Abstract
The AML/CBFα runt transcription factors are key regulators of hematopoietic and bone tissue-specific gene expression. These factors contain a 31-amino acid nuclear matrix targeting signal that supports association with the nuclear matrix. We determined that the AML/CBFα factors must bind to the nuclear matrix to exert control of transcription. Fusing the nuclear matrix targeting signal to the GAL4 DNA binding domain transactivates a genomically integrated GAL4 responsive reporter gene. These data suggest that AML/CBFα must associate with the nuclear matrix to effect transcription. We used fluorescence labeling of epitope-tagged AML-1B (CBFA2) to show it colocalizes with a subset of hyperphosphorylated RNA polymerase II molecules concentrated in foci and linked to the nuclear matrix. This association of AML-1B with RNA polymerase II requires active transcription and a functional DNA binding domain. The nuclear matrix domains that contain AML-1B are distinct from SC35 RNA processing domains. Our results suggest two of the requirements for AML-dependent transcription initiation by RNA polymerase II are association of AML-1B with the nuclear matrix together with specific binding of AML to gene promoters.
Many observations suggest a linkage between nuclear architecture and the regulation of gene expression. These include drastic changes in nuclear morphology often seen in differentiation and in transformation to malignancy. The filamentous ribonucleoprotein network known as the nuclear matrix is a major component of nuclear structure (1–5). It serves to localize gene sequences and may maintain the distribution of regulatory factors throughout the nuclear space (6–19).
The AML/CBFα transcription factors are key regulators of hematopoietic and bone-tissue-specific gene expression (20–23). We have recently shown that the transcriptionally active AML factors contain a specific sequence that targets them to the nuclear matrix (24). In contrast, several inactive forms of AML that lack this targeting signal do not associate with the matrix (24). The targeting sequence resides in a 31-aa segment (nuclear matrix targeting signal, NMTS; aa 351–381) within the C-terminal domain. This NMTS is physically distinct from the nuclear localization signal and functions autonomously to direct AML-1B (CBFA2) to the nuclear matrix (24). These findings suggest that association with nuclear structure may be an important aspect of the mechanisms of gene regulation.
In this report we begin to examine the significance of the AML-1B transcription factor binding to the nuclear matrix. We show that AML-1B localizes to sites where there is also active transcription. We demonstrate that the NMTS, which directs AML-1B to the nuclear matrix, functions as a transactivation domain when interacting with an appropriate promoter. Furthermore, we show that the AML-1B transcription factor is targeted to discrete sites on the nuclear matrix. These sites also contain the hyperphosphorylated active form of RNA polymerase II. Colocalization of AML transcription factors with RNA polymerase II on the nuclear matrix is dependent on ongoing transcription and requires the promoter-recognition function of the runt homology domain of AML/CBFα.
MATERIALS AND METHODS
Transient Transfections.
The AML-1B expression vector was based on pcDNA/ampI and contains the cytomegalovirus (CMV) promoter and the human AML-1B cDNA linked to the hemagglutinin (HA)-tag fused in frame with aa 27 of AML-1B (CMV/HA/AML-1B construct). DEAE-dextran-mediated transfection was performed with 9 μg of plasmid DNA in 3 ml of F12 serum-free medium containing 0.2 mg/ml DEAE-dextran and 0.05 mg/ml chloroquine. Aliquots of 0.5 ml of this mixture were added to each well of a six-well plate and incubated for 1.5–2.0 hr at 37°C. SAOS cells were subject to a 15% glycerol shock for 2 min, washed with PBS, refed, and harvested at 24 hr following transfection. Identical results were obtained by transfection with lipofectamine using a protocol provided by GIBCO/BRL.
In Situ Nuclear Matrix Isolation and Indirect Immunofluorescence Analysis.
In situ nuclear matrices were prepared as described (25). Cells on coverslips were washed in PBS and extracted twice in CSK buffer (25) for 15 min each. DNase digestion was performed twice in digestion buffer (CSK buffer with 50 mM NaCl) containing 100 μg/ml DNase I for 30 min, followed by extraction in digestion buffer containing 0.25 M (NH4)2SO4 for 10 min. The coverslips were fixed in 4% formaldehyde in PBS. The primary antibody was incubated for 1–1.5 hr at 37°C. The primary antibodies were anti-HA (1:1,500 dilution, 12CA5 mouse mAb, a gift from M. Czech, University of Massachusetts Medical Center, Worcester; or a 1:1000 dilution of polyclonal anti-HA antibodies from Santa Cruz Biotechnology), anti-RNA polymerase II0 (B3 mouse IgM mAb) that recognizes the hyperphosphorylated large subunit (250 kDa) of RNA polymerase II (26), anti-SC35 and anti-BrdUrd (Sigma). The secondary antibody was incubated for 1 hr at 37°C and was either a fluorescein isothiocynate-conjugated goat anti-rabbit antibody (1:400, Jackson ImmunoResearch), a Texas red-conjugated donkey anti-mouse antibody (1:400, Jackson ImmunoResearch), or a rhodamine-conjugated goat anti-mouse IgM antibody. DNA content was evaluated by 4′,6-diamidino-2-phenylindole staining (5 μg/ml 4′,6-diamidino-2-phenylindole in PBS containing BSA and 0.05% Triton X-100). Cells were mounted in Vectashield H-1000. Microscopic images were obtained by using a CCD camera interfaced with a digital microscope system or a Bio-Rad MRC 1000 confocal microscope. Images were displayed by using the Adobe photoshop program.
RNA Synthesis Inhibition and BrUTP Labeling.
Cells were transfected as above for 21 hr. Actinomycin D (5 μg/ml) or 5,6-dichlorobenzimidazole riboside (40 μg/ml) RNA synthesis inhibitors were added for 3 hr. The cells were harvested after 24 hr. BrUTP labeling was conducted on Triton X-100 permeabilized cells as described (27, 28).
Transactivation Assays.
Transactivation assays were performed with HeLa GAL-5-Luc, a stable cell line carrying an integrated luciferase gene with five tandem GAL4 binding sites. This cell line was transfected with pCMV driven GAL4/AML-1B fusion proteins (250 ng pDNA per well) by using the polycation Superfect (Qiagen, Chatsworth, CA). Construct pCMV Sport β-galactosidase (GIBCO/BRL) was used as a control for transfection efficiency. Luciferase activity was used to calculate the average fold-induction relative to control (pCMV). Statistical comparisons were performed by using the unpaired Student’s t test (statview ii, MacIntosh).
RESULTS
The NMTS of the AML Transcription Factor Acts as a Context-Dependent Activation Domain.
We have shown previously that the C terminus of AML-1B contains a 31-aa NMTS (aa 351–381), as well as a transactivation domain (aa 432–480) that functions within the context of the TCRβ promoter (24, 29). Transcriptional activation controlled by AML depends on promoter context and interactions with additional factors (22, 23, 29). We tested whether either the C-terminal AML domain (aa 432–480) or the NMTS (aa 351–381) will transactivate gene transcription when interacting with a heterologous promoter. We fused each of these two segments separately to the GAL4 DNA binding domain (aa 1–147). The fusion proteins expressed by these two constructs have very different activities when interacting with a promoter containing multiple GAL4 binding sites.
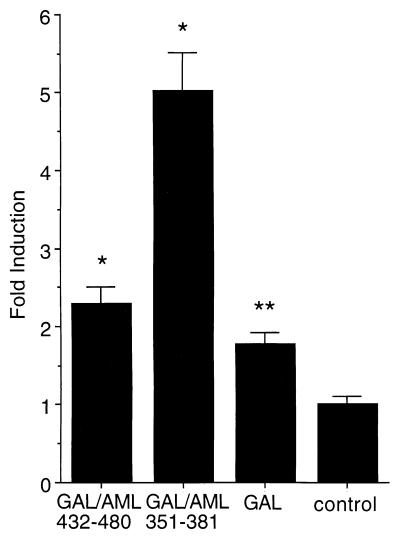
The transactivating activities of the two resulting GAL4/AML-1B fusion proteins were assayed by transfection and expression in a genetically modified HeLa cell line. This cell line contains a stably integrated reporter construct with five GAL4 responsive sites upstream of the TATA box that together drive the luciferase (LUC) gene. Fig. 1 shows the experimental results. Expression of the GAL4 (aa 1–147) protein alone results in, at most, a modest 1.5–2-fold increase over basal LUC activity (control). Fusing the C-terminal AML (aa 432–480) segment to GAL4 has almost no effect on promoter activity although this segment is known to be a powerful activation domain when present in the TCRβ promoter (24, 29). Conversely, although the NMTS targeting sequence does not enhance TCRβ promoter activity (24, 29), it does function as a potent transactivation domain when assayed here with the GAL4 promoter (Fig. 1). The results show that LUC activity is enhanced 5-fold relative to the control (Fig. 1). The NMTS appears to be a context-dependent transactivation domain.
Figure 1.
The NMTS of AML-1B trans-activates heterologous reporter gene expression. The reporter gene is chromosomally integrated and is composed of multimerized GAL4-sites fused to the Adenovirus minimal E1A promoter that drives expression of luciferase activity. Data are expressed as fold-induction in response to CMV driven coexpression of fusion-proteins between the GAL4 DNA binding domain (aa 1–147; GAL) and C-terminal segments of AML-1B (GAL/AML 432–480 and GAL/AML 351–381), relative to cells transfected with the CMV-vector alone (control). Each bar represents the data from 5 experiments (2–3 replicates). The asterisks indicates statistically significant differences relative to the control (*P ≤ 0.0005; **P ≤ 0.005).
AML Regulatory Factors Associate with a Subset of RNA Polymerase II Domains on the Nuclear Matrix.
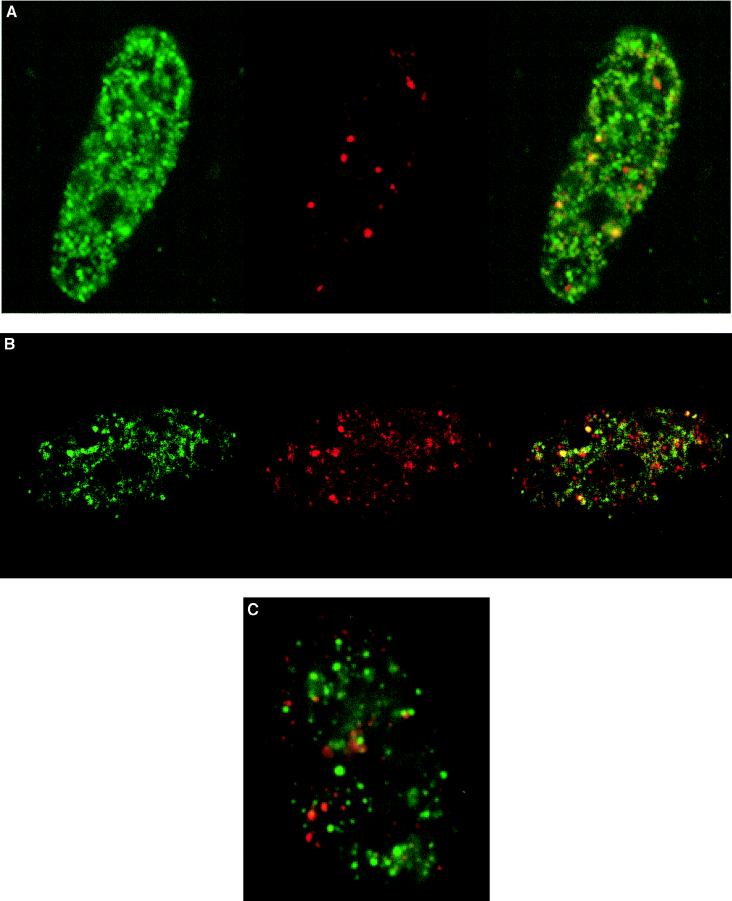
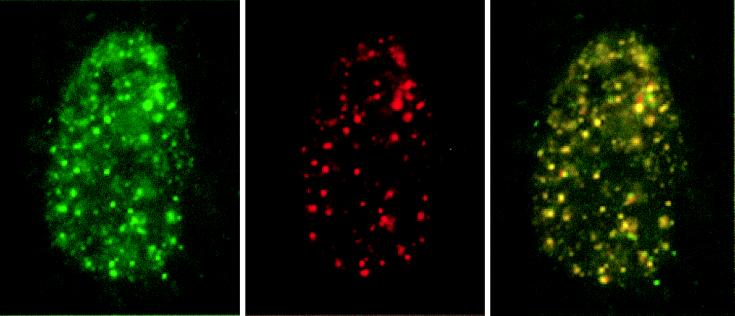
The interesting finding that the NMTS (aa 351–381) domain affects both nuclear matrix targeting and transcriptional activation suggests that these two functions are causally related. One consequence of such a linkage would be the association of actively transcribing RNA polymerase II with AML-1B. We performed double label immunofluorescence experiments to ascertain the extent of colocalization of nuclear matrix-associated AML-1B with the hyperphosphorylated form of RNA polymerase II (pol II0) (Fig. 2). We confirmed that RNA polymerase II domains associated with the nuclear matrix are transcriptionally active by immunofluorescence analysis of BrUTP-labeled RNA transcripts and RNA polymerase II in SAOS cells. The majority (>95%) of RNA polymerase II-containing foci are labeled with BrUTP (Fig. 3) consistent with previous reports (9, 26). Therefore, the RNA polymerase II0 domains detected in SAOS cells with the B3 antibody used in this study represent sites of ongoing transcription.
Figure 2.
AML-1B is targeted to transcriptionally active nuclear matrix domains. (A and B) A subset of AML-1B and RNA polymerase II are colocalized in the nuclear matrix. Human osteosarcoma SAOS-2 cells were transiently transfected with a construct expressing HA-tagged CBFα2/AML-1B. In situ nuclear matrix samples were prepared 24 hr after transfection as described in Materials and Methods. HA/AML-1B was detected with an antibody against HA (green) (Left) and RNA polymerase II0 with the B3 anti-RNA polymerase II antibody (red) (Center). The merged image is shown in the Right panel; yellow indicates colocalization of AML-1B with RNA polymerase II0. The colocalization of AML-1B with RNA polymerase II0 was evaluated by digital immunofluorescence microscopy (A) and by confocal microscopy (B) with a series of optical sections (0.5 μm intervals) through a single cell. B shows single-color and merged images (green, HA/AML-1B; red, RNA polymerase II; yellow, colocalization) for one of the optical sections. (C) Promoter recognition is required for the colocalization of AML-1B with RNA polymerase II. SAOS-2 cells were transfected with the HA-tagged L148D mutant that contains a point mutation in the runt homology domain and lacks DNA-binding activity. In situ nuclear matrix preparations were visualized as described in A (green, HA/AML-1B; red, RNA polymerase II; yellow, colocalization). The images shown were obtained by digital fluorescence microscopy. Similar images were obtained by directly photographing immunofluorescence signals (data not shown).
Figure 3.
AML-1B/RNA polymerase II0 sites are coupled with RNA synthesis. The HA/AML-1B construct was transfected into SAOS and ROS 17/2.8 cells. BrUTP labeling was performed with Triton X-100 permeabilized cells. Cells were double-stained with the B3 anti RNA polymerase II antibody and the BU33 anti-BrdUrd antibody. BrUTP (green) and RNA polymerase II0 (red) single-color channels and the merged image are each shown, with yellow indicating colocalization of BrUTP and RNA polymerase II0. The same extent of colocalization was observed in mock-transfected and untransfected cells (data not shown).
Human SAOS osteosarcoma cells were transfected with AML-1B tagged with the HA epitope (Fig. 2). The cells were extracted to obtain the nuclear matrix, which was then labeled with antibodies to HA and to RNA polymerase II0. Fig. 2 shows that AML-1B and RNA polymerase II0 molecules are bound to the nuclear matrix and distributed throughout the nuclear space. AML-1B is localized in a few prominent foci but most are more broadly distributed. RNA polymerase II0 shows a similar but not identical distribution in large foci and in smaller domains throughout the nucleus. The number and size of RNA pol II0 foci in SAOS cells showed cell-to-cell variation, possibly related to cell cycle stage and/or metabolic state. Strikingly, AML-1B and RNA polymerase II0 colocalize at some of the most prominent foci in the nuclear matrix as well as at many of the smaller, more numerous sites. The colocalization of AML-1B and RNA polymerase II0 suggests that nuclear matrix association of AML-1B is related to RNA polymerase II activity.
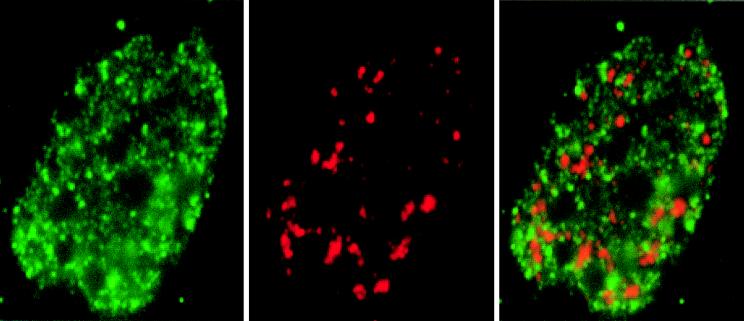
The foci labeled by fluorescent antibodies to AML-1B and polymerase II0 are distinct from the RNA processing domains. We compared the immunofluorescence pattern of AML-1B to that of SC-35, a spliceosome assembly factor that is associated with domains enriched in splicing factor (2, 18, 30). The results show that immunofluorescence signals for SC-35 and HA/AML-1B antibodies do not overlap (Fig. 4). Hence, AML-1B nuclear matrix domains clearly do not coincide with SC-35 domains.
Figure 4.
AML-1B does not colocalize with SC-35 RNA processing domain. SAOS cells were transfected with HA/AML-1B and evaluated by immunofluorescence analysis of the nuclear matrix in situ by using the HA antibody (green, AML-1B) and the SC-35 antibody (red, SC-35) that recognizes SC-35 RNA processing domain. In some cells, proximity of AML-1B and SC-35 domains is reflected by limited yellow staining.
Association of AML/CBFα to RNA Polymerase II Domains in the Nuclear Matrix Requires a Functional runt Homology DNA-Binding Domain.
In our earlier studies we have shown that the NMTS in AML-1B is by itself sufficient to insure the association of the factor with the nuclear matrix (24). However, auxiliary signals may be required for colocalization of AML with transcriptionally active RNA polymerase II0. The role of the runt homology domain in establishing effective interactions of AML-1B with RNA polymerase II0 at specific sites within the nucleus was assessed by analyzing the distribution of an AML-1B mutant (L148D). This mutant protein contains an amino acid substitution in the runt homology domain that abrogates recognition of gene-specific promoters. Fig. 2C shows the distribution of this mutant AML-1B protein. There is no longer overlap with the RNA polymerase II0 domains. Clearly, a functional runt homology domain is essential for AML-1B to colocalize with RNA polymerase II0. This result reinforces the proposal that promoter recognition plays an essential role in forming foci containing both AML-1B and active RNA polymerase II. Also, loss of AML-1B and RNA polymerase II0 colocalization occurs following inhibition of transcription with either actinomyocin D or dichlorobenzimidazole riboside, although the mechanism remains to be determined (data not shown).
DISCUSSION
Association of genes and cognate factors with the nuclear matrix may support the formation and/or activities of nuclear domains that facilitate transcriptional control (1–13). Our findings indicate that the association of a transcription factor with the nuclear matrix is obligatory for its activity. The results presented here provide insight into control of tissue-specific gene expression in situ. First, we show that active transcription is required for colocalization of AML-1B and RNA polymerase II at the nuclear matrix. Second, we show that the promoter-recognition function of the runt homology domain of AML-1B, and thus the consequential interactions with AML responsive genes is essential for formation of transcriptionally active foci containing AML and RNA polymerase II in the nuclear matrix.
Several sites on the nuclear matrix have been implicated in gene expression and DNA replication, including the nucleolus, RNA polymerase II-containing transcriptional domains, SC-35 domains, coiled bodies, and PML domains (16–19, 27, 28, 30). Furthermore, many regulatory and catalytic components of macromolecular complexes that mediate transcription and DNA replication are associated with the nuclear matrix (6–14, 31, 32). To function, transcription factors that concentrate in the nuclear matrix must colocalize with coactivators and RNA polymerase II at nuclear sites that support RNA synthesis. We propose that control of gene expression requires a molecular mechanism that targets regulatory proteins to specific spatial domains within the nucleus.
We have previously demonstrated that targeting of AML-1B to specific sites within the nucleus involves at least two trafficking signals, one of which mediates nuclear import and the other promotes association with the nuclear matrix (24). Analogous but distinct trafficking signals dictate association of proteins with DNA replication foci (31, 32). This multiplicity of determinants for intranuclear targeting presumably provides some of the requisite complexity to direct proteins to specific sites within the nucleus in response to diverse biological signals. In this study we have established that AML-1B and RNA polymerase II colocalize at defined nuclear foci. Apart from a nuclear localization signal and the NMTS of AML-1B, this colocalization requires a third molecular determinant, the runt homology domain. This domain is responsible for DNA binding and interacts with the CBFβ partner protein (22, 23). Our findings suggest that promoter recognition by nuclear matrix associated AML-1B, recruitment of RNA polymerase II0 and the ensuing formation of nascent RNA transcripts are functionally linked.
We note that association with the nuclear matrix is necessary but not sufficient to support AML-1B enhanced transcription. Only a subset of AML-1B is associated with RNA polymerase II0 in the nuclear matrix. Those AML sites that are not colocalized with RNA polymerase II0 may represent locations of AML responsive genes that are poised but not fully competent to be transcribed. Alternatively, these AML domains may represent inactive storage sites.
AML factors transactivate a broad spectrum of genes with different promoter organizations that must be regulated under diverse biological conditions (22, 23). Consistent with this concept, we find that both a previously identified C-terminal transactivation domain (aa 432–480) (24, 29) and the NMTS (aa 351–381) (24) support transactivation when associated with an appropriate promoter. However, there are key distinctions in the manner by which the NMTS (aa 351–381) and the C-terminal AML(432–480) region activate transcription. In contrast to the AML C-terminal region (432–480), the transcriptional activity of the NMTS depends on association with the nuclear matrix.
In summary, our data provide evidence that targeting of AML/CBFα transcription factors to the nuclear matrix is important for their function in gene transcription. Further studies will be necessary to identify components of the nuclear matrix that function as acceptor sites.
Acknowledgments
The authors thank Elizabeth Bronstein for help in preparation of the manuscript. These studies were supported by the National Institutes of Health (AR42262).
ABBREVIATIONS
- NMTS
nuclear matrix targeting signal
- CMV
cytomegalovirus
- HA
hemagglutinin
- AML
acute myelogenous leukemia.
References
- 1.Nickerson J A, Blencowe B J, Penman S. In: The Architectural Organization of Nuclear Metabolism. Berezney R, Jeon K W, editors. 162A. NY: Academic Press; 1995. pp. 67–123. [DOI] [PubMed] [Google Scholar]
- 2.Stein G S, Stein J L, Lian J B, van Wijnen A J, Montecino M. J Cell Biochem. 1996;62:198–209. doi: 10.1002/(SICI)1097-4644(199608)62:2%3C198::AID-JCB8%3E3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 3.Berezney R, Mortillaro M, Ma H, Meng C, Samarabandu J, Wei X, Somanathan S, Liou W S, Pan S J, Cheng P C. J Cell Biochem. 1996;62:223–226. doi: 10.1002/(SICI)1097-4644(199608)62:2%3C223::AID-JCB10%3E3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 4.Jackson D A. Mol Biol Rep. 1997;24:209–220. doi: 10.1023/a:1006873614521. [DOI] [PubMed] [Google Scholar]
- 5.Davie J R. Mol Biol Rep. 1997;24:197–207. doi: 10.1023/a:1006811817247. [DOI] [PubMed] [Google Scholar]
- 6.Guo B, Odgren P R, van Wijnen A J, Last T J, Nickerson J, Penman S, Lian J B, Stein J L, Stein G S. Proc Natl Acad Sci USA. 1995;92:10526–10530. doi: 10.1073/pnas.92.23.10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merriman H L, van Wijnen A J, Hiebert S, Bidwell J P, Fey E, Lian J, Stein J, Stein G S. Biochemistry. 1995;34:13125–13132. doi: 10.1021/bi00040a025. [DOI] [PubMed] [Google Scholar]
- 8.Lindenmuth D M, van Wijnen A J, Hiebert S, Stein J L, Lian J B, Stein G S. J Cell Biochem. 1997;66:123–132. [PubMed] [Google Scholar]
- 9.Grande M A, van der Kraan I, de Jong L, van Driel R. J Cell Sci. 1997;110:1781–1791. doi: 10.1242/jcs.110.15.1781. [DOI] [PubMed] [Google Scholar]
- 10.Alvarez M, Long H, Onyia J, Hock J, Xu W, Bidwell J. Endocrinology. 1997;138:482–489. doi: 10.1210/endo.138.1.4852. [DOI] [PubMed] [Google Scholar]
- 11.Chen H Y, Sun J M, Hendzel M J, Rattner J B, Davie J R. Biochem J. 1996;320:257–265. doi: 10.1042/bj3200257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nardozza T A, Quigley M M, Getzenberg R H. J Cell Biochem. 1996;61:467–477. doi: 10.1002/(SICI)1097-4644(19960601)61:3%3C467::AID-JCB14%3E3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 13.Spelsberg T C, Lauber A H, Sandhu N P, Subramaniam M. Recent Prog Horm Res. 1996;51:63–96. [PubMed] [Google Scholar]
- 14.Reyes J-C, Muchardt C, Yaniv M. J Cell Biol. 1997;137:263–274. doi: 10.1083/jcb.137.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tawfic S, Davis A T, Faust R A, Gapany M, Ahmed K. J Cell Biochem. 1997;64:499–504. [PubMed] [Google Scholar]
- 16.Iborra F J, Pombo A, Jackson D A, Cook P R. J Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- 17.Bregman D B, Du L, van der Zee S, Warren S L. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xing Y, Johnson C V, Moen P T J, McNeil J A, Lawrence J. J Cell Biol. 1995;131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grande M A, van der Kraan I, van Steensel B, Schul W, de The H, van der Voort H T, de Jong L, van Driel R. J Cell Biochem. 1996;63:280–291. doi: 10.1002/(sici)1097-4644(19961201)63:3<280::aid-jcb3>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 20.Banerjee C, McCabe L R, Choi J-Y, Hiebert S W, Stein J L, Stein G S, Lian J B. J Cell Biochem. 1997;66:1–8. doi: 10.1002/(sici)1097-4644(19970701)66:1<1::aid-jcb1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 21.Ducy P, Zhang R, Geoffroy V, Ridall A L, Karsenty G. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 22.Meyers S, Hiebert S W. Crit Rev Eukaryotic Gene Expression. 1995;5:365–383. doi: 10.1615/critreveukargeneexpr.v5.i3-4.70. [DOI] [PubMed] [Google Scholar]
- 23.Speck N A, Stacy T. Crit Rev Eukaryotic Gene Expression. 1995;5:337–364. doi: 10.1615/critreveukargeneexpr.v5.i3-4.60. [DOI] [PubMed] [Google Scholar]
- 24.Zeng C, van Wijnen A J, Stein J L, Meyers S, Sun W, Shopland L, Lawrence J B, Penman S, Lian J B, Stein G S, et al. Proc Natl Acad Sci USA. 1997;94:6746–6751. doi: 10.1073/pnas.94.13.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fey E G, Wan K M, Penman S. J Cell Biol. 1984;98:1973–1984. doi: 10.1083/jcb.98.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortillaro M J, Blencowe B J, Wei X, Nakayasu H, Du L, Warren S L, Sharp P A, Berezney R. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wansink D G, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wansink D G, Manders E E, van der Kraan I, Aten J A, van Driel R, de Jong L. J Cell Sci. 1994;107:1449–1456. doi: 10.1242/jcs.107.6.1449. [DOI] [PubMed] [Google Scholar]
- 29.Hiebert S W, Sun W, Davis J N, Golub T, Shurtleff S, Buijs A, Downing J R, Grosveld G, Roussell M F, Gilliland D G, et al. Mol Cell Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jimenez-Garcia L F, Spector D L. Cell. 1993;73:47–59. doi: 10.1016/0092-8674(93)90159-n. [DOI] [PubMed] [Google Scholar]
- 31.Leonhardt H, Cardoso M C. Int Rev Cytol. 1995;162B:303–335. doi: 10.1016/s0074-7696(08)62620-0. [DOI] [PubMed] [Google Scholar]
- 32.Leonhardt H, Page A W, Weier H U, Bestor T H. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]