Abstract
The production of new chemicals for industrial or therapeutic applications exceeds our ability to generate experimental data on their biological fate once they are released into the environment. Typically, mixtures of organic pollutants are freed into a variety of sites inhabited by diverse microorganisms, which structure complex multispecies metabolic networks. A machine learning approach has been instrumental to expose a correlation between the frequency of 149 atomic triads (chemotopes) common in organo-chemical compounds and the global capacity of microorganisms to metabolise them. Depending on the type of environmental fate defined, the system can correctly predict the biodegradative outcome for 73–87% of compounds. This system is available to the community as a web server (http://www.pdg.cnb.uam.es/BDPSERVER). The application of this predictive tool to chemical species released into the environment provides an early instrument for tentatively classifying the compounds as biodegradable or recalcitrant. Automated surveys of lists of industrial chemicals currently employed in large quantities revealed that herbicides are the group of functional molecules more difficult to recycle into the biosphere through the inclusive microbial metabolism.
Keywords: BDPServer, biodegradation, chemotopes, machine learning, REACH
Introduction
The number of new molecules generated by the chemical and pharmaceutical industry has boomed in the last few years owing to the emergence of combinatorial chemistry along with the demand for novel industrial, agricultural and therapeutic products (Dolle, 2004). The number of natural or man-made organic compounds present in the biosphere is somewhere between 8 and 16 million molecular species, of which as many as 40 000 are predominant in our daily lives (Hou et al, 2003). Microorganisms are key players in determining the environmental fate of novel compounds because they can be used as carbon and energy sources (Mishra et al, 2001). Microbial metabolism may not only cause the complete elimination of a given chemical compound but it can also generate chemical species that are as toxic or as persistent as the original ones. In the case of complete metabolism, microbial biodegradation can be exploited for waste treatment and used in directed bioremediation processes in situ or ex situ (Diaz, 2004). Therefore, knowing whether a novel chemical compound is likely to be metabolised by microorganisms is crucial for assessing the environmental risks associated to its production, transportation, utilisation and disposal (Wackett and Ellis, 1999; Wackett, 2004b). However, after 50 years of research on microbial biodegradation, detailed knowledge about biodegradative pathways is available for only about 900 chemical species (Urbance et al, 2003; Ellis et al, 2006). New pesticides and pharmaceuticals are being produced at rates that cannot be matched by experimental attempts to determine the outcome when spilled or released into the environment. This makes essential to develop systems that can predict the fate of chemical compounds (Wackett and Hershberger, 2001; Wackett, 2004b) before experimentally assessing the capacity of the microbiota to degrade them.
Although hydrophobicity, water solubility and the presence of xenophores (Klopman et al, 1992; Wackett and Ellis, 1999; Hou et al, 2003) have been invoked for assessing the biodegradability of given compounds, there are many examples in which the presence/absence of certain functional groups do not match the experimental results. As an alternative, we have approached the problem of predicting the environmental fate of chemical species from an experience-based perspective, using a (micro)biological logic rather than a purely (bio)chemical appraisal, for example, making the most out of available information about known microbial catabolic reactions on organic pollutants. To this end, we have exploited the wealth of knowledge on the genetic and biochemical basis of microbial metabolism available at the University of Minnesota Biodegradation and Biocatalysis Database (UMBBD; Ellis et al, 2003, 2006) and the Biodegrative Strain Database of the Michigan State University (BSD; Urbance et al, 2003) to train a rule-based classification system (Quinlan, 1993) for detecting the association between certain chemical compound descriptors and environmental fates. Such descriptors are based on the deconstruction of chemical structures in atomic triads (also referred to as chemotopes). A machine learning system (Quinlan, 1993) was then used to identify explicit rules that associate compound vectors to environmental fates as inferred from the analysis of the metabolic network that represents the global biodegradative potential of microorganisms. Finally, a scheme to predict the fate of new chemical compounds, using the previously identified rules, was implemented as a web server. The results obtained include the evaluation of the prediction capacity of the system and its application to several sets of compounds provided by the European Chemicals Bureau or obtained from the database PubChem Compound—for most of which there are no data on their biological fate. Herbicides seem to be the group of functional molecules that have less favourable prospects of recycling through the global microbial biodegradation network.
Results
Deconstructing organic chemicals into atomic triad-based compound vectors
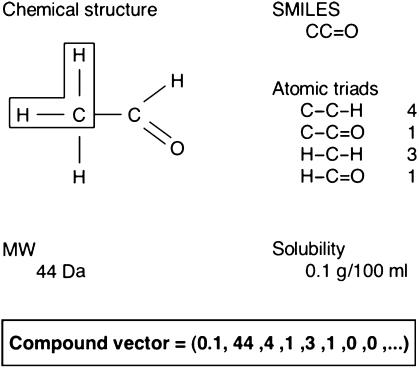
At the time of starting this work, the UMBBD contained information on 850 compounds and 903 reactions (Ellis et al, 2003, 2006). The first issue at stake was whether structural features of the target molecules could be significantly correlated to their known environmental fate. To this end, we resorted to describing each chemical structure as a whole of 152 descriptors that represented atomic triad frequencies, molecular weight (MW) and water solubility, the latter expressed both quantitatively and qualitatively. Such atomic triads (or chemotopes) included 149 groups of three consecutive, connected atoms that can be identified on the structure of a compound, taking into account the type of connecting chemical bonds. For example, the atomic triad C–C–H is different from C=C–H, whereas C=C–H is equal to H–C=C (Figure 1). The choice of atomics triads instead of focusing on reactive groups or functional motives reflected the tradeoff between having significant structural information and the handling of a minimal number of attributes (see the Discussion section). Deconstruction of each compound in this way is achieved by first translating the SMILES (Weininger, 1988) representation of each molecule, which is available from UMBBD, into other forms of chemical depiction that include explicit information regarding atom connectivity and chemical bond types. Then, the frequency in which the different atomic triads appear for each compound is recorded. MW is also available from UMBDD and compound solubility is, in some cases, accessible through links to the corresponding entry in ChemFinder (Figure 2A). The collection of atomic triad frequencies, the MW and the solubility were then assembled to generate molecular descriptors, henceforth referred to as compound vectors (Figures 1 and 2A).
Figure 1.
Deconstruction of acetaldehyde into its constituent atomic triads (chemotopes). The figure shows a simple example of generation of the compound vectors mentioned in the text. To this end, the chemical structure of acetaldehyde is shown along with its corresponding SMILES string and its composition in terms of atomic triads. One instance of the atomic triad H–C–H is boxed on the chemical structure of the molecule. The vector representing the properties of acetaldehyde regarding its degradability is assembled from its solubility, MW and the corresponding set of atomic triad frequencies as indicated.
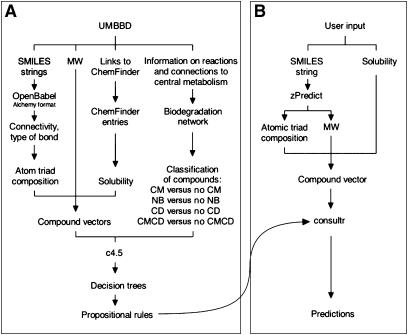
Figure 2.
Rationale for developing an experience-based biodegradation prediction system. (A) represents the strategy to generate environmental fate classifiers with the learning machine c4.5, in the form of sets of propositional rules, starting from information gathered from the Biodegradation database UMBBD. (B) Sketches the functioning and queries of BDPServer.
Through these criteria, nine compounds out of the 850 listed were not associated to any vector because they had less than three atoms or because their entries in UMBBD did not include SMILES strings. About 718 distinct vectors represented the remaining set of 841 compounds, indicating that the correspondence between compounds and vectors is not equipotent. A one-to-one relation between compounds and vectors existed for 625 compounds, whereas 93 vectors described the remaining 216 compounds. Many-to-one relations between compounds and vectors is explained by the fact that positional isomers in which functional groups have changed between equivalent positions may share the same pattern of atomic triad frequencies even if they do have different connectivity and different SMILES strings. That is the case for pyrogallol versus phloroglucinol, and also the case of 2-formil-1-indanone versus 1-formil-2-indanone. In addition, as stereoisomers have the same atomic connectivity and identical composition in terms of atomic triads, they are encoded by the same vectors. This kind of information was expected to enter some noise in the predictive system, although (as explained below) not as important as one could anticipate. Description of chemicals as compound vectors of this sort (Figures 1 and 2A) was used to feed the training algorithm for classification of the molecule according to its fate in the global network shaped by the global microbial metabolism (see below).
Classification of compounds according to their environmental fate
Once each compound had been expressed in a vector form, the reactions in which the chemical is known to take part, as a substrate or as product, were retrieved from the database (Ellis et al, 2003, 2006; Pazos et al, 2005). To categorise the environmental outcome of the complete list of 850 chemical species under study, we exploited all known metabolic reactions for organic chemicals (independent of their specific bacterial host) to delineate a global network of microbial catalysis (Pazos et al, 2003). Such an inclusive biodegradation network has been described before as an entity with topological properties that resemble single-cell metabolic transactions. Although a network of this kind includes interconnected pathways that may not stand alone in a single organism (MacNaughton et al, 1999; Pelz et al, 1999; Whiteley and Bailey, 2000; Koizumi et al, 2002; Dennis et al, 2003; Zhou, 2003), it does represent the known biodegradative potential of microbial communities at a global scale (Pazos et al, 2003). Such a pooled biodegradation network (Pazos et al, 2003, 2005) was employed to pinpoint the channelling of every compound into one of three final destinations (Figure 3) as follows.
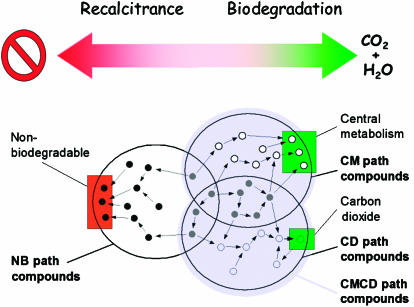
Figure 3.
Categorisation of the three partially overlapping sets of metabolic pathways that form the global biodegradation network. The sets of chemicals and their metabolic products were defined according to their final environmental sinks: (i) NBs, chemicals that cannot be degraded (nonbiodegradable) and metabolic precursors of molecules that cannot be degraded; (ii) CMs, chemicals that belong to the central metabolism and precursors that are biologically processed to central metabolites; (iii) CDs, molecules that are directly channelled to production of CO2; (iv) CMCDs, the sum of CMs and CDs. The general trend of these types of compounds towards recalcitrance or biodegradation is sketched on top.
The first sink was composed of 38 compound entries in UMBBD that were annotated as belonging to the central metabolism. We extended this category of chemicals by including all molecules that participated in pathways through the network leading them to the central metabolism. In this way, a group of 533 chemical species were defined as central metabolism path compounds (CMs). On the other hand, we labelled as recalcitrant, nonbiodegradable compounds those that do not participate as substrates in any reaction documented in UMBBD, and thus can never reach the central metabolism or being biodegraded otherwise. After scrutiny of the global biodegradation network, 108 compounds of the database unequivocally fulfilled that criterion. In addition, two pairs of somewhat special compounds (arsenate/arsenite, benzyldisulphide/benzylmercaptane) that were linked by bidirectional reactions but had no other outgoing connections were also classified as nonbiodegradable. The operative list of recalcitrant compounds included, therefore, 112 compounds. Yet, as before, we extended the nonbiodegradable whole to those molecular species that were directly or indirectly connected to recalcitrant compounds as precursors of ultimately intractable chemicals (Figure 3). The extended set of such molecules included 353 specimens, which were operatively tagged as nonbiodegradable path compounds (NB). This set of compounds did overlap by 112 compounds with the previously defined set of CM compounds. This indicated that many chemicals can either be degraded upon being channelled into the central metabolism or accumulated in the environment if diverted into nonproductive reactions.
The nonredundant set that contained all CM and NB compounds included 774 chemical species. The 76 remaining molecules were not connected to either central metabolism or nonbiodegradable compounds. Instead, they belong to various pathways that go straight into carbon dioxide and water, without converting into any of the typical intermediates of the central metabolism. Although they are of course biodegradable, the lack of connections to the central metabolism rules out their classification as CMs. On the other hand, they cannot be classified as NB compounds either, as CO2 is not a bona fide recalcitrant, terminal molecule: it can be captured back to metabolism by a formylmethanofuran dehydrogenase reaction or (in practice) by many other CO2-fixing microbial processes. We thus established a separate, extended type of compounds, which were directly or indirectly positioned in pathways leading to CO2. This group, which includes 329 molecular specimens, was termed as carbon dioxide path compounds (CDs). One further extension of this criterion was to take CO2 and central metabolism as the same final fate, and group all compounds connected to them. The resulting set thus comprises central metabolism and carbon dioxide path compounds (CMCDs) and includes 634 chemicals. CMCDs correspond to what can be considered intuitively as the set of biodegradable compounds. In summary, as shown in Figure 3, each compound can be ascribed to each of three environmental fates (CM, NB and CDs), in which the sum of CMs and CDs forms the operative biodegradable (CMCD) category.
The four types of biodegradative fates (CM, NB, CD and CMCD, Figure 3) did overlap to a significant extent. To refine further the sorting of the chemicals and to generate better classifiers for the compounds, we established four separate, binary categorisation schemes that would label out each chemical as belonging to each of the groups or to the cognate negated classes. Accordingly, we defined the following four classification classes: (i) CM or No CM, (ii) NB or No NB, (iii) CD or No CD and (iv) CMCD or No CMCD. Obviously, the most important categorisation for our purposes is the last (CMCD or no CMCD), as it reflects either eventual recalcitrance or amenability to biological recycling. Yet, the other classifiers do hold a considerable practical value as well (see the Discussion section).
Matching compound vectors to environmental fates
Once a vectorial description of each chemical of the UMBB had been established and a clear classification of outcomes through the global metabolism delineated, we set out to discover relationships between them. As mentioned above, one early difficulty to this end is that one-to-one relations between compounds and vectors existed for only 625 compounds, whereas 93 vectors redundantly described the remaining 216 compounds. A similar scenario occurs with stereoisomers, which are encoded by the same vector compound but may differ in their accessibility to biodegradation. To assess the importance of these cases in the global process, we determined the number of instances in which compounds that share the same frequencies of atomic triads happen to have the same environmental fate. Starting with the 216 compounds that were associated to 93 vectors, we identified all possible pairs of compounds that had the same pattern of atomic triad distribution. Out of the resulting 163 cases, the number of pairs consisting of two compounds with identical fate in the different classification schemes, was as follows: 141 (87%) for CM or No CM; 126 (77%) for NB or No NB; 111 (68%) for CD or No CD; and 142 (87%) for CMCD or No CMCD. These results indicated that in most cases (average, 80%), the environmental fate of structural isomers and stereoisomers after passing them through the global biodegradation network is the same—although in some cases, the specific reactions involved might be different.
A second consideration was related to the structural similarity between the different types of compounds. One could suspect that chemicals belonging to the same group (CM, NB, CD, CMCD, or the corresponding negated classes) might share some structural features, especially if they are part of the same metabolic pathway. To examine rigorously this issue, chemical compound similarity was estimated for each pair of compounds using their atomic triad frequencies for calculating a modified version of the Tanimoto association coefficient τ (Holliday et al, 2002). This coefficient reflects the ratio between the number of atomic triads that two compounds have in common and the number of atomic triads that they do not have in common, and can be used as a measure of the distance between compounds, in respect to their chemical similarity. The distribution of such distances for the whole of compound pairs (Supplementary Figure S1A), indicated that the collection of chemicals was quite diverse. Although 90% of the pairs had τ values <50, only 1% of the pairs had τ figures ⩾80. When the distribution of distances was calculated for pairs of compounds that belong to the same classes of environmental fates, we found that all groups were equally diverse (data not shown). The degree of clusterisation, however, varied among the different groups, as measured by their average clustering coefficient (Cv; Supplementary Figure S1B).
Production of classifiers
Having categorised the compounds according to the four binary classification schemes mentioned above, and having defined compound vectors that describe their composition, topology, MW and solubility, the machine learning algorithm c4.5 (Quinlan, 1993) was used to generate classifiers in the form of sets of rules. This program uses an inductive decision tree process that generates classification schemes matching the attributes of the training examples to given classes. These schemes can later be used for assigning new (unseen) examples to such classes (Figure 2B). Each classification involves a tree structure, which can be also be expressed as a set of rules, in which internal nodes represent a test condition (formulated in terms of the attributes), whereas the terminal (leaf) nodes represent classes. Such an approach was preferred over other available learning methods, for example, neural networks, because (i) the machine learning of choice can handle missing values (such as water solubility, unknown for some compounds), and (ii) the explicit rules created by c4.5 can be easily interpreted by the user. In addition, such rules produce generalised models that become instrumental to make predictions on molecules not previously visited by the learning machine. Rules take the form of propositions with two sides: the left-hand side contains a conjunction of attribute-based tests, and the right-hand side is a class. For example, the rule in Table I states that a compound with more than 19 triads of the type C–C–C, more than one triad of the type O–C–C and three or less triads of the type O–C–C, should belong to the NB class, with a support confidence of 94%.
Table 1.
Example of propositional rule generated for the classification of compounds in the scheme NB or No NB
| Rule 55: IF | –C–C–C >19 | |
| –O–C–C >1 | ||
| –O–C–C ⩽3 | ||
| THEN, the compound belongs to the NB class (Confidence 90.6%) Examples (14 cases) | ||
| No. |
Class |
Compound |
| 451 | NB | 1-Methoxyphenanthrene |
| 454 | NB | 9-Phenanthrol |
| 389 | NB | 9-Fluorenol |
| 535 | NB | 1-Phenanthrylsulfate |
| 493 | NB | 4-Phenanthrol |
| 525 | NB | 2-Phenanthrol |
| 513 | NB | 2,2′-Biphenyldimethanol |
| 539 | NB | 4-Phenanthrylsulfate |
| 538 | NB | 3-Phenanthrylsulfate |
| 537 | NB | 2-Phenanthrylsulfate |
| 529 | NB | 9-Phenanthrylsulfate |
| 494 | NB | 3-Phenanthrol |
| 450 | NB | 1-Phenanthrol |
| 390 | NB | 9-Fluorenone |
NB, nonbiodegradable path compound.
Each of the four final classifiers was composed of a set of 16–23 rules, and each rule was composed, in average, of 3.3 attribute-based tests (standard deviation 2.07, range 1–12). To gain some insight on the relationship between chemical structure (i.e., the frequency of triplets) and environmental fate, the rules were reanalysed to assess the weight of each of the attributes. Out of such 152 traits (149 frequencies of atomic triads, MW, quantitative solubility and qualitative solubility), only 52 were included as part of the propositional rules of all classifiers. These attributes are listed in Supplementary Figure S2, together with a graphical depiction of the frequencies in which each of them appears in the rules. For example, attribute-based tests referring to the frequency of atomic triad O–C=O come out in about 45% of the rules that conform the classifier for the scheme CD or No CD. Also, although MW and solubility are taken into account by the classifier NB or No NB, they are useless for the classifier CM or No CM, (Supplementary Figure S2). This reflects that not all attributes have the same importance for each of the environmental fates.
To assess the predictive capacity of the system, we followed a fivefold cross-validation strategy. For this, the data set was divided into five blocks; four of them were used as a training set, to generate the classifiers (rules), and the remaining block was used as a test set. This allowed measuring the ability of the classifiers for predicting the environmental fate of chemicals not included in the training set. The process was repeated five times, changing the block that was used as a test set. The accuracy of the system (i.e., the percentage of compounds correctly classified as belonging to any of the NB, CM, CD, CMCD classes, or their negation) was averaged for each classification scheme. The resulting averaged accuracies ranged from 73 to 87%, for the different classification schemes (Table II). A more detailed picture of the predictive capacity of the system was obtained by calculating its sensitivity and specificity for the prediction of specific classes. The values for sensitivity (fraction of compounds correctly classified as belonging to a specific class, relative to the total number of cases of that particular class) and specificity (fraction of compounds correctly classified as belonging to a specific class, relative to the total number of predictions for that class) ranged from 50 to 93%, and from 66 to 85%, respectively, in the different classification schemes (Table II). In general, the classification scheme CM or No CM is the one for which the best predictive performance was achieved. This could be explained in part by the fact that CM is the group with the highest average clustering coefficient, Cv, making it easier for the c4.5 algorithm to generate rules that represent the more similar compounds of the group (Supplementary Figure S1B). Consistently with this explanation, we have observed that the relationship between the average clustering coefficient and the sensitivity of the predictions for any given class can be adjusted to a regression line with a correlation coefficient r=0.53 (Supplementary Figure S1B).
Table 2.
Predictive performance in fivefold cross-validation experiments
| Classification schemea | CM or No CM | NB or No NB | CD or No CD | CMCD or No CMCD |
|---|---|---|---|---|
| Accuracy (%) | 87±4 | 77±4 | 73±5 | 82±3 |
| Significance respect to random—P(N) | 1.1 × 10−39 | 1.2 × 10−24 | 2.4 × 10−16 | 6.4 × 10−26 |
| Default class | CM | No NB | No CD | CMCD |
| Majority class | CM | No NB | No CD | CMCD |
| No. of cases | 533 | 496 | 513 | 634 |
| Sensitivity (%) | 93±4 | 86±4 | 78±7 | 92±3 |
| Specificity (%) | 83±3 | 78±2 | 78±3 | 85±2 |
| Minority class | ||||
| No. of cases | 308 | 345 | 328 | 207 |
| Sensitivity (%) | 67±5 | 64±5 | 66±9 | 50±11 |
| Specificity (%) | 85±8 | 77±4 | 66±5 | 71±5 |
CM, central metabolism path compound; NB, nonbiodegradable path compound; CD, carbon dioxide path compound; CMCD, central metabolism and carbon dioxide path compounds. Accuracy is the percentage of compounds correctly classified. Sensitivity is the percentage of compounds correctly classified as belonging to a specific class, relative to the total number of cases of that particular class. Specificity is the percentage of compounds correctly classified as belonging to a specific class, relative to the total number of predictions for that particular class. Accuracy, sensitivity and specificity are indicated in the Table as the average±s.d. for the five iterations in the cross-validation experiment. The statistical significance of the observed difference in the performance of the c4.5-based system compared with a random prediction is indicated by the P(N) of a sign test.
Evaluation of classifiers and confirmation of their predictive value
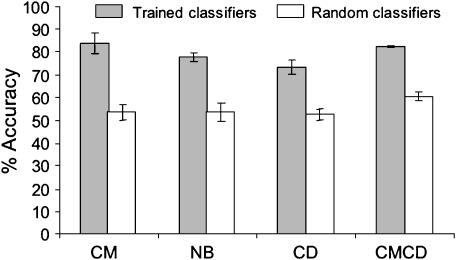
To authenticate the significance of the figures generated above and uncover possible biases of the dataset on the predictive value of the classifiers, we compared the performance of our c4.5-based system with one employing random predictors. Possible biases due to overrepresentation of classes were corrected by having such predictors assigning compounds randomly to one of the two fates for each classification scheme, with a probability that was proportional to the population of each of the classes. Figure 4 shows the average accuracy obtained for the different classes (versus their negated classes) for the real and the randomised dataset. It can be seen that the real dataset produces a considerably higher accuracy for all the classes, which is more pronounced in the CM and NB groups. Details of the analyses of the randomised dataset (sensitivity or specificity) are shown in the Supplementary Figure S3. To assess the statistical significance of the differences between the c4.5-based system and the random predictors, the dataset was subject to a sign test. This analysis compares the performance of two methods based on the number of cases that one of them provides a correct response, whereas the other fails and vice versa. A P(N) is thereby obtained which can be interpreted as the probability for the null–hypothesis, that is that the observed differences are happening by chance. These probabilities are shown in Table II. The result clearly demonstrates the superiority of the c4.5-based predictors as compared to their equivalent random counterparts with values of P(N) in the order 10−16–10−39.
Figure 4.
Comparison of the prediction accuracies in cross-validation tests with trained classifiers and random classifiers. Fivefold cross-validation tests were conducted, for each of the considered classification schemes, using both the original classifiers and the equivalent random classifiers, which assign compounds arbitrarily to classes with a probability that is proportional to the size of the classes (Table II for the statistical significance of these differences between the predictors and their random counterparts). The averaged accuracy of the five iterations of the cross-validation experiment and the corresponding standard deviation are represented. Note that the dataset was extracted from UMBBD, which is overrepresented with biodegradable compounds. This makes the accuracy of the predictive scheme reflected in the figure (as well as the related specificities and sensitivities; Supplementary Figure S3) to be an underestimation of the actual prognostic power of the system for new chemicals.
Once the predictive capacity of the system had been established in the cross-validation experiments above, we set out to obtain the final components of the prediction system. To this end, new rule-based classification models were generated from training sets that included all compounds. Because they had been trained with a larger data set than the preliminary classifiers obtained in cross-validation experiments, it could be expected that the final classifiers had a higher capacity to predict the environmental fate of new compounds, as long as the new compounds do not diverge too much from the models.
To compare the efficacy of our predictive system with experimental data, we made use of an additional set of compounds that had not been included as part of the training set as they were not available at the time of its setup. To this end, we took the 147 compounds entered in the UMBBD along with fresh information on its biodegradability from 17 November 2003 (when the predictive system was first set) to 27 November 2006. As before, the structures of these new compounds were translated into SMILES formats, used to generate compound vectors and fed as inputs for each of the four classifiers established previously. The complete set of biochemical reactions involved in their biodegradation (as of 27 November 2006) was collected as well, thereby allowing a refinement of the global biodegradation network (Pazos et al, 2003) and the assignment of the corresponding metabolic sink. Table III compares the actual classification of each of the compounds to the predictions emitted by the system. The accuracy values for the four classification schemes ranged from 40 to 69%, lower than those of the fivefold cross-validation tests reported above with the original dataset (73–87%). However, the sensitivity values for the major classes ranged from 72 to 91%, which are comparable to those of the cross-validation experiment (78–93%). The best accuracy value was that for the classification scheme CMCD or no CMCD (69%). Consistently with this, the prediction of compounds that belong to the CMCD class achieved the highest sensitivity value, 91% of the compounds actually belonging to this category being classified as such. The least sensitivity was associated to the prediction of compounds belonging to No CM. Only 16% of these compounds were predicted as such. This is not unexpected, as this set of compounds is very heterogeneous, holding the lowest clustering coefficient (Supplementary Figure S1B).
Table 3.
Predictive performance in validation tests with recent experimental dataa
| Classification schemeb | CM or No CM | NB or No NB | CD or No CD | CMCD or No CMCD |
|---|---|---|---|---|
| Accuracy (%) | 58 | 40 | 51 | 69 |
| Default class | CM | No NB | No CD | CMCD |
| Majority class | CM | No NB | No CD | CMCD |
| Sensitivity (%) | 78 | 72 | 75 | 91 |
| Specificity (%) | 66 | 16 | 49 | 71 |
| Minority class | No CM | NB | CD | No CMCD |
| Sensitivity (%) | 16 | 35 | 30 | 22 |
| Specificity (%) | 27 | 88 | 58 | 57 |
Data on 147 compounds entered in the UMBBD between 17 November 2003 and 27 November 2006 along with the set of biochemical reactions involved in their biodegradation.
CM, NB, CD, CMCD, accuracy, sensitivity, specificity: as in footnote to Table II.
Implementation of the prediction system: the BDPServer
To put the prediction system into operation as a user-friendly resource, it was implemented as a public web server called Biodegradation Prediction Server (BDPServer, http://www.pdg.cnb.uam.es/BDPSERVER). The input for the BDPServer (Figure 2B) requires the expression of the formula of the chemical under study in SMILES format, although an integrated Java applet allows the user to draw the chemical structures directly, instead of typing SMILES strings. Quantitative and qualitative solubility information can also be entered (Supplementary Table S1), but it is optional because the learning machine c4.5 can deal with missing values. The prediction engine within the BDPServer (a Perl script called zPredict, Figure 2B) uses OpenBabel for adding hydrogen bonds and translating SMILES strings into BS and Alchemy formatted reports. The connectivity between atoms is extracted from such reports, which includes the type of chemical bonds between atom pairs. zPredict then calculates the composition of each compound in terms of atomic triads. OpenBabel is also used to calculate the MW of compounds. A vector that conveys the compound descriptors is then generated. These include atomic triad frequencies, solubility (if provided by the user), and MW. Predictions are generated by the consultr module of the c4.5 package, using the classification models mentioned above. The output of the system consists of four independent predictions, associated to confidence factors that are calculated by consultr. If a user-provided SMILES string happens to match another string in the database, the server returns the actual classification of the compound, and predictions are not shown unless the user chooses to force the forecast.
An example of how the BDPServer works and its predictive ability operates is shown in the exercise summarised in the Supplementary Figure S4. In this case, we set out to classify a collection of compounds that belong to well-characterised microbial pathways for biodegradation of toluene—which were part of the training data. Such pathways, which include a total of 42 different compounds, were taken from the MetaRouter web server (Pazos et al, 2005) by querying the database for all reactions connecting toluene to the central bacterial metabolism. SMILES strings of each of the 42 chemicals were submitted to the BDPServer and the predictions compared to the actual classification of the compounds, according to the four binary schemes defined previously. The number of compounds that belong to each of the classes, and the absolute number of successful BDPServer predictions, within each of the classification schemes, are shown in Supplementary Figure S4. All compounds were correctly categorised for the classification schemes CM or No CM and CMCD or No CMCD. Suboptimal—but still significant—results were obtained for the classes CD or No CD, and NB or No NB in which 37 out of 42 and 40 out of 42 compounds, respectively, were classified correctly. Given the fact that the pathways for degradation of toluene were included in the training of the system, it is likely that these figures overestimate the capacity of the system. Yet, they represent the type of result and error margin that one would expect from the analysis of compounds that participate in full metabolic routes.
An additional exercise was designed to test the ability of the system for recognising compounds that are clearly linked to the central metabolism. To this end, the KEGG database was used to generate a collection of 733 molecules that fulfilled the following conditions: (i) they were components of Escherichia coli metabolic pathways; (ii) they were not part of metabolic pathways involving xenobiotic or recalcitrant compounds; (iii) they were composed of more than three atoms; and (iv) they had an associated description in MOL format that could be converted into SMILES format. The BDPServer was then used to generate biodegradability predictions for such compound set. As the result of this, the BDPServer predicted that 662 KEGG compounds (90.31%) could be assigned to the CMCD class (central metabolism plus carbon dioxide sinks), that is, the class that defines the group of bona fide biodegradable chemicals. This figure is consistent with the prediction that 597 (81.44%) out of the 733 KEGG compounds belong to the class of chemicals not connected to recalcitrant compounds (No NB class) and that 501 molecules (68.34%) could be directed to central metabolism (CM). As a control, random predictions were generated as above, that is, assigning compounds arbitrarily to one of the two fates for each classification scheme, proportionally to the population of each of the classes in the original training set. The accuracies for these random predictors are 76.8% for CMCD, 58.66 for No NB and 60.84 for CM. These differences in performance are statistically supported by the associated values of P(N): 1.52 × 10−11 (CMCD), 9.45 × 10−20 (No NB) and 3.3 × 10−3 (CD).
Early diagnosis of degradability of new chemicals
With the tools described above in hand, and after having evaluated the reliability of the system in different sets of chemicals with a known biodegradative fate, we set out to produce global predictions for compounds found in lists that are subject to regulations through the European Chemical Bureau. Such lists include (i) 3365 dangerous substances incorporated to directive 67/548 of the European Commission, which regulates the classification, packaging and labelling of hazardous chemicals, last updated in April 2004 (the so-called Annex-I); (ii) 2747 High Production Volume Chemicals (HPVCs, defined by directive 793/93 as molecules that are produced or imported in quantities exceeding 1000 tons per year); and (iii) 7829 Low Production Volume Chemicals (LPVCs, between 10 and 1000 tons per year). As each of the three catalogs contain many substances of poorly defined composition (such as petroleum derivatives) that cannot be analysed by our system, we filtered the lists with the SMILECAS database, to obtain refined inventories of defined compounds with associated SMILES strings. The curated lists contained 1766, 1653 and 5645 compounds, respectively. The overlap between those from Annex-I with HPVCs and LPVCs included 595 compounds in one case and 366 chemicals in the other. Upon blind testing of such molecules with the BDPServer (Supplementary Table S2), about 5% of these compounds were automatically rejected because they had less than three atoms or because their SMILES entry was not correctly interpreted by OpenBabel. Yet, the system predicted that, for any of the three lists, ∼60% of the compounds would be connected to central metabolism (CM), ∼20% would be linked to carbon dioxide (CD) and ∼70% would be connected to either central metabolism or carbon dioxide (CMCD). Therefore, more than two thirds of the compounds could be in principle biodegraded. However, about 47% of the compounds of any list were either recalcitrant or could evolve into nonbiodegradable compounds (NB). The highest percentage of NBs (55%) was found in the subset of compounds that are part of the curated Annex-1 but not of HPVCs (data not shown).
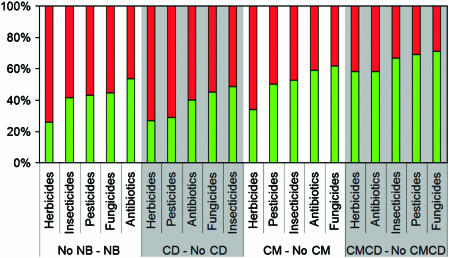
In a subsequent step, we analysed sets of chemical species of the PubChem database that were explicitly labelled as pesticides (1707 compounds), herbicides (199), fungicides (169), insecticides (279), antibiotics (1365) and flame retardants (6). As indicated in Figure 5, our system exposed that the percentage of CM (∼54%) or CMCD (∼70%) compounds within these lists were roughly similar to those of the species listed by the European Chemical Bureau. However, the percentage of compounds connected to CO2 (CD, ∼34%) and to nonbiodegradable end products (NBs, ∼59%) was significantly higher. The highest percentage of predicted difficult-to-degrade compounds was observed in the collection of herbicides included in PubChem (NB ∼74%). The much feared flame retardants (Darnerud, 2003) incorporated in the study turned out to be in principle amenable to microbial degradation leading to central metabolism or carbon dioxide (CMCD, 100%), although four of them were also classified as precursors of eventually nonbiodegradable compounds (NB, ∼66%). The detailed predictions for all the sets of compounds mentioned in this section are available on-line at http://www.pdg.cnb.uam.es/BDPSERVER.
Figure 5.
Prediction of environmental fates of selected groups of functional chemical compounds extracted from the PubChem Compound database. The y axis indicate the percentage of compounds within each category (versus the corresponding negated class) of the lists that are predicted to belong to any of the classes: CM, central metabolism path compound; NB, nonbiodegradable path compounds; CD, carbon dioxide path compound; CMCDs, central metabolism and carbon dioxide path compounds. The red coloured is a signal of recalcitrance, whereas the green is an indication of degradability. The category of flame retardants was excluded from this analysis, because only a few (six chemical specimens) were listed in the accessed dabases. Note that, by any criterion, the herbicides form the most-difficult-to-degrade group of chemicals.
Discussion
The growing production of new chemicals make the early diagnoses of their environmental fate and their microbial metabolism necessary (McShan et al, 2003; Wackett, 2004b). Previous attempts to predict biodegradability (for instance, UMBBDpredict) have focused on the identification of specific metabolic pathways that a compound might follow on the basis of the presence of predefined functional groups (Ellis et al, 2003; Hou et al, 2004). The user must choose one of the possible transformations to generate, iteratively, a virtual degradation route. Such a manual, iterative approach lengthens the procedure if a large number of chemicals are being analysed. In addition, strategies that focus on functional groups have the limitation of being restricted to predefined structures that have been manually collected. Other schemes such as Meteor and Catabol evaluate only the pathways that are most likely to occur, instead of predicting all possible pathways. In these cases, transformation rule libraries have to be constructed manually from the literature and generalised through chemical criteria (Dimitrov et al, 2002; Button et al, 2003).
As an alternative, we have tackled the problem from an experience-based perspective, using a computational machine learning approach trained with all known microbial catabolic reactions on organic pollutants (Urbance et al, 2003; Ellis et al, 2006). This approach allows the combination of continuous and discrete attributes (descriptors), permits dealing with missing values, and generates classification rules in human-readable forms endowed with biological meaning. This is an important difference with other machine learning techniques (such as neural networks), which generate classifications with opaque black box rules. The application of such predictive system to lists of chemicals released into the environment thus represents an early tool for tentatively classifying the compounds as biodegradable or recalcitrant. The pivotal feature of our predictive system is the vectorial representation of chemical compounds as sets of 152 descriptors that express atomic composition and topology in terms of atomic triad frequencies (chemotopes), plus MW and water solubility. This approach is related to some QSAR-type systems that use, as chemical descriptors, all possible subfragments of connected atoms that can be obtained from a compound (Damborsky, 1996; Damborsky and Schultz, 1997; Tong et al, 1998; Dimitrov et al, 2002; Blinova et al, 2003; Sutherland et al, 2004). In our hands, deconstruction of any given chemical as an assembly of atomic triads was a far superior descriptor of the molecules, biodegradation-wise, than any other representation tried. This codification seems to represent an optimal tradeoff between significant structural/chemical properties and the processing of a minimal number of attributes by the machine learning sytem employed. Indeed, because the reactivity properties of any given atom generally depends on its neighbours up to a distance of 2, triplets do hold a considerable information on the Chemistry of the molecule while keeping low the number of descriptors (possible atomic triads) per molecule. But is such a correlation casual or does it embody a biological meaning? We argue that the frequency of atomic triad presets the susceptibility of the compounds to the global biodegradation network. In fact, the outcome of the approaches presented in this paper suggest that enzymatic activities of catabolic pathways coevolve to target discrete molecular motifs which can be shared by many chemicals, rather than adapting to deal with one specific molecule, with obvious consequences for the evolution of the substrate recognition sites of the enzyme pool (Wackett, 2004a). It is thus reasonable that confrontation of a diverse microbial community with a mix of chemical compounds (i.e., the most frequent environmental pollution scenario) results in the encounter of a multispecies biodegradation network with a landscape of chemotopes—rather than dealings of single type of bacteria with an unique chemical species. The consequences of such a situation for surveying the degradation gene scenery through experimental and computational means deserve further research.
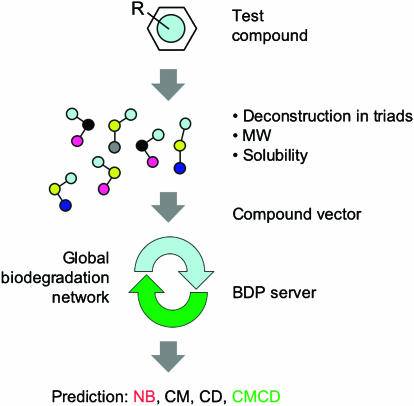
The strategy sketched in Figure 6 has two major incentives. First, it is fully automated and, therefore, it can be quickly applied to massive lists of compounds, as we have shown above. Second, it is not restricted to known functional groups and, therefore, it may provide hints about the environmental fate of compounds that contain undocumented structures, allowing an early prediction of their environmental fate before releasing them into the environment. Its simplicity makes it suitable as a screening predictive tool and provides and early rationale for putting interventions into practice and setting priority procedures. These applications will probably be intensified by the growingly restrictive European Union Regulatory Framework for Chemicals (REACH; ec.europa.eu/enterprise/reach) and other international rules, for example, the Pollution Prevention Framework (www.epa.gov/oppt/p2home). In the meantime, our analysis (Figure 5) indicates that hundreds (if not thousands) of the compounds which are produced in large quantities by the chemical industry may not have a chance of ever being biologically degraded—at least as understood with our current level of knowledge of the microbial metabolism. In this respect, although our prognostic system says nothing on the possible kinetics of degradation of specific compounds, we expect predictive approaches of the sort presented in this paper to inform decisions about acceptability of the release of current and future chemicals into the environment.
Figure 6.
Schematic flowchart of the BDP System. The figure summarises the steps of the prediction process, including the automated deconstruction of any given chemical formula into frequencies of each of the 149 possible chemical triads and its combination with MW and water solubility data for assembling the vector compound. This is fed into the BDPServer which, to an extent, simulates the full potential of the global microbial biodegradation network.
Materials and methods
Databases
UMBBD (www.umbbd.msi.umn.edu) regularly compiles information on experimentally characterised biodegradative reactions. MetaRouter (Pazos et al, 2005) is a system mainly based on UMBBD (Ellis et al, 2003, 2006) designed to maintain heterogeneous sets of data related to biodegradation and bioremediation. ChemFinder (http://chemfinder.cambridgesoft.com) is a database that contains a variety of information about all types of chemical compounds. The lists of compounds known as Annex-I, HPVC and LPVC were kindly provided by Rémi Allanou, of the European Chemicals Bureau (www.ecb.jrc.it). The three lists included compound names and Chemical Abstract Service (CAS) Registration Numbers. The SMILECAS database contains SMILES strings for more than 100 000 compounds that are referred to by their names and CAS Registration Number, and was kindly provided by Bill Meylan, from Syracuse Research Corporation (www.syrres.com). SMILES strings (Weininger, 1988) are linear text representations of the atomic structure of molecules. Atoms are represented with the standard nomenclature and specific signs are used to express different types of chemical bonds and to denote branching, cycles, and other molecular features (http://daylight.com/smiles/index.html). Although SMILES strings unambiguously represent the structure and connectivity of any given compound, each chemical may have several alternative SMILES strings. PubChem Compound (http://pubchem.ncbi.nlm.nih.gov) is a database maintained by the NCBI that contains information about more than five million unique chemical structures, including their SMILES strings.
Software
OpenBabel is a program and library designed to interconvert file formats used in molecular modelling and computational chemistry (http://openbabel.sourceforge.net/). c4.5 is a machine-learning algorithm for the construction of decision trees and rule-based classifiers (Quinlan, 1993). JME (www.molinspiration.com/jme) is a Java applet that generates SMILES strings from drawings of chemical compounds produced with a graphical interface, and was kindly provided by Peter Ertl from Novartis AG. BioLayout JAVA is a program designed for the visualisation of biological networks (Enright and Ouzounis, 2001). All data preparation and manipulation was carried out by means of ad hoc scripts written in Perl language.
Definition of the global biodegradation network
Each of the 903 reactions described in UMBBD as in November 17 2003, was deconstructed into all possible pairs of compounds that sustain a substrate–product relation. For example, for the reaction A → B+C, the following pairs of connected compounds would be generated: A → B, A → C. By assembling a single, nonredundant list of compound pairs, a directed graph representing the global biodegradation network was defined in which nodes correspond to compounds, and edges to reactions, as described previously (Pazos et al, 2003).
Calculation of atomic triad frequencies
Compositional and topological information about each chemical was expressed as series of atomic triad frequencies. Triads were preferred over other possibilities (pairs, tetrads, etc.) because the two nearest neighbours of any given atom in a molecule determine intrinsic reactivity the most. To deconstruct given compounds into such atomic triad series, the SMILES strings associated with each chemical was processed with the OpenBabel sofware (see above), which adds hydrogen atoms not explicitly represented in SMILES strings, and translates the results into BS and Alchem formatted reports. Atom names and connectivity information were extracted from such BS reports in the form of adjacency lists, whereas bond types were extracted from Alchemy reports. The frequencies of atomic triads were then calculated from the information on their connectivity. With such criteria, 149 different atomic triads were identified and categorised for each of the 850 chemical compounds included in the 17 November 2003 update of UMBBD (http://umbbd.msi.umn.edu), and their absolute frequencies were determined for every target molecule.
Compound solubility
Water solubility figures were obtained from the ChemFinder database. Because solubility can be expressed either qualitatively or quantitatively, both types of records were mined from the database and converted into the reciprocal form according to a scale of solubility. Apart of the numerical data on solubility, an operative qualitative scale was set by examining the distribution of solubility values and classes found in the collection of compounds as indicated in Supplementary Table S1. Alas, information on solubility was available for only 214 of the studied compounds. For the rest, the solubility values were left as missing information, which is a circumstance that can be handled by c4.5 (Quinlan, 1993).
Measure of chemical similarity
A modified version of the Tanimoto association coefficient (τ) was used for expressing the degree of chemical similarity between compounds in a fashion that was coherent with their description as series of atomic triads. This coefficient is particularly well suited for dealing with molecular representations consisting of strings of binary descriptors that may indicate, for example, whether predefined substructures are present or absent in a compound (Holliday et al, 2002). In our case, the Tanimoto association coefficient was calculated with nonbinary data (i.e., atomic triad frequencies) by means of the following formula:
where A and B are the number of atomic triads in two compounds, and C is the number of those that they have in common. The Tanimoto coefficient ranges between 0 and 100, and its value can be interpreted as the degree of identity between compounds relative to their atomic triad composition. Once chemical similarity for each pair of compounds had been defined, we studied the distribution of chemical distances for the whole set of compound pairs (Supplementary Figure S1A) and for pairs of compounds that belong to specific classes of environmental fate. We also examined to what extent the whole set of compounds and the environmental fate classes of chemicals involved clusters of similar molecules. To this end, we calculated the clustering coefficient (Cv) for each compound (v) with the rule:
where Kv is the number of compounds that are connected to v, and Nv is the number of connections between the compounds that are linked to v (Watts and Strogatz, 1998). To define whether two compounds were connected or not, we considered two different thresholds for the Tanimoto coefficient: τ⩾50% and ⩾80%. The lower and upper limits of Cv are 0 and 1, respectively: compounds that are not connected to any other molecule are considered to have a clustering coefficient of 0, whereas those that belong to clusters in which many of the members are linked have clustering coefficients closer to 1. Finally, we calculated the average clustering coefficient for the whole set of compounds and for the specific environmental classes of compounds (Supplementary Figure S1B). The average clustering coefficient of a given class represents the cliquishness of that set of compounds (Watts and Strogatz, 1998).
Production of classifiers and evaluation
The machine learning algorithm c4.5 (Quinlan, 1993) was employed to generate rule-based classifiers that associate the properties of chemical compounds with one of the two predictable fates for each of the four independent binary classification schemes defined upon the analyses of the biodegradation network (see the Results section). To assess the predictive capacity of the system, we followed a fivefold cross-validation strategy, in which the data set was divided into five blocks; four of them were used as a training set, to generate the classifiers (rules). The remaining block was used as a test set, for measuring the ability of the classifiers in predicting the environmental fate of chemicals not included in the training set. The process was repeated five times, changing the block that was used as a test set, in such a way that all compounds were part of both sets, at least once. Only those compounds with an associated vector were taken into account (841 out of the original set of 850 compounds). As a basic measure of the predictive capacity of the classifiers, we averaged the robustness of the predictions for the five iterations of the cross-validation experiment. In this context, accuracy was defined as the percentage of correctly classified cases, relative to the total number of them taking together the majority and the minority classes. On the contrary, sensitivity is the fraction of compounds correctly classified as belonging to a specific class, relative to the total number of cases of that particular class. Specificity is the fraction of compounds correctly classified as belonging to a specific class, relative to the total number of predictions for that particular class. Therefore, the last two features (sensitivity and specificity) were independently calculated for the majority and the minority classes. This was made because the generalisation process carried out by c4.5 produces classification models in which one of the classes is defined as the default one. This class is usually the most frequent in the training set, as it happens with the four classifiers generated by this work. When the models are used to classify new cases, those that are not covered by any rule are assigned to the default class. Therefore, by calculating the sensitivity and specificity of the predictions for each distinct class, it is possible to generate a more detailed estimation of the predictive performance of the system, than that represented by accuracy only.
Supplementary Material
Supplementary Tables and Figures
Acknowledgments
We are grateful to the UMBBD and its scientific leader, L Wackett, as the main source of curated public information on Biodegradation and Biotransformations. The Syracuse Research Corporation deserves also a special acknowledgement for providing the SMILECAS database. Peter Ertl, Bill Meylan, Rémi Allanou and Stéphane Brion are kindly acknowledged for providing useful resources and databases. This work was supported by the 6th Framework Programme of the EC. MJG has been beneficiary of an I3P contract from the CSIC and an INTA Postdoctoral Fellowship.
References
- Blinova VG, Dobrynin DA, Finn VK, Kuznetsov SO, Pankratova ES (2003) Toxicology analysis by means of the JSM-method. Bioinformatics 19: 1201–1207 [DOI] [PubMed] [Google Scholar]
- Button WG, Judson PN, Long A, Vessey JD (2003) Using absolute and relative reasoning in the prediction of the potential metabolism of xenobiotics. J Chem Inf Comput Sci 43: 1371–1377 [DOI] [PubMed] [Google Scholar]
- Damborsky J (1996) A mechanistic approach to deriving quantitative structure–activity relationship models for microbial degradation of organic compounds. SAR QSAR Environ Res 5: 27–36 [DOI] [PubMed] [Google Scholar]
- Damborsky J, Schultz TW (1997) Comparison of the QSAR models for toxicity and biodegradability of anilines and phenols. Chemosphere 34: 429–446 [DOI] [PubMed] [Google Scholar]
- Darnerud PO (2003) Toxic effects of brominated flame retardants in man and in wildlife. Environ Int 29: 841–853 [DOI] [PubMed] [Google Scholar]
- Dennis P, Edwards EA, Liss SN, Fulthorpe R (2003) Monitoring gene expression in mixed microbial communities by using DNA microarrays. Appl Environ Microbiol 69: 769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz E (2004) Bacterial degradation of aromatic pollutants: a paradigm of metabolic versatility. Int Microbiol 7: 173–180 [PubMed] [Google Scholar]
- Dimitrov S, Breton R, Macdonald D, Walker JD, Mekenyan O (2002) Quantitative prediction of biodegradability, metabolite distribution and toxicity of stable metabolites. SAR QSAR Environ Res 13: 445–455 [DOI] [PubMed] [Google Scholar]
- Dolle RE (2004) Comprehensive survey of combinatorial library synthesis: 2003. J Comb Chem 6: 623–679 [DOI] [PubMed] [Google Scholar]
- Ellis LB, Hou BK, Kang W, Wackett LP (2003) The university of minnesota biocatalysis/biodegradation database: post-genomic data mining. Nucleic Acids Res 31: 262–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis LB, Roe D, Wackett LP (2006) The University of Minnesota Biocatalysis/Biodegradation Database: the first decade. Nucleic Acids Res 34: D517–D521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Ouzounis CA (2001) BioLayout—an automatic graph layout algorithm for similarity visualization. Bioinformatics 17: 853–854 [DOI] [PubMed] [Google Scholar]
- Holliday JD, Hu CY, Willett P (2002) Grouping of coefficients for the calculation of inter-molecular similarity and dissimilarity using 2D fragment bit-strings. Comb Chem High Throughput Screen 5: 155–166 [DOI] [PubMed] [Google Scholar]
- Hou BK, Ellis LB, Wackett LP, Kang W, Handelsman J (2004) Encoding microbial metabolic logic: predicting biodegradation. J Ind Microbiol Biotechnol 10: 10. [DOI] [PubMed] [Google Scholar]
- Hou BK, Wackett LP, Ellis LB (2003) Microbial pathway prediction: a functional group approach. J Chem Inf Comput Sci 43: 1051–1057 [DOI] [PubMed] [Google Scholar]
- Klopman G, Wang S, Balthasar DM (1992) Estimation of aqueous solubility of organic molecules by the group contribution approach. Application to the study of biodegradation. J Chem Inf Comput Sci 32: 474–482 [DOI] [PubMed] [Google Scholar]
- Koizumi Y, Kelly JJ, Nakagawa T, Urakawa H, El-Fantroussi S, Al-Muzaini S, Fukui M, Urushigawa Y, Stahl DA (2002) Parallel characterization of anaerobic toluene- and ethylbenzene-degrading microbial consortia by PCR-denaturing gradient gel electrophoresis, RNA-DNA membrane hybridization, and DNA microarray technology. Appl Environ Microbiol 68: 3215–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNaughton SJ, Stephen JR, Venosa AD, Davis GA, Chang Y-J, White DC (1999) Microbial population changes during bioremediation of an experimental oil spill. Appl Environ Microbiol 65: 3566–3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShan DC, Rao S, Shah I (2003) PathMiner: predicting metabolic pathways by heuristic search. Bioinformatics 19: 1692–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra V, Lal R, Srinivasan C (2001) Enzymes and operons mediating xenobiotic degradation in bacteria. Crit Rev Microbiol 27: 133–166 [DOI] [PubMed] [Google Scholar]
- Pazos F, Guijas D, Valencia A, de Lorenzo V (2005) MetaRouter: bioinformatics for bioremediation. Nucleic Acids Res 33: D588–D592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos F, Valencia A, de Lorenzo V (2003) The organization of the microbial biodegradation network from a systems-biology perspective. EMBO Rep 4: 994–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelz O, Tesar M, Wittich RM, Moore ER, Timmis KN, Abraham WR (1999) Towards elucidation of microbial community metabolic pathways: unravelling the network of carbon sharing in a pollutant-degrading bacterial consortium by immunocapture and isotopic ratio mass spectrometry. Environ Microbiol 1: 167–174 [DOI] [PubMed] [Google Scholar]
- Quinlan JR (1993) c4.5: Programs for Machine Learning. San Mateo, CA: Morgan-Kaufmann [Google Scholar]
- Sutherland JJ, O'Brien LA, Weaver DF (2004) A comparison of methods for modeling quantitative structure-activity relationships. J Med Chem 47: 5541–5554 [DOI] [PubMed] [Google Scholar]
- Tong W, Lowis DR, Perkins R, Chen Y, Welsh WJ, Goddette DW, Heritage TW, Sheehan DM (1998) Evaluation of quantitative structure–activity relationship methods for large-scale prediction of chemicals binding to the estrogen receptor. J Chem Inf Comput Sci 38: 669–677 [DOI] [PubMed] [Google Scholar]
- Urbance JW, Cole J, Saxman P, Tiedje JM (2003) BSD: the biodegradative strain database. Nucleic Acids Res 31: 152–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett LP (2004a) Evolution of enzymes for the metabolism of new chemical inputs into the environment. J Biol Chem 279: 41259–41262. [DOI] [PubMed] [Google Scholar]
- Wackett LP (2004b) Prediction of microbial biodegradation. Environ Microbiol 6: 313 [Google Scholar]
- Wackett LP, Ellis LB (1999) Predicting biodegradation. Environ Microbiol 1: 119–124 [PubMed] [Google Scholar]
- Wackett LP, Hershberger CD (2001) Biocatalysis and Biodegradation: Microbial Transformation of Organic Compounds. Washington: American Society for Microbiology [Google Scholar]
- Watts DJ, Strogatz SH (1998) Collective dynamics of small-world networks. Nature 393: 440–442 [DOI] [PubMed] [Google Scholar]
- Weininger D (1988) SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J Chem Inf Comput Sci 28: 31–36 [Google Scholar]
- Whiteley AS, Bailey MJ (2000) Bacterial community structure and physiological state within an industrial phenol bioremediation system. Appl Environ Microbiol 66: 2400–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JH (2003) Microarrays for bacterial detection and microbial community analysis. Curr Opin Microbiol 6: 288–294 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables and Figures