Abstract
Ubiquitin-protein ligases (E3s) are responsible for target recognition and regulate stability, localization or function of their substrates. However, the substrates of most E3 enzymes remain unknown. Here, we describe the development of a novel proteomic in vitro ubiquitination screen using a protein microarray platform that can be utilized for the discovery of substrates for E3 ligases on a global scale. Using the yeast E3 Rsp5 as a test system to identify its substrates on a yeast protein microarray that covers most of the yeast (Saccharomyces cerevisiae) proteome, we identified numerous known and novel ubiquitinated substrates of this E3 ligase. Our enzymatic approach was complemented by a parallel protein microarray protein interaction study. Examination of the substrates identified in the analysis combined with phage display screening allowed exploration of binding mechanisms and substrate specificity of Rsp5. The development of a platform for global discovery of E3 substrates is invaluable for understanding the cellular pathways in which they participate, and could be utilized for the identification of drug targets.
Keywords: endocytosis, Nedd4, post-translational modification, proteomics, ubiquitin ligase
Introduction
Substrates of the ubiquitin pathway are covalently modified by the attachment of a small protein called ubiquitin and as a result are targeted for degradation or other cellular fates (Pickart, 2001; Glickman and Ciechanover, 2002; Hicke and Dunn, 2003). Ubiquitination involves the sequential action of three enzymes: E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme) and E3 (ubiquitin-protein ligase) (Pickart, 2001). The E3 enzyme, which is responsible for the specificity of the reaction, associates with substrates (Hershko and Ciechanover, 1998; Pickart, 2001; Fang and Weissman, 2004), and defects in this interaction have been implicated in numerous diseases (Abriel et al, 1999; Dawson and Dawson, 2003; Liu, 2004; Nakayama and Nakayama, 2006).
A significant fraction of the proteome is regulated by the ubiquitin pathway and eukaryotic genomes express hundreds of E3 ligases to coordinate the ubiquitination of cellular proteins (Peng et al, 2003; Willems et al, 2004). Currently, most E3 enzymes have not been linked to any specific substrate and any platform that would allow for the systematic discovery of enzymatic E3 substrates would be tremendously useful for advancing our understanding of the ubiquitin pathway. Rsp5 is a yeast E3 ubiquitin ligase that belongs to the Nedd4 family (Rotin et al, 2000). It contains a C2 domain, a catalytic HECT domain and three WW domains that can bind substrates directly by recognizing a (L/P)PxY sequence (PY motif) (Kanelis et al, 2001, 2006; Kasanov et al, 2001; Hu et al, 2004; Shcherbik et al, 2004). Ubiquitination of proteins by the Nedd4 E3 family has been implicated in numerous cellular functions, including endocytosis, sorting and trafficking (Rotin et al, 2000; Horak, 2003; Ingham et al, 2004). For example, Nedd4 (or Nedd4-2), the human Rsp5 homologue, ubiquitinates the epithelial sodium channel (ENaC) to regulate its endocytosis, and mutations that inhibit the Nedd4-2:ENaC interaction cause Liddle syndrome, a hereditary hypertension (Staub et al, 1996; Abriel et al, 1999; Lifton et al, 2001). Similarly, Rsp5 was demonstrated to regulate endocytosis and sorting of several yeast plasma membrane proteins (Horak, 2003; Dupre et al, 2004). Moreover, Rsp5 has been implicated in the regulation of several other cellular functions, including mitochondrial inheritance, drug resistance, intracellular pH, fatty acid biosynthesis and transcriptional control (see below).
Despite the biological importance of the Nedd4/Rsp5 family of E3 ligases, only a few substrates have been identified to date for this ubiquitin ligase family. Thus, our goal was to globally identify Rsp5 substrates in the yeast proteome. For that, we chose to use protein microarray technology as our experimental platform. The arrays used in this study contain thousands of purified proteins (most of the Saccharomyces cerevisiae proteome) immobilized at a high spatial density on standard sized slides and can be readily used to probe the yeast proteome using traditional biochemical approaches (Zhu et al, 2001; Schweitzer et al, 2003; Zhu and Snyder, 2003; Bertone and Snyder, 2005; Ptacek et al, 2005; Smith et al, 2005).
To date, few studies have assayed enzymatic activities using this technology. In the current study, we have successfully used yeast (S. cerevisiae) protein microarrays to assay the enzymatic (ubiquitination) activity and binding of Rsp5 to its substrates, and we have identified previously reported and novel ubiquitinated substrates and interacting partners of this E3 ligase.
Our results also demonstrate how this approach can yield informative data regarding the binding mechanisms and substrate specificity of an E3 enzyme.
Results
Identification of proteins ubiquitinated by Rsp5 on a proteome array
For this study, ubiquitinated Rsp5 substrates were identified using commercially available yeast protein microarrays (Invitrogen ProtoArray® Yeast Proteome Microarray). These protein microarrays are based on technology described previously (Zhu et al, 2001) and contain more than 4000 GST- and 6 × HIS-tagged yeast proteins from S. cerevisiae spotted in duplicate on nitrocellulose slides (ProtoArray® Yeast Proteome Microarray nc v1.1).
Before assaying for ubiquitinated proteins on the protein microarray, we developed conditions in which Rsp5 could ubiquitinate one of its known substrates, the C-terminal domain of Rpb1 (CTD) (Beaudenon et al, 1999). The ubiquitination of CTD was dependent on the budding yeast E1 enzyme, an E2 enzyme (Ubc4), ubiquitin, Rsp5 and ATP, and was visualized by Western blotting. This control reaction was used to optimize conditions for Rsp5-dependent ubiquitination on nitrocellulose-coated glass slides. In these experiments, the CTD and other proteins were robotically spotted onto slides and incubated with a reaction mixture containing Rsp5 and FITC-labeled ubiquitin. The proteome array was then assayed for Rsp5-dependent ubiquitination using the optimized conditions (Figure 1A).
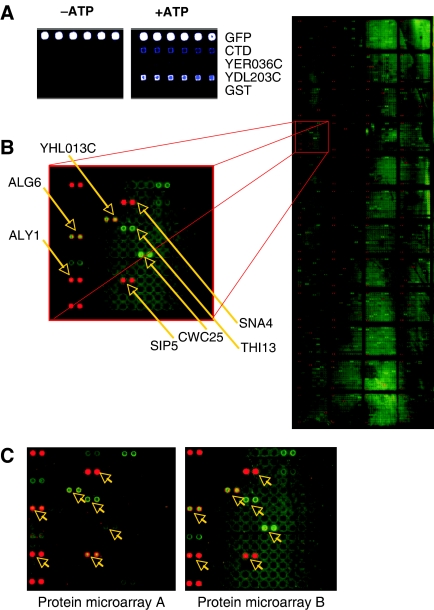
Figure 1.
Rsp5-mediated ubiquitination of the yeast proteome. (A) Assay development. To optimize ubiquitination conditions using protein microarrays, known substrates of Rsp5 (CTD and Ydl203c) and proteins not ubiquitinated by Rsp5 in vitro (Yer036c and GST alone) were robotically printed on slides and incubated in ubiquitination reactions containing Rsp5 and FITC-labeled ubiquitin. The fluorescent signal demonstrates CTD and Ydl203c ubiquitination in the presence of ATP (right panel), while negative control proteins are not ubiquitinated (left panel). Blue color represents ubiquitination (detected with FITC-Ub). GFP was used as a positive control, as it has the same excitation wavelength as FITC. The colors associated with the protein microarray spots indicate of the intensity of the signal (with light blue<bright blue<white). (B) Image of a scanned ubiquitinated protein microarray with an enlargement of one grid. All proteins are printed in duplicate and arrows indicate proteins that were identified as substrates after quantitative data analysis. Alexa dyes are spotted as controls in the left-hand corners of each grid. (C) Reproducibility. Two protein microarrays were ubiquitinated in separate experiments and the same grid from each array is shown. Arrows point to ubiquitinated proteins that were identified as substrates. Most spots producing significantly higher signal than background can be seen on both arrays, suggesting high reproducibility between slides.
Following the reaction, protein microarray slides were washed, scanned and proteins modified by ubiquitin were identified by quantifying the intensity of the FITC signal produced compared with the background (Figure 1B). Although detection of protein–protein interactions on microarrays is generally highly reproducible (Zhu et al, 2001; Hesselberth et al, 2006) (Figure 1A and B), we repeated the Rsp5 ubiquitination reaction on two separate protein microarray slides to increase the quality of our data set. Based on selection criteria for identifying positive hits described in Materials and methods, we generated a data set of 150 Rsp5 substrates (henceforth referred to as the ‘relaxed Rsp5 substrate set'). From this set of substrates, we selected a ‘high-confidence' data set, which comprises the 40 proteins that produce the strongest signal. These proteins were considered for further study (Table I and Supplementary Table SI).
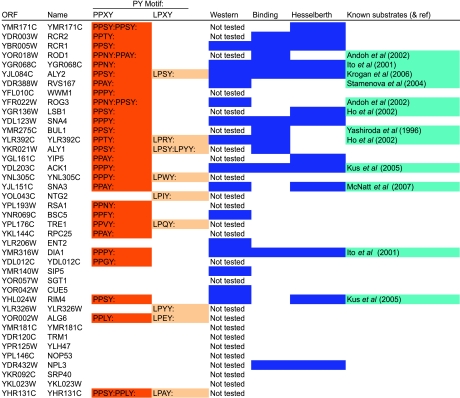
Table 1.
High-confidence Rsp5 substrate (unbiquitination) data set
High-confidence Rsp5 substrate data set. The top 40 proteins and their PY motifs identified as Rsp5 substrates using the protein microarray are listed. A blue color in the column labeled ‘Western' indicates proteins that were ubiquitinated in a Western blot. Proteins identified by protein microarray as interacting partners of Rsp5 in this study are shaded in blue in the column labeled ‘Binding'. Boxes shaded in blue in the column labeled ‘Hesselberth' indicate that the protein was identified in the microarray screen by Hesselberth and co-workers as an Rsp5 binding partner. The column labeled ‘Known substrates' contains proteins that were previously described as Rsp5 substrates.
Properties of the high-confidence Rsp5 substrate set
PY motifs
Since Rsp5-WW domains are known to bind PY motifs, we looked for the presence of these motifs in the sequences of proteins belonging to the Rsp5 substrate sets. As expected, proteins containing PY motifs were significantly enriched in the Rsp5 high-confidence substrate set (P<0.01—exact randomization test). In the yeast proteome, approximately 4% of proteins contain PPxY motifs, and 7% contain LPxY motifs. In the Rsp5 high-confidence substrate set, 72% of proteins had at least one of these motifs. Proteins with PPxY and LPxY motifs were significantly enriched both in the high-confidence and relaxed Rsp5 substrate data sets (P<0.01 for both—exact randomization test).
Identification of known substrates
Rsp5 has been implicated in a wide range of cellular pathways and a number of its substrates have previously been described. Eleven proteins in the high-confidence Rsp5 substrate set, and 17 proteins in the relaxed substrate set, have previously been identified in Rsp5 pathways through genetic and biochemical methods (Table I). The relatively large number of previously described Rsp5 substrates identified in this study suggests that the proteome microarray experimental approach is capable of discovering proteins ubiquitinated by Rsp5 in vivo and that many of the proteins in the high-confidence substrate set and the relaxed substrate set are likely novel biologically relevant substrates of Rsp5.
Detection of substrate ubiquitination using Western blotting
We used an established ubiquitination assay to confirm that the proteins identified as Rsp5 substrates on the protein microarray are modified by this E3. Traditional approaches for monitoring ubiquitination involve subjecting specific purified proteins to ubiquitination by an E3 in vitro and using a Western blot approach to visualize ubiquitination.
Fifteen proteins from the Rsp5 high-confidence substrate set, and six proteins that were not identified as substrates of Rsp5, were purified from yeast using glutathione affinity purification, incubated in ubiquitination reactions containing Rsp5 and the above described E1 and E2 (Ubc4), and assayed for ubiquitination using anti-ubiquitin antibodies and Western blots. All of the proteins whose ubiquitination was detected on the protein microarray were verified to be ubiquitinated by Western blot analysis (Figure 2A; Table I). Most of the proteins were efficiently polyubiquitinated or ubiquitinated on multiple lysines. In contrast, the six proteins tested whose ubiquitination was not detected on the protein microarray did not appear to be ubiquitinated after Western blot analysis (Figure 2B), confirming that the enzymatic activity detected is specific and that the data generated by the protein microarray approach are consistent with established methods of detecting ubiquitination.
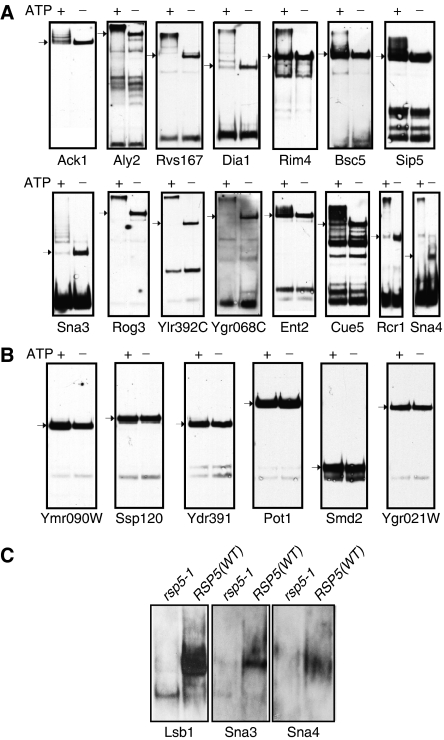
Figure 2.
Validation of substrate ubiquitination in vitro and in vivo. (A, B) In vitro ubiquitination: (A) 15 proteins identified as ‘high-confidence' Rsp5 substrates using protein microarrays were expressed (fused to GST) in yeast, purified and incubated in ubiquitination reactions containing Rsp5. (B) Six randomly selected proteins that were not identified as Rsp5 substrates in the protein microarray experiments were used as negative controls. All 15 of the ‘high-confidence' Rsp5 substrates and none of the negative control proteins were visibly ubiquitinated in the Western blots with anti-GST antibodies (arrows indicate the original size of the protein in the absence of ubiquitination (i.e. without ATP)). (C) In vivo ubiquitination: example of three Rsp5 substrates from the protein microarray exhibiting ubiquitination in vivo. The three proteins (HA tagged) were expressed in RSP5 (WT) or rsp5-1 mutant yeast cells. Following a shift to the non-permissive temperature (37°C), proteins were immunoprecipitated with anti-HA antibodies and immunoblotted with anti-ubiquitin antibodies. Note ubiquitination in the RSP5-WT but not the rsp5-1 cells.
To further validate our in vitro data, we tested for in vivo ubiquitination of several putative substrates (known or suspected to be involved in sorting/endocytosis), by comparing ubiquitination of these proteins expressed in RSP5 (WT) or rsp5-1 mutant yeast cells. rsp5-1 is a temperature-sensitive mutant that reduces Rsp5 expression upon temperature shift to 37°C (an rsp5-null mutant is lethal). As shown in Figure 2C, Lsb1 and Sna3 (both known interactors or substrates of Rsp5; Ho et al, 2002; McNatt et al, 2007; Oestreich et al, 2007), as well as Sna4, were ubiquitinated in vivo by Rsp5. Although the function of Sna4 is unknown, it is a vacuolar resident protein, much like Sna3, and we thus anticipate that it too utilizes interactions with Rsp5 for vacuolar targeting. Our preliminary data also revealed in vivo ubiquitination of other substrates by Rsp5 (e.g. Yip5, Rcr1 and Rcr2—data not shown).
Identification of Rsp5 interacting proteins
To directly test Rsp5 substrate binding using the protein microarrays, and to compare these data to the ubiquitination data sets above, we screened the protein microarrays for proteins that bind Rsp5. Purified Rsp5 was labeled with Alexa 647 and incubated with the protein microarray in two separate experiments. After washing and scanning the slides, the data were analyzed and a data set of 155 Rsp5 binding proteins was generated (Table II and Supplementary Table SII).
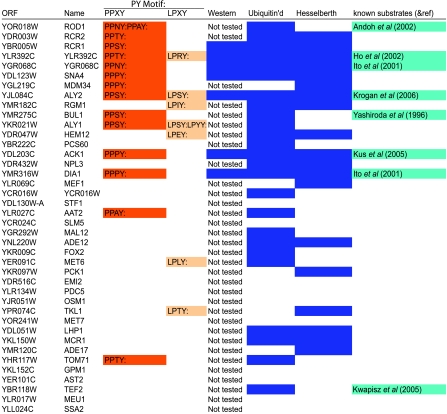
Table 2.
High-confidence Rsp5 interaction data set
High-confidence Rsp5 interaction data set. The top 40 proteins and their PY motifs identified as Rsp5 interacting partners using the protein microarray are listed. The columns are the same as in Table I, except the column labeled ‘Ubiquitin'd' contains proteins that were identified in both ubiquitination and binding protein microarray assays.
A sequence search revealed that the Rsp5 binding set was significantly enriched for proteins containing PY motifs (P<0.01—exact randomization test). Ten proteins in the Rsp5 binding set have previously been identified in Rsp5 pathways.
Comparison between the Rsp5 substrate set and binding set
Twelve proteins in the high-confidence Rsp5 substrate set and 52 proteins in the relaxed Rsp5 substrate set were also present in the Rsp5 interaction set. Conversely, 34% of the proteins in the Rsp5 interaction set were ubiquitinated by Rsp5. Eleven of the 12 proteins that both bound to and were ubiquitinated by Rsp5 contain PY motifs.
Pro, Ser and Ala residues are enriched at the ‘x' position of (L/P)PxY motifs
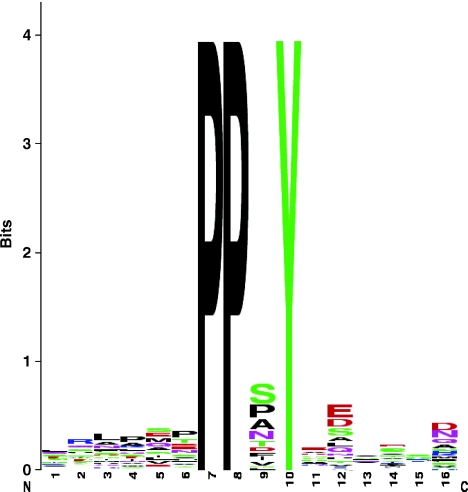
We examined the amino-acid sequences of Rsp5 substrates to determine whether additional amino-acids residues in the PY motif ((L/P)PxY) may contribute to substrate specificity. A total of 38 PY motifs were present in 29 proteins of the high-confidence subset. Considering the third (x) position in the motif, the most frequent motifs were PPSY (ten), PPAY (five) and PPPY (five). Comparisons with sets of randomly selected proteins containing PY motifs showed that Ser and Ala (but not Pro) were both significantly overrepresented at the third position within our experimentally determined Rsp5 substrates (P<0.001; randomized exact test) (Figure 3).
Figure 3.
Sequence logos for substrates of Rsp5. PPxY motifs, together with six residues upstream and downstream of the motif from proteins identified as substrates of Rsp5, were aligned and used to generate sequence logos (Crooks et al, 2004). In each logo, stacks of letters indicate the relative frequency of certain amino acids at each position in the sequence. The overall height provides a guide to the level of sequence conservation associated at that position.
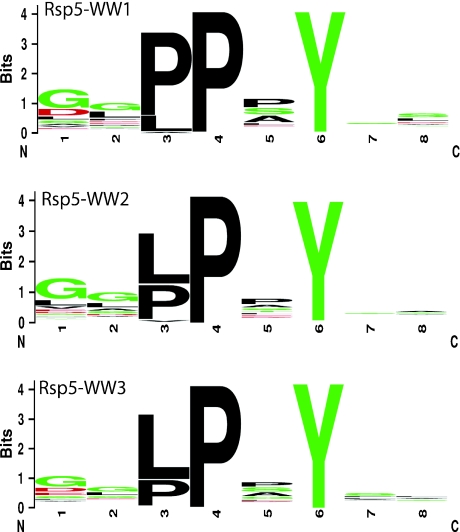
To further confirm these findings, we performed a modified phage display screen to explore substrate specificity of each of the three WW domains of Rsp5. All peptides identified through this screen (over 300) were found to contain a PY motif. Consistent with data presented above, Ser and Ala were both found to be preferred at the third position (Figure 4). More interestingly, the most common amino-acid residue associated with the third position was Pro, suggesting an important biological role for this residue.
Figure 4.
Phage display logos. Peptides identified as substrates of Rsp5, using the phage display system, were aligned and are graphically displayed as logos, as shown in Figure 3 above.
Discussion
In this report, we demonstrate that protein microarrays can be used to identify, on a global scale, ubiquitinated substrates and binding partners of a yeast E3 ubiquitin ligase, Rsp5. A combination of techniques was used to validate the protein microarray data and contributes to our understanding of Rsp5 substrate interaction mechanisms.
The high-confidence Rsp5 substrate set contains 12 proteins previously reported in Rsp5 pathways. Six of these (Ygr068c, Aly2, Lsb1, Ylr392c, Dia1 and Rim4) were reported in other HTP screens (Ito et al, 2001; Ho et al, 2002; Kus et al, 2005; Krogan et al, 2006), while the remaining six (Rod1, Rog3, Rvs167, Bul1, Sna3 and Ack1) were validated as substrates using a combination of genetic and biochemical approaches (Yashiroda et al, 1996; Andoh et al, 2002; Stamenova et al, 2004; Kus et al, 2005).
Most of the high-confidence Rsp5 substrates contained at least one PY motif, usually PPxY (Table I). However, a few substrates did not (e.g. Sgt1, Cue5, Sip5). Sgt1, Cue1 and Sip5 are known to be involved in the ubiquitin pathway. The precise role of Sgt1 is not clear, but the association of this protein with Rsp5 is interesting, since it has been implicated as an activator of SCF E3 enzymes (Kitagawa et al, 1999; Spiechowicz and Filipek, 2005). Cue1 has a ubiquitin binding motif and its affinity for ubiquitin may facilitate its monoubiquitination (Kang et al, 2003). Alternatively, it is possible that it might have bound FITC-ubiquitin non-covalently; however, this is unlikely because its ubiquitination was also detected on a Western blot. Sip5 may not be an Rsp5 substrate, since it has a RING/U-box domain and likely produced a positive signal in the screen because it used the ubiquitin machinery present during the reaction for autoubiquitination.
In addition to the known Rsp5 substrates described above, the relaxed Rsp5 substrate set contains six other proteins that were previously identified as Rsp5 substrates or implicated in Rsp5 pathways. These include, Rpb7 (Kus et al, 2005), Tef2 (Kwapisz et al, 2005), Ubi4, Uba1 (Huibregtse et al, 1995), Rpl40B (Kabir et al, 2005) and Rpl40A (Kabir et al, 2005; Kwapisz et al, 2005).
The identification of 18 proteins known to participate in Rsp5 pathways or to be ubiquitinated directly by Rsp5 suggests that the protein microarray experimental approach is a valid tool for the discovery of ubiquitinated E3 substrates, and that this approach is capable of discovering physiological substrates of Rsp5.
Not all known Rsp5 substrates were identified in our screen. First, some of the known substrates were not printed on the array (e.g. Mga2, Rpb1, Hpr1, Bsd2 and Pma1). Second, our approach is likely to have missed Rsp5 substrates that do not bind Rsp5 directly or require cofactors for their interaction. This is a plausible explanation because some of the known substrates that were not identified on the array (Gap1, Fur4, Rfa1, Zrt1 and Tat2) do not have PY motifs, do not bind Rsp5 directly, and may require adaptor proteins (e.g. Bul1 and Bul2; Helliwell et al, 2001) to bind Rsp5. Third, it is impossible to tell how the purification and printing process used to make the array may have affected the accessibility of some substrates to bind Rsp5.
In the high-confidence data set, 20 novel Rsp5 substrates were identified. Consistent with the well-established role of Rsp5 in ubiquitinating proteins at the plasma membrane or Golgi and affecting their sorting to vesicles, endosomes and to the vacuole, there are nine proteins in the high-confidence Rsp5 substrate list (Ymr171c, Rcr1, Rcr2, Sna3, Sna4, Yip5, Ydl012C and Alg6) that localize to either the plasma membrane, the vacuole, or have otherwise been implicated in the secretory pathway. Further characterization of these substrates is necessary in order to better understand their role in Rsp5-dependent cellular pathways.
Twelve proteins in the high-confidence Rsp5 substrate set, and 52 proteins in the relaxed Rsp5 substrate set, were also present in the Rsp5 interaction set. Of the 12 proteins that bound Rsp5 in the high-confidence substrate set, seven had been previously described in Rsp5 pathways, and four are novel substrates.
A recent study used the same protein microarray to find binding substrates for individual WW domains of Rsp5 (Hesselberth et al, 2006). Their network of interactions identified 124 interactions, of which eight had previously been reported as Rsp5 substrates (compared with 10 known substrates in our Rsp5 interaction data set). Fourteen proteins in this Hesselberth data set overlapped with our high-confidence Rsp5 substrate set and 58 overlapped with the relaxed Rsp5 ubiquitinated substrate set. Fifty-eight proteins from the Hesselberth data set are also present in the Rsp5 interaction set.
The number of known substrates identified by probing the protein microarray for enzymatic (ubiquitination) substrates as opposed to binding partners of Rsp5 suggests that this experimental approach results in a much higher quality data set. One explanation for this is that the binding affinity for WW domains and PY motifs is relatively weak (low to mid micromolar range) (Kanelis et al, 2001, 2006) and transient Rsp5 substrate interactions may therefore be missed when probing the array for interactions. If these transient interactions result in the enzymatic transfer of ubiquitin, however, it will likely be detected because ubiquitin becomes covalently, and therefore permanently bound to its substrate. Furthermore, polyubiquitination (or multi-monoubiquitination) of substrates on the proteome microarray may result in the amplification of fluorescent signal and thus a higher sensitivity may be achieved. Finally, in addition to being more sensitive, probing for ubiquitination is a more direct assay of Rsp5's biological role and therefore more likely to yield information that is more physiologically relevant.
As expected, the high-confidence substrate set was significantly enriched for proteins containing (L/P)PxY motifs. Statistical analysis of proteins identified from binding and ubiquitination protein microarrays and the protein screens from phage display experiments, reveal a preponderance of Pro, Ala and Ser residues in the third (x) position of the PY motif. These findings are in accordance with a survey of previously reported Rsp5 substrates containing PY motifs; Ala and Ser are found in the PY motifs of six proteins (Sna3, Bul1, Bul2, Rod1, Rog3 and Rvs167) and absent in two (Spt23 and Mga2; Hoppe et al, 2000). Furthermore, it was shown that the WW domains from three distinct proteins (KIAA0082, Ras GTPase-activating-like protein IQGAP1 and Transcription Factor CA150) have a binding preference to motifs with Pro and Ala at the third position (Hu et al, 2004). Finally, an earlier phage display study, which characterized the binding preferences of WW domains from Rsp5 and other proteins, had also demonstrated that Ala and Ser residues are favored within the PY motif of the first domain, whereas Pro was found to be most abundant in the second and third domains (Kasanov et al, 2001).
The mechanism of how Rsp5 recognizes PY motifs in substrates is not well understood. By highlighting the preference for a limited number of residues within the (L/P)PxY motif, these analyses may provide additional insights into the mechanism of substrate recognition by Rsp5 and ultimately a better general understanding of WW domain–substrate interactions.
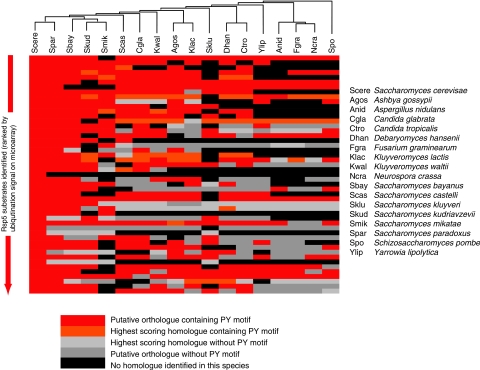
The availability of complete genomes for an additional 17 species of related fungi (Ascomycetes) was exploited to explore the evolution of the Rsp5 substrates identified in S. cerevisiae. Orthologous sets of proteins for each Rsp5 substrate were generated and used to determine the presence of conserved (L/P)PxY motifs (Figure 5 and Supplementary Figure S1A). Conservation of the PY motif was observed in the majority of orthologues, particularly within the closely related Saccharomyces senso stricto species. Comparisons of Ka/Ks ratios for the orthologues identified across other fungal species found that the nucleotide sequence underlying the (L/P)PxY motif is under stronger purifying selection (and hence more highly conserved) than the neighboring residues (Ka/Ks <0.1; Supplementary Figure S1B)), reflecting their functional importance. In addition, this was associated with the conservation of Ser, Pro and Ala at the third position (position ‘x' in the (L/P)PxY motif) in all but two orthologous sets (Supplementary Figure S1C), in agreement with the data shown in Figures 3 and 4. These data are consistent with the hypothesis that the sequence (L/P)P(S/P/A)Y is important to maintain the interaction between the substrate and the WW domain of the Rsp5 orthologue in each species.
Figure 5.
Conservation of Rsp5 substrates across fungal species. BLAST analyses were used to identify potential orthologues and homologues of the top 49 most highly ubiquitinated Rsp5 substrates, together with the presence of a (L/P)PxY motif in 17 species of ascomycetes. Putative orthologues were defined as those demonstrating reciprocal best BLAST matches, and best scoring homologues were defined as those matches with the highest BLAST bit scores, which were considered homologous through manual inspection of sequence alignments (see Materials and methods). The cladogram at the top of the figure shows the phylogenetic relationships of the 18 fungal species (including S. cerevisiae) considered.
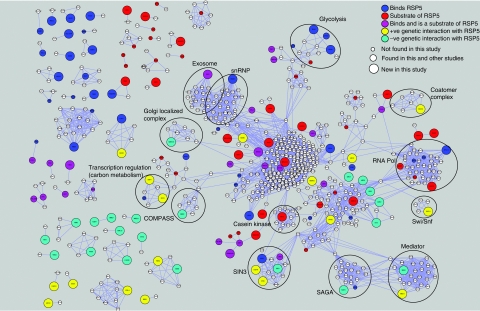
To help assess the physiological relevance of the ubiquitination and in vitro binding assays, we integrated the data from this current study with recently generated physical and genetic interaction datasets, to derive an Rsp5 interaction network (Figure 6) (Schuldiner et al, 2005; Gavin et al, 2006; Krogan et al, 2006; Collins et al, 2007b). Consistent with a role in ubiquitination, both positive (alleviating) and negative (aggravating) genetic interactions were observed with the deubiquitinating enzymes Ubp13 and Ubp3/Bre5, suggesting that these enzymes may share similar substrates. Further inspection of the Rsp5 map reveals that a large number of physical and genetic interactions place Rsp5 into pathways responsible for the regulation of chromatin function and transcription. Consistent with this, Rsp5 was originally isolated in a genetic screen for suppressors of mutations in Spt3 (Wang et al, 1999), a subunit of the SAGA (Spt/Ada/Gcn5/acetyltransferase) complex (Eisenmann et al, 1992; Roberts and Winston, 1997), which modulates the transcription activity of RNA polymerase II (RNAPII). Although it is known that Rsp5 ubiquitinates the large subunit of RNAPII (Rpb1) under conditions of UV irradiation, its exact role in transcriptional regulation is not well understood. From the interaction network it is clear that Rsp5 is physically and genetically linked to several complexes involved in chromatin remodeling and/or transcription regulation, including the histone deacetylase Rpd3C(L) complex, the histone Snf1 kinase complex, SAGA, SWI/SNF and Mediator. In addition to transcriptional regulation, Rsp5 seems to be functionally linked to several other processes including rRNA metabolism and mRNA splicing. For example, Dis3 has previously been shown to be the key regulator of the exosome (responsible for degradation of snoRNAs, mRNAs and rRNAs) (Dziembowski et al, 2007). Here, we have shown that Rsp5 is both capable of binding and ubiquitinating Dis3, suggesting that one mode of its (and hence the exosomes) regulation may occur through the ubiquitin pathway. Finally, two previously uncharacterized proteins, Yjl084c and Ykr021w, were not only found to bind to and are substrates of Rsp5 but also were recently shown to be physically associated with Rsp5 in vivo. (Krogan et al, 2006; Collins et al, 2007a). Interestingly, these are two out of the four budding yeast proteins that contain an arrestin N domain, which in metazoans is linked to inactivation of G protein-coupled receptors and cross-talk with other signaling pathways (Modzelewska et al, 2006). These relatively specific connections suggest this region may be mediating the binding and/or activity of Rsp5. It is expected that the interaction network presented in Figure 6 will facilitate the generation of many more testable hypotheses of Rsp5 function.
Figure 6.
Network diagram describing Rsp5 functional pathways. Proteins identified from this study, together with previously published studies, were used to generate a protein–protein interaction network describing functional pathways associated with Rsp5. Two studies have recently attempted to comprehensively define the physical interactome in budding yeast through systematic affinity tagging, purification and mass spectrometry (Gavin et al, 2006; Krogan et al, 2006). A more recent study that combines these data sets to produce a single high-quality integrated data set (Collins et al, 2007a, 2007b) was used to identify proteins with physical interactions to: 42 (of 150) proteins identified in the current study as being a substrate of Rsp5; 40 (of 155) proteins identified in this study as binding partners of Rsp5; 24 (of 59) proteins identified in other small-scale experiments as being substrates of Rsp5; 38 (of 110) proteins identified in a previous high-throughput screen of Rsp5 binding partners (Hesselberth et al, 2006) and 39 (of 79) proteins, whose genes have been found to either positively or negatively genetically interact with Rsp5. Genetic interactions were obtained from an rsp5-DamP hypomorphic allele (Schuldiner et al, 2005; Collins et al, 2007b) derived from a recently generated E-MAP (epistatic miniarray profile), using a normalized score of >1.5 for positive interactions and <−1.5 for negative interactions. Physical interactions were obtained using a purification enrichment (PE) score of >3.2 (Collins et al, 2007a, 2007b). In total, 1258 interactions (light blue lines) between 694 proteins were identified. Colors and sizes of nodes are described in the inset key.
In summary, by applying the ubiquitination assay to the protein microarray platform, we have developed a sensitive assay that can be used to discover numerous substrates simultaneously, covering the whole proteome. Our approach should support the screening of other E3 systems both in humans and in yeast. Furthermore, once proteins on the array are ubiquitinated by a particular E3, deubiquitinating enzymes could be screened for specific substrates. In such experiments, loss of signal would indicate that a particular protein has been deubiquitinated.
Materials and methods
Purification of yeast E2 enzymes
Yeast E2 gene UBC4 was expressed in Escherichia coli strain BL21 (DE3) from pET15b plasmids as described previously (Kus et al, 2004). Transformed cells were grown at 37°C to an absorbance of A590 of 0.6 in 2 l of Luria broth and expression was induced by addition of 1 mM isopropyl-β-1-thio-D-galactopyranoside (IPTG). After 12 h of induction at 16°C, the cells were harvested and lysed by sonication in binding buffer (20 mM HEPES, pH 8.0, 500 mM NaCl, 10% glycerol, 10 μM ZnCl2, 0.5 mM tris(2-chloroethyl) phosphate (TCEP)) and protease inhibitor tablets (one tablet per 50 ml of buffer, Roche Applied Science) containing 5 mM imidazole. Lysates were clarified by centrifugation at 100 000 g for 1 h at 4°C and His-tagged proteins were purified from the clarified lysate on a 2-ml nickel–nitrilotriacetic acid superflow agarose column (Qiagen). Bound proteins were washed with binding buffer supplemented with 30 mM imidazole and eluted with binding buffer supplemented with 500 mM imidazole.
Purification of yeast E3 Rsp5
The GST-Rsp5 expression plasmid (pGEX-6P2-RSP5) (Kus et al, 2004) was used to express GST-Rsp5 in E. coli using the same method as described for the E2 enzyme, except that imidazole was omitted from the binding buffer. The recombinant proteins were purified from the cell lysate on a column containing 3 ml of glutathione–Sepharose resin (Amersham Biosciences), washed once with 50 ml of binding buffer, followed by a wash with 25 ml of PreScission cleavage buffer (PCB: 50 mM Tris–HCl, pH 7.0, 150 mM NaCl, 1 mM TCEP, 10% glycerol). Rsp5 was proteolytically cleaved from the GST moiety by incubating the resin for 4 h with 1 ml of PCB containing 40 U of PreScission protease (Amersham Biosciences).
Labeling of Rsp5
Rsp5 was purified as described above, except that 50 mM Tris–HCl was replaced with 50 mM HEPES in the PCB. Purified Rsp5 (1 mg/ml) was labeled with AlexaFluor 647 using the Microscale Protein Labeling kit (Molecular Probes) according to manufacturers' instructions. A 50 μg weight of Rsp5 was used for the reaction. Final concentration of Alexa647-Rsp5 was 0.1 mg/ml in a volume of 100 μl.
Purification of yeast GST-CTD
The GST-CTD expression vector (pET21a-GST-TEV-CTD) was constructed by Dr N Fong and generously provided by Dr D Bentley. GST-CTD was expressed in E. coli and purified using the same method as described for the GST-Rsp5, except that proteins were eluted with binding buffer containing 15 mM glutathione.
Purification of yeast GST-tagged substrates
The collection of yeast strains expressing GST proteins was a generous gift from Dr M Snyder. Culture of the yeast strains and expression of recombinant GST proteins were carried out as described previously (Kus et al, 2005). Proteins were purified from 50 ml of growth media using 100 μl of glutathione–Sepharose resin and eluted in 100 μl of binding buffer containing 15 mM glutathione (Amersham Biosciences). The final yield of purified proteins varied from 10 to 200 μg.
Protein analysis
All purified proteins were resolved on 12% SDS–polyacrylamide gels and visualized by Coomassie Blue staining (Sigma B-7920). Immunoblotting was performed using a mouse monoclonal anti-GST antibody (B-14, Santa Cruz Biotechnology). Purified proteins were frozen in ethanol and dry ice and stored at −80°C.
In vitro ubiquitination
Reactions contained 3 μl of 5 × assay buffer (250 mM HEPES, pH 7.4, 25 mM MgOAc, 2.5 mM TCEP, 500 mM NaCl and 50% glycerol), 1 μg of ubiquitin (b-Ub), 0.16 μg of yeast E1, 3.8 μg of Ubc4 E2, 1.2 μg of Rsp5 E3, 8 pmol of GST-tagged substrate and 3.3 mM ATP (Sigma). E1 and b-Ub were purchased from Boston Biochem. Water was added to each reaction to bring the final volume of all reactions to 15 μl. ATP was either omitted or added last in order to minimize autocatalytic ubiquitination reactions by the ubiquitination enzymes. Reactions were allowed to proceed for 4 h at room temperature and stopped by boiling in 5 μl of sample buffer. Detection of a shift to high MW, indicative of ubiquitination, was performed by immunoblotting with anti-GST antibodies.
In vivo ubiquitination experiments
The rsp5-1 and the corresponding wild-type (RSP5) strains expressing the desired HIS-tagged proteins were grown to log phase in Ura− synthetic drop-out media containing 2% raffinose. The temperature was then changed to 37°C; the expression of the proteins was induced by the addition of galactose to 2% and growth was continued at the restrictive temperature for 2 h. The cells were then lysed and the proteins were purified from the cells using anti-HA antibodies, and immunoblotted with anti-ubiquitin antibodies (Covance).
Production of protein microarrays
For the purpose of developing the ubiquitination assay using protein microarrays, protein samples were spotted onto ArrayIt™ or eight-pad FAST™ nitrocellulose slides at various concentrations using a Piezorray™ (Perkin Elmer) platform. Following printing, the slides were stored at −20°C. Invitrogen ProtoArray® Yeast Proteome Microarray was purchased for subsequent experiments.
Ubiquitination on protein microarrays
Slides were removed from the freezer, briefly allowed to thaw, rinsed with 0.5%. PBST and then blocked for an hour in 5% skim milk prepared with 0.5% PBST. Following blocking, the slide was washed three times for 5 min each with 0.5% PBST. After washing, 0.6 ml of a ubiquitination reaction mixture (50 mM HEPES, 5 mM MgCl2·6H2O, 0.5 mM TCEP, 10% glycerol, 4 μg of FITC-labeled ubiquitin (Boston Biochem) 0.64 μg of E1, 15.2 μg of E2 (Ubc4), 4.8 μg of E3 Rsp5) was gently pipetted onto the surface of the slide after 2 μl of 100 mM ATP was added to start the ubiquitination reaction. The reaction mixture on the slide was kept humid using wet filter paper and was allowed to proceed for 3 h. Following the reaction, the slide was briefly rinsed with 0.5% PBST followed by three 10-min washes with 0.5% PBST. The slide was dried by centrifuging for 4 min at 1000 g and visualized by fluorescent laser scanning at 10 μm resolution using a 488 nm laser on a ProScan Array HT™ scanner (Perkin Elmer). Printed slides containing fluorescent proteins and dyes were kept in the dark for the duration of the experiment.
Binding assay using protein microarrays
Slides were removed from the freezer, briefly allowed to thaw, rinsed with PBS, followed by a rinse in 0.1%. PBST, and then blocked for one hour in 5% skim milk made with 0.1% PBST for 2 h. Probe solution (2.5 μl of Alexa647-Rsp5 (0.1 mg/ml) diluted in 0.75 ml of reaction buffer (50 mM HEPES, 5 mM MgCl2·6H2O, 0.5 mM TCEP, 10% glycerol) was carefully pipetted over the entire area of the slide, kept humid with a wet filter paper and the allowed to incubate for 1.5 h. The slide was washed 3 × in 0.1% PBST for 10 min and dried by centrifugation (4 min at 200 g). The slide was then scanned at 10 μm resolution using a 633 nm laser on a ProScan Array HT™ (Perkin Elmer) scanner. Printed slides containing fluorescent proteins and dyes were kept in the dark for the duration of the experiment.
Data analysis
Data from the two ubiquitination assay slides were analyzed using ProScan Array HT™ (Perkin Elmer) software. Spots on which 50% of the pixels produced signal greater than two standard deviations above the background were identified as ‘hits'. These proteins were eliminated from the Rsp5 substrate list unless both the duplicated spots met this criterion on both of the assayed slides (i.e. all four spots had to meet the criteria). Once the Rsp5 substrate list was generated, the spots were ranked according to their signal intensity calculated as (signal intensity=mean signal on the spot−background)/concentration of the protein on the spot). Once this list was generated, all the proteins with a signal intensity >2 (40 proteins) were chosen as ‘high-confidence' Rsp5 substrates. Data analysis to generate the Rsp5 interaction list was performed using the same quantitative parameters.
Cross-species comparisons
The protein complements for the 17 yeast species were obtained from the following online databases: Saccharomyces cerevisiae, S. bayanus, S. castelli, S. kluyveri, S. kudriavzevii, S. mikatae and S. paradoxus—Saccharomyces genome database (http://www.sgd.org); Candida glabrata, Debaromyces hansenii, Kluyveromyces lactis, Yarrowia lipolytica—Genolevures (http://cbi.labri.fr/Genolevures/) (Sherman et al, 2006); Candida tropicalis, Aspergillus nidulans, Fusarium graminearum, Kluyveromyces waltii, Neurospora crassa—the fungal genome initiative at the Broad Institute (http://www.broad.mit.edu/annotation/fgi/); Ashbya gossypii—National Center for Biotechnology Information (ftp://ftp.ncbi.nih.gov/genomes/Fungi); Schizosaccharomyces pombe—The Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/Projects/Fungi/).
Orthology was determined using a modified implementation of the widely used reciprocal best BLAST hits approach (Tatusov et al, 1997). In brief, BLASTP (Altschul et al, 1997) comparisons of protein complements were performed for every pair of species. For each query protein, the top 10 hits (ranked on the basis of sequence similarity) for a particular species were extracted. Each hit was then subjected to a more sensitive Smith–Waterman alignment against the original query protein (Rice et al, 2000) and hits ranked according to their bit scores. Those proteins that were found to be the best reciprocal hits (in terms of the bit scores) were then defined as ‘putative orthologues'. For species in which no such ‘putative orthologue' could be determined, proteins, which displayed the highest bit score to the query protein in the S. cerevisiae query protein, were manually inspected and compared with ‘putative orthologues' identified in other species. These proteins were defined as ‘highest scoring homologues'.
Calculation of Ka/Ks ratios
Multiple sequence alignments were generated for groups of orthologue proteins using the program—MUSCLE (Edgar, 2004). The nucleotide sequences were mapped onto this alignment using a script written in-house and the resultant alignments used for the calculating the Ka/Ks values using the codeml program available through the PAML software suite (Yang, 1997).
Isolation of WW domain binding peptides
A library of random dodecapeptides fused to the N-terminus of the M13 gene-8 major coat protein was constructed and cycled through rounds of binding selections with the bacterially expressed WW domain immobilized on 96-well Maxisorp immunoplates (NUNC), as described previously (Sidhu et al, 2000; Laura et al, 2002). Phage were propagated in E. coli XL1-blue (Stratagene) in medium supplemented with M13-KO7 helper phage (New England Biolabs) to facilitate phage production and 10 μM IPTG to induce expression of the library. After four rounds of selection, individual phage were isolated and analyzed in a phage ELISA. Phages that bound to the WW domain were subjected to DNA sequence analysis. Unique binding sequences were aligned to derive a specificity profile.
Supplementary Material
Supplementary Material
Supplementary Table SI
Supplementary Table SII
Supplementary Data Sets 1
Supplementary Data Sets 2
Acknowledgments
We thank R Kardish for protein printing on array slides and C Boone for helpful comments on the manuscript. Computational analyses were performed at the Center for Computational Biology, Hospital for Sick Children, Toronto. This work was supported by grants from the Canadian Institute of Health Research (to DR), the Canadian Foundation for Innovation (SIDNET) to DR and the Natural Sciences and Engineering Research Council of Canada to JP. JW and BK were supported by the Hospital for Sick Children (Toronto, Ontario, Canada) Research Training Centre. JP is supported by a New Investigators award from the Canadian Institute of Health Research. DR is a recipient of a CRC chair (Tier I). NJK is Sandler Family Fellow.
References
- Abriel H, Loffing J, Rebhun JF, Pratt JH, Schild L, Horisberger JD, Rotin D, Staub O (1999) Defective regulation of the epithelial Na+ channel by Nedd4 in Liddle's syndrome. J Clin Invest 103: 667–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andoh T, Hirata Y, Kikuchi A (2002) PY motifs of Rod1 are required for binding to Rsp5 and for drug resistance. FEBS Lett 525: 131–134 [DOI] [PubMed] [Google Scholar]
- Beaudenon SL, Huacani MR, Wang G, McDonnell DP, Huibregtse JM (1999) Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol Cell Biol 19: 6972–6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Snyder M (2005) Advances in functional protein microarray technology. FEBS J 272: 5400–5411 [DOI] [PubMed] [Google Scholar]
- Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ (2007a) Towards a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics 6: 439–450 [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, Han J, Ingvarsdottir K, Cheng B, Andrews B, Boone C, Berger SL, Hieter P, Zhang Z, Brown GW, Ingles CJ, Emili A, Allis CD, Toczyski DP, Weissman JS, Greenblatt JF, Krogan NJ (2007b) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TM, Dawson VL (2003) Molecular pathways of neurodegeneration in Parkinson's disease. Science 302: 819–822 [DOI] [PubMed] [Google Scholar]
- Dupre S, Urban-Grimal D, Haguenauer-Tsapis R (2004) Ubiquitin and endocytic internalization in yeast and animal cells. Biochim Biophys Acta 1695: 89–111 [DOI] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Seraphin B (2007) A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol 14: 15–22 [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F (1992) SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev 6: 1319–1331 [DOI] [PubMed] [Google Scholar]
- Fang S, Weissman AM (2004) A field guide to ubiquitylation. Cell Mol Life Sci 61: 1546–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636 [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A (2002) The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373–428 [DOI] [PubMed] [Google Scholar]
- Helliwell SB, Losko S, Kaiser CA (2001) Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J Cell Biol 153: 649–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hesselberth JR, Miller JP, Golob A, Stajich JE, Michaud GA, Fields S (2006) Comparative analysis of Saccharomyces cerevisiae WW domains and their interacting proteins. Genome Biol 7: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L, Dunn R (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol 19: 141–172 [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S (2000) Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102: 577–586 [DOI] [PubMed] [Google Scholar]
- Horak J (2003) The role of ubiquitin in down-regulation and intracellular sorting of membrane proteins: insights from yeast. Biochim Biophys Acta 1614: 139–155 [DOI] [PubMed] [Google Scholar]
- Hu H, Columbus J, Zhang Y, Wu D, Lian L, Yang S, Goodwin J, Luczak C, Carter M, Chen L, James M, Davis R, Sudol M, Rodwell J, Herrero JJ (2004) A map of WW domain family interactions. Proteomics 4: 643–655 [DOI] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM (1995) A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA 92: 2563–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham RJ, Gish G, Pawson T (2004) The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene 23: 1972–1984 [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir MA, Kaminska J, Segel GB, Bethlendy G, Lin P, Della Seta F, Blegen C, Swiderek KM, Zoladek T, Arndt KT, Sherman F (2005) Physiological effects of unassembled chaperonin Cct subunits in the yeast Saccharomyces cerevisiae. Yeast 22: 219–239 [DOI] [PubMed] [Google Scholar]
- Kanelis V, Bruce MC, Skrynnikov NR, Rotin D, Forman-Kay JD (2006) Structural determinants for high-affinity binding in a Nedd4 WW3* domain–Comm PY motif complex. Structure 14: 543–553 [DOI] [PubMed] [Google Scholar]
- Kanelis V, Rotin D, Forman-Kay JD (2001) Solution structure of a Nedd4 WW domain–ENaC peptide complex. Nat Struct Biol 8: 407–412 [DOI] [PubMed] [Google Scholar]
- Kang RS, Daniels CM, Francis SA, Shih SC, Salerno WJ, Hicke L, Radhakrishnan I (2003) Solution structure of a CUE–ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell 113: 621–630 [DOI] [PubMed] [Google Scholar]
- Kasanov J, Pirozzi G, Uveges AJ, Kay BK (2001) Characterizing class I WW domains defines key specificity determinants and generates mutant domains with novel specificities. Chem Biol 8: 231–241 [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P (1999) SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell 4: 21–33 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MH, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O'Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- Kus B, Gajadhar A, Stanger K, Cho R, Sun W, Rouleau N, Lee T, Chan D, Wolting C, Edwards A, Bosse R, Rotin D (2005) A high throughput screen to identify substrates for the ubiquitin ligase Rsp5. J Biol Chem 280: 29470–29478 [DOI] [PubMed] [Google Scholar]
- Kus BM, Caldon CE, Andorn-Broza R, Edwards AM (2004) Functional interaction of 13 yeast SCF complexes with a set of yeast E2 enzymes in vitro. Proteins 54: 455–467 [DOI] [PubMed] [Google Scholar]
- Kwapisz M, Cholbinski P, Hopper AK, Rousset JP, Zoladek T (2005) Rsp5 ubiquitin ligase modulates translation accuracy in yeast Saccharomyces cerevisiae. RNA 11: 1710–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laura RP, Witt AS, Held HA, Gerstner R, Deshayes K, Koehler MF, Kosik KS, Sidhu SS, Lasky LA (2002) The Erbin PDZ domain binds with high affinity and specificity to the carboxyl termini of delta-catenin and ARVCF. J Biol Chem 277: 12906–12914 [DOI] [PubMed] [Google Scholar]
- Lifton RP, Gharavi AG, Geller DS (2001) Molecular mechanisms of human hypertension. Cell 104: 545–556 [DOI] [PubMed] [Google Scholar]
- Liu YC (2004) Ubiquitin ligases and the immune response. Annu Rev Immunol 22: 81–127 [DOI] [PubMed] [Google Scholar]
- McNatt MW, McKittrick I, West M, Odorizzi G (2007) Direct binding to Rsp5 mediates ubiquitin-independent sorting of Sna3 via the multivesicular body pathway. Mol Biol Cell 18: 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modzelewska A, Filipek S, Palczewski K, Park PS (2006) Arrestin interaction with rhodopsin: conceptual models. Cell Biochem Biophys 46: 1–15 [DOI] [PubMed] [Google Scholar]
- Nakayama KI, Nakayama K (2006) Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer 6: 369–381 [DOI] [PubMed] [Google Scholar]
- Oestreich AJ, Aboian M, Lee J, Azmi I, Payne J, Issaka R, Davies BA, Katzmann DJ (2007) Characterization of multiple multivesicular body sorting determinants within Sna3: a role for the ubiquitin ligase Rsp5. Mol Biol Cell 18: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP (2003) A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 21: 921–926 [DOI] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, McCartney RR, Schmidt MC, Rachidi N, Lee SJ, Mah AS, Meng L, Stark MJ, Stern DF, De Virgilio C, Tyers M, Andrews B, Gerstein M, Schweitzer B, Predki PF, Snyder M (2005) Global analysis of protein phosphorylation in yeast. Nature 438: 679–684 [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A (2000) EMBOSS: the European molecular biology open software suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Roberts SM, Winston F (1997) Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147: 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D, Staub O, Haguenauer-Tsapis R (2000) Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J Membr Biol 176: 1–17 [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, Weissman JS, Krogan NJ (2005) Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123: 507–519 [DOI] [PubMed] [Google Scholar]
- Schweitzer B, Predki P, Snyder M (2003) Microarrays to characterize protein interactions on a whole-proteome scale. Proteomics 3: 2190–2199 [DOI] [PubMed] [Google Scholar]
- Shcherbik N, Kee Y, Lyon N, Huibregtse JM, Haines DS (2004) A single PXY motif located within the carboxyl terminus of Spt23p and Mga2p mediates a physical and functional interaction with ubiquitin ligase Rsp5p. J Biol Chem 279: 53892–53898 [DOI] [PubMed] [Google Scholar]
- Sherman D, Durrens P, Iragne F, Beyne E, Nikolski M, Souciet JL (2006) Genolevures complete genomes provide data and tools for comparative genomics of hemiascomycetous yeasts. Nucleic Acids Res 34: D432–D435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu SS, Lowman HB, Cunningham BC, Wells JA (2000) Phage display for selection of novel binding peptides. Methods Enzymol 328: 333–363 [DOI] [PubMed] [Google Scholar]
- Smith MG, Jona G, Ptacek J, Devgan G, Zhu H, Zhu X, Snyder M (2005) Global analysis of protein function using protein microarrays. Mech Ageing Dev 126: 171–175 [DOI] [PubMed] [Google Scholar]
- Spiechowicz M, Filipek A (2005) The expression and function of Sgt1 protein in eukaryotic cells. Acta Neurobiol Exp (Wars) 65: 161–165 [DOI] [PubMed] [Google Scholar]
- Stamenova SD, Dunn R, Adler AS, Hicke L (2004) The Rsp5 ubiquitin ligase binds to and ubiquitinates members of the yeast CIN85–endophilin complex, Sla1-Rvs167. J Biol Chem 279: 16017–16025 [DOI] [PubMed] [Google Scholar]
- Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J, Rotin D (1996) WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J 15: 2371–2380 [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ (1997) A genomic perspective on protein families. Science 278: 631–637 [DOI] [PubMed] [Google Scholar]
- Wang G, Yang J, Huibregtse JM (1999) Functional domains of the Rsp5 ubiquitin-protein ligase. Mol Cell Biol 19: 342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems AR, Schwab M, Tyers M (2004) A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta 1695: 133–170 [DOI] [PubMed] [Google Scholar]
- Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13: 555–556 [DOI] [PubMed] [Google Scholar]
- Yashiroda H, Oguchi T, Yasuda Y, Toh EA, Kikuchi Y (1996) Bul1, a new protein that binds to the Rsp5 ubiquitin ligase in Saccharomyces cerevisiae. Mol Cell Biol 16: 3255–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, Mitchell T, Miller P, Dean RA, Gerstein M, Snyder M (2001) Global analysis of protein activities using proteome chips. Science 293: 2101–2105 [DOI] [PubMed] [Google Scholar]
- Zhu H, Snyder M (2003) Protein chip technology. Curr Opin Chem Biol 7: 55–63 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplementary Table SI
Supplementary Table SII
Supplementary Data Sets 1
Supplementary Data Sets 2