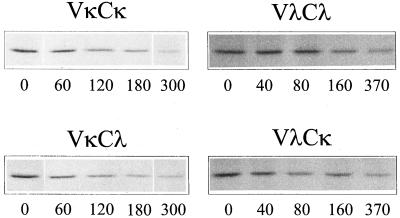
Next, we asked whether the half-life of an Ig L chain is determined by one of the Ig L chain domains. In addition to the stable X63Ag8.653 cell lines described above, we established a second set of transfectants expressing the prototypical, κNS1or λFS62, or the respective chimeric Ig L chains (named VλCκ and VκCλ, respectively) by using the H62 hybridoma line. Pulse–chase labeling and immunoprecipitation experiments were performed to determine the biological stability of the various Ig L chains (Fig. 3). The half-life of λFS62 Ig L chains was between 3 and 4 hr in both transfectants (see Table 1), which is in accordance with their half-life in NS1 cells (16). The VλCκ chimeric Ig L chain had approximately the same half-life as the λFS62 Ig L chains in X63Ag8.653 cells and appeared to be somewhat more stable in H62 cells. In both the X63Ag8.653 and H62 cells transfected with κNS1, the L chain was degraded with a half-life of 2 hr, which was about twice as long as observed for the same Ig L chain in NS1 cells (15) but still perceptibly shorter than the λFS62 Ig L chains in the same cells. When the VκCλ chimeric chain was expressed in X63Ag8.653 and H62 cells, we found that they also had a half-life of 2 hr. Thus, the half-life of a given Ig L chain depended on the V region present and appeared independent of which C domain it was combined to.
Figure 3.
Half-life determination of wt and chimeric Ig L chains. Stably transfected X63Ag8.653 cells were starved for 2.5 hr in medium lacking methionine and cysteine and labeled with 100 μCi 35S-Translable for 20 min when the cells expressed Ig L chains bearing the V domain of κNS1 (VκCκ and VκCλ, respectively) or 2.5 hr when the cells expressed Ig L chains bearing the V domain of λFS62 (VλCλ and VλCκ, respectively). Cells were replated in complete medium, chased for the indicated times, and then lysed. Ig L chains were immunoprecipitated by using antisera that recognize the C domain.
Table 1.
Half-life of various Ig L chains in different cell lines
| Cell line/ Ig L chain | NS1* | X63Ag8.653 | H62 |
|---|---|---|---|
| VκCκ (κNS1) | 0.8 | 2.0; 2.1 | 2.0 |
| VκCλ | 1.8; 1.7 | 2.3 | |
| VλCλ (λFS62) | 3.5 | 3.2 | 3.3; 3.7 |
| VλCκ | 3.0; 4.0 | >6; >6 |
DISCUSSION
Recognition and subsequent degradation of malfolded polypeptides are important physiological processes required to maintain the functional integrity of a cell. A quality control system acting in the ER prevents the transport of polypeptides that have not acquired a mature state with regard to assembly and/or three-dimensional conformation. We previously showed that Ig L chains depend on assembly with heavy chains or into homodimers for complete folding and secretion (18). Ig L chains that are unable to form homodimers remain associated with BiP and are eventually degraded in the ER (16, 18). In the case of secreted Ig L chains, BiP binds transiently to the reduced and partially oxidized Ig L chains but is released to allow complete folding of the L chains (15, 31). In the present study, we demonstrate that in the case of two different nonsecreted Ig L chains, it is the V domain that is not folded and BiP binds to this unfolded V domain. Furthermore, we show that the half-life of the Ig L chains is dictated by this unfolded, BiP-bound domain and the stability of BiP binding is directly related to the stability of the Ig L chain.
We do not know whether all nonsecreted Ig light chains fail to fold their V region, but a number of nonsecreted Ig L chains are known to possess mutations in the V region (20, 32–34). It is possible that the V region is often less readily able to fold because of the variability in sequences that it must accommodate whereas the C domain (which always has the same sequence) has evolved to fold well. In support of this idea, we found that wild-type secreted Ig L chains that bind BiP transiently do so through a single domain (31) which, as was most recently demonstrated, is also the V domain (R. Hellman, M. Vanhove, and L.M.H., unpublished data). Having shown that the V region is not folded, it is not entirely surprising that BiP binds to the V region in unassembled Ig L chains (Fig. 2), because earlier studies demonstrated that dnaK, the bacterial hsp70 homologue, preferentially binds to unfolded peptides (35). However, we were somewhat surprised to find that the half-life of the individual Ig L chains was entirely controlled by the unfolded V domain. When chimeric proteins were made between the more rapidly degraded and the more stable Ig L chain, exchange of the C domain did not affect the protein’s half-life, whereas swapping the V domain associated with a given constant region changed the half-life of the new chimeric Ig L chain to the same as that of the V domain with its original C domain. In this way a more stable Ig L chain could be rendered more susceptible to intracellular degradation by combining its C domain with the V region of the more short-lived molecule. Although little is known of how proteins that are retained in the ER are recognized and targeted for degradation, our results would suggest this process is controlled by the availability of unfolded regions on the protein. Even more interesting in this respect is the correlation of stability of BiP binding to a given V domain and the half-life of the protein. Although in both cases, the incompletely folded V domain was bound to BiP in the cell, we found that the BiP:λFS62 complex was much more stable during immunoprecipitation than the BiP:κNS1 complex. And correspondingly, Ig L chains bearing the λFS62 V domain had a much longer half-life than the Ig L chains containing the κNS1 V domain. Although experiments presented here do not allow us to conclude that the BiP:κNS1 complex is also less stable in vivo, it is tempting to speculate that turnover of these BiP-associated proteins is related to their release from BiP.
How could the physical stability of a BiP/ligand complex relate to the physiological process determining the half-life of the ligand? Previous work demonstrated that the binding of ATP to BiP causes a conformational change in BiP sufficient to induce release of bound ligand (36). This seems to reflect a general principle of hsp70 function because ATP binding is also sufficient to induce dissociation of proteins complexed to the bacterial hsp70 molecule DnaK (37). Bukau and coworkers showed that the nucleotide-binding state of DnaK does not alter its affinity for the substrate but rather affects the association and thus also the dissociation kinetics (38). If this also applies for BiP, a polypeptide exhibiting a higher affinity for this chaperone would statistically remain bound to BiP for a longer period of time as compared with a molecule with low BiP-binding affinity, irrespective of the nucleotide-binding state of the hsp70 protein. Thus, the strength of BiP interaction might determine how fast the molecule is delivered to the degradation machinery.
Assuming that Ig L chain degradation occurs in the cytosol, as was reported for other ER degradation substrates in mammalian cells (12, 13, 39–42), unassembled Ig L chains have to re-traverse the ER membrane. It is a matter of debate whether soluble polypeptides retained in the ER remain bound to the ER translocation channel because of incomplete folding. Unassembled Ig L chains, however, have probably completely entered the lumen of the ER as the C domain that comprises the C-terminal half of the molecule assumes the mature Ig domain fold stabilized by the disulfide bond. Thus, the rate-limiting step in the degradation of unassembled Ig L chains in mammalian cells could be the BiP-mediated delivery to the cytosol. In yeast, functional interaction of BiP with the DnaJ-like translocon component Sec63 is required for Sec61-mediated co- and posttranslational translocation into the ER (43–45). A similar BiP:Sec63 interaction may serve to retro-translocate ER degradation substrates because these two proteins are involved in the proteasome-dependent degradation of mutated lumenal yeast carboxypeptidase (8). In mammalian cells, BiP could directly participate in the delivery of ER degradation substrates to the cytosol, possibly via an interaction with the DnaJ-like membrane protein MTJ1 that has been identified in mouse cells (46). However, mammalian BiP might functionally differ from the yeast homolog, as it has not been possible to demonstrate a role for BiP in the translocation of proteins into the mammalian ER (47, 48). If mammalian BiP is not part of the translocation machinery, our data could suggest that BiP must release from an unfolded protein for that protein to engage the translocon and be translocated into the cytosol for degradation. In this context, it would be important to know whether degradation of substrates that do not accumulate as BiP-bound molecules in the ER, like transport impaired variants of alpha-1-antitrypsin (9, 49), also require functional BiP for degradation (50).
Acknowledgments
With a feeling of great indebtedness, and with the support of all authors, I.G.H. dedicates this work to her former mentor, Tommy Meo, who recently died in Paris. We thank E. Ivessa and H. Tschochner for their critical comments, which greatly helped to improve the quality of the manuscript. We are grateful to S. Dirks for his help in DNA cloning and to S. Daugalies for expert technical help. I.G.H. received a Heisenberg award from the Deutsche Forschungsgemeinschaft. This work was supported through Grant SFB352/B5 of the Deutsche Forschungsgemeinschaft awarded to I.G.H., and GM54068 from the National Institutes of Health awarded to L.M.H.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: ER, endoplasmic reticulum; H, heavy; L, light; V, variable; C, constant; NEM, N-ethylmaleimide.
References
- 1.Leitzgen, K. & Haas, I. G. (1998) Chemtracts, in press.
- 2.Haas I G. Experientia. 1994;50:1012–1020. doi: 10.1007/BF01923455. [DOI] [PubMed] [Google Scholar]
- 3.Hurtley S M, Helenius A. Annu Rev Cell Biol. 1989;5:277–307. doi: 10.1146/annurev.cb.05.110189.001425. [DOI] [PubMed] [Google Scholar]
- 4.Hammond C, Helenius A. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 5.Klausner R D. New Biol. 1989;1:3–8. [PubMed] [Google Scholar]
- 6.Brodsky J L, McCracken A A. Trends Cell Biol. 1997;7:151–156. doi: 10.1016/S0962-8924(97)01020-9. [DOI] [PubMed] [Google Scholar]
- 7.Pilon M, Schekman R, Romisch K. EMBO J. 1997;16:4540–4548. doi: 10.1093/emboj/16.15.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plemper R K, Bohmler S, Bordallo J, Sommer T, Wolf D H. Nature (London) 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- 9.Graham K S, Le A, Sifers R N. J Biol Chem. 1990;265:20463–20468. [PubMed] [Google Scholar]
- 10.Schmitz A, Maintz M, Kehle T, Herzog V. EMBO J. 1995;14:1091–1098. doi: 10.1002/j.1460-2075.1995.tb07092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamande S R, Chessler S D, Golub S B, Byers P H, Chan D, Cole W G, Sillence D O, Bateman J F. J Biol Chem. 1995;270:8642–8649. doi: 10.1074/jbc.270.15.8642. [DOI] [PubMed] [Google Scholar]
- 12.Yu H, Kaung G, Kobayashi S, Kopito R R. J Biol Chem. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
- 13.Huppa J B, Ploegh H L. Immunity. 1997;7:113–122. doi: 10.1016/s1074-7613(00)80514-2. [DOI] [PubMed] [Google Scholar]
- 14.Beggah A, Mathews P, Beguin P, Geering K. J Biol Chem. 1996;271:20895–20902. doi: 10.1074/jbc.271.34.20895. [DOI] [PubMed] [Google Scholar]
- 15.Knittler M R, Haas I G. EMBO J. 1992;11:1573–1581. doi: 10.1002/j.1460-2075.1992.tb05202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knittler M R, Dirks S, Haas I G. Proc Natl Acad Sci USA. 1995;92:1764–1768. doi: 10.1073/pnas.92.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner A M, Aviel S, Argon Y. J Biol Chem. 1993;268:25940–25947. [PubMed] [Google Scholar]
- 18.Leitzgen K, Knittler M R, Haas I G. J Biol Chem. 1997;272:3117–3123. doi: 10.1074/jbc.272.5.3117. [DOI] [PubMed] [Google Scholar]
- 19.Köhler G, Howe S C, Milstein C. Eur J Immunol. 1976;6:292–295. doi: 10.1002/eji.1830060411. [DOI] [PubMed] [Google Scholar]
- 20.Dul J L, Argon Y. Proc Natl Acad Sci USA. 1990;87:8135–8139. doi: 10.1073/pnas.87.20.8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cremer A, Knittler M R, Haas I G. In: Glyco- and Cell Biology. Wieland F, Reutter W, editors. Heidelberg: Springer; 1994. pp. 171–184. [Google Scholar]
- 22.Freiden P J, Gaut J R, Hendershot L M. EMBO J. 1992;11:63–70. doi: 10.1002/j.1460-2075.1992.tb05028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pedrazzini E, Giovinazzo G, Bielli A, de Virgilio M, Frigerio L, Pesca M, Faoro F, Bollini R, Ceriotti A, Vitale A. Plant Cell. 1997;9:1869–1880. doi: 10.1105/tpc.9.10.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kearney J F, Radbruch A, Liesegang B, Rajewsky K. J Immunol. 1979;123:1548–1550. [PubMed] [Google Scholar]
- 25.Haas I G, Wabl M R. Proc Nat Acad Sci USA. 1984;81:7185–7188. doi: 10.1073/pnas.81.22.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allen D, Simon T, Sablitzky F, Rajewsky K, Cumano A. EMBO J. 1988;7:1995–2001. doi: 10.1002/j.1460-2075.1988.tb03038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gluzman Y. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 28.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1989. [Google Scholar]
- 29.Simon T, Rajewsky K. EMBO J. 1990;9:1051–1056. doi: 10.1002/j.1460-2075.1990.tb08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jäck H M, Sloan B, Grisham G, Reason D, Wabl M. J Immunol. 1993;150:4928–4933. [PubMed] [Google Scholar]
- 31.Hendershot L, Wei J, Gaut J, Melnick J, Aviel S, Argon Y. Proc Natl Acad Sci USA. 1996;93:5269–5274. doi: 10.1073/pnas.93.11.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosmann T R, Williamson A R. Cell. 1980;20:283–292. doi: 10.1016/0092-8674(80)90614-5. [DOI] [PubMed] [Google Scholar]
- 33.Wu G E, Hozumi N, Murialdo H. Cell. 1983;33:77–83. doi: 10.1016/0092-8674(83)90336-7. [DOI] [PubMed] [Google Scholar]
- 34.Dul J L, Burrone O R, Argon Y. J Immunol. 1992;149:1927–1933. [PubMed] [Google Scholar]
- 35.Landry S J, Jordan R, McMacken R, Gierasch L M. Nature (London) 1992;355:455–457. doi: 10.1038/355455a0. [DOI] [PubMed] [Google Scholar]
- 36.Wei J Y, Gaut J R, Hendershot L M. J Biol Chem. 1995;270:26677–26682. doi: 10.1074/jbc.270.44.26677. [DOI] [PubMed] [Google Scholar]
- 37.Palleros D R, Reid K L, Shi L, Welch W J, Fink A L. Nature (London) 1993;365:664–666. doi: 10.1038/365664a0. [DOI] [PubMed] [Google Scholar]
- 38.McCarty J S, Buchberger A, Reinstein J, Bukau B. J Mol Biol. 1995;249:126–137. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- 39.Ward C L, Omura S, Kopito R R. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 40.Jensen T J, Loo M A, Pind S, Williams D B, Goldberg A L, Riordan J R. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 41.Wiertz E, Tortorella D, Bogyo M, Yu J, Mothes W, Jones T R, Rapoport T A, Ploegh H L. Nature (London) 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 42.Qu D, Teckman J H, Omura S, Perlmutter D H. J Biol Chem. 1996;271:22791–22795. doi: 10.1074/jbc.271.37.22791. [DOI] [PubMed] [Google Scholar]
- 43.Brodsky J L, Goeckeler J, Schekman R. Proc Natl Acad Sci USA. 1995;92:9643–9646. doi: 10.1073/pnas.92.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lyman S K, Schekman R. J Cell Biol. 1995;131:1163–1171. doi: 10.1083/jcb.131.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corsi A K, Schekman R. J Cell Biol. 1997;137:1483–1493. doi: 10.1083/jcb.137.7.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brightman S E, Blatch G L, Zetter B R. Gene. 1995;153:249–254. doi: 10.1016/0378-1119(94)00741-a. [DOI] [PubMed] [Google Scholar]
- 47.Gorlich D, Rapoport T A. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 48.Dierks T, Volkmer J, Schlenstedt G, Jung C, Sandholzer U, Zachmann K, Schlotterhose P, Neifer K, Schmidt B, Zimmermann R. EMBO J. 1996;15:6931–6942. [PMC free article] [PubMed] [Google Scholar]
- 49.Cresteil D, Ciccarelli E, Soni T, Alonso M A, Jacobs P, Bollen A, Alvarez F. FEBS Lett. 1990;267:277–280. doi: 10.1016/0014-5793(90)80944-e. [DOI] [PubMed] [Google Scholar]
- 50.McCracken A A, Karpichev I V, Ernaga J E, Werner E D, Dillin A G, Courchesne W E. Genetics. 1996;144:1355–1362. doi: 10.1093/genetics/144.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]