Abstract
Although there exists a large family of nuclear transport receptors (Karyopherins), the majority of known import cargoes use an adapter protein, Importin-α (Impα), which links the cargo to a karyopherin, Importin-β (Impβ). The reason for the existence of transport adapters is unknown. One hypothesis is that, as Impα re-export is coupled to GTP hydrolysis, it can drive a higher concentration of nuclear cargo than could be achieved by direct cargo binding to Importin-β. However, computer simulations predicted the opposite outcome, and showed that direct transport is faster than adapter-mediated transport. These predictions were validated experimentally. The data, together with previous analyses of nuclear protein import, suggest that the use of adapters such as importin-α provides the cell with increased dynamic range for control of nuclear import rates, but at the expense of efficiency.
Keywords: cell biology, computer modeling, nuclear transport
Introduction
Nuclear transport serves as a key regulatory step in signal transduction, cell cycle progression, and mRNA processing (Gorlich and Kutay, 1999; Macara, 2001; Weis, 2002). Access to the nucleus is provided by nuclear pore complexes that allow passive diffusion by small molecules, but restrict translocation by molecules larger than ∼40 kDa (Fahrenkrog and Aebi, 2003; Suntharalingam and Wente, 2003). Entry or exit of large molecules is usually mediated by soluble receptors such as the karyopherins, which are associated with the nucleoporin proteins that line the pores. Importins bind cargo in the cytoplasm and release it in the nucleus, whereas exportins reverse this process (Chook and Blobel, 2001).
Transport is driven in both directions by the high concentration of the GTPase Ran bound to GTP in the nucleus. Importins can only bind cargo in the absence of RanGTP. The association of RanGTP with an importin induces cargo release (Gorlich et al, 1996b). The RanGTP gradient across the NPC is maintained by the restriction of RCC1 (a guanine nucleotide exchange factor, RanGEF) to the nucleus and of the GTPase-activating protein, RanGAP (Bischoff and Ponstingl, 1991), to the cytoplasm and NPC. With each transport cycle, one RanGTP is exported and one RanGDP is returned to the cytoplasm by the transport receptor NTF2 (Ribbeck et al, 1998; Smith et al, 1998).
The majority of known import cargoes contain a nuclear localization signal (NLS) (Conti et al, 1998; Herold et al, 1998). This ‘bar-code' can be a monopartite stretch of seven basic amino acids or a longer, bipartite sequence. The NLS is recognized by an adapter protein, importin-α (Impα), which binds to the karyopherin importin-β (Impβ). In the nucleus, RanGTP binding to Impβ disassociates the complex. RanGTP–Impβ then translocates back to the cytoplasm, where RanGAP (assisted by RanBP1) hydrolyzes RanGTP, releasing Impβ for another round of transport (Bischoff and Gorlich, 1997). Impα in the nucleus is exported by a specific exportin called CAS, which also promotes release of cargo from the adapter (Floer et al, 1997; Kutay et al, 1997; Petersen et al, 2000).
Some import cargoes bind Impβ directly. Without the need to export Impα from the nucleus, this import pathway uses only one GTP cycle rather than two. Because the Impα pathway utilizes more energy and more protein production, strong selective pressure must have driven its evolution in the cell. One possibility is the flexibility that adapter proteins provide to allow specific populations of cargoes to be imported at different times or different cellular states. At least five isoforms of Impα exist in mammalian cells, and some isoforms show specificity for particular protein cargoes. However, budding yeast expresses only a single Impα. Another possibility is that, as Impα-mediated transport is coupled to the hydrolysis of two GTP molecules, it might drive a higher nuclear/cytoplasmic cargo gradient than direct Impβ-mediated import.
We now demonstrate, using a combined in silico/experimental approach, that contrary to expectations, Impα-mediated transport is actually less efficient than direct import by Impβ. Direct import is faster, and can drive a higher nuclear/cytoplasmic cargo gradient. In addition, we show that a bipartite NLS can accumulate in the nucleus to a higher concentration than a monopartite NLS, as predicted by our computer model. However, an in silico sensitivity analysis shows that Impα provides a greater dynamic range of control over import than Impβ. To test this prediction in vivo, we use a combination of recombinant protein co-injection and siRNA knockdown.
Results
Cargo gradients in adapter-mediated and direct import
To investigate cargo gradients in both types of import, we developed a 3-compartment in silico transport model (Figure 1). Details of the model are in Materials and methods, and the complete schematic for the cargo import, Ran transport, and Karyopherin transport modules can be found in the Supplementary Data by Riddick and Macara (2005). Addition of either type of cargo to the cytoplasm was simulated by instantaneously stepping its concentration from 0 to 4 μM and measuring nuclear accumulation over 1800 s. Unexpectedly, cargo imported directly by Impβ had a greater initial rate and a higher steady-state nuclear accumulation than cargo imported via the adapter, Impα (Figure 2A). This difference results from the greater reaction rate for a bimolecular interaction, faster cycling time of Impβ between the nucleus and the cytoplasm, and the slightly higher permeability for the Impβ–cargo complex through the NPC, as compared to the Impα/β–cargo complex.
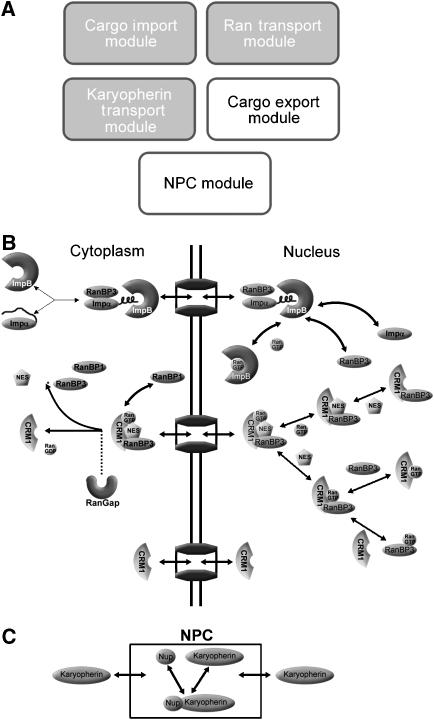
Figure 1.
Overview of the CRM1/RanBP3 export model. (A) The entire nuclear transport model includes modules for cargo import, Karyopherin transport, Ran transport, cargo export, and the NPC. (B) Detail of the cargo export module. RanBP3, CRM1, RanGTP, and the NES cargo combine to form an export complex. After translocating through the NPC, RanBP1 binds to the complex and allows RanGAP to hydrolyze RanGTP to RanGDP, releasing the cargo and disassociating the complex. RanBP3 contains an NLS and is imported into the nucleus by Impα/Impβ. (C) Detail of the Nuclear Pore Complex module. The NPC is represented as a single separate compartment containing nucleoporins that bind the cargo complex.
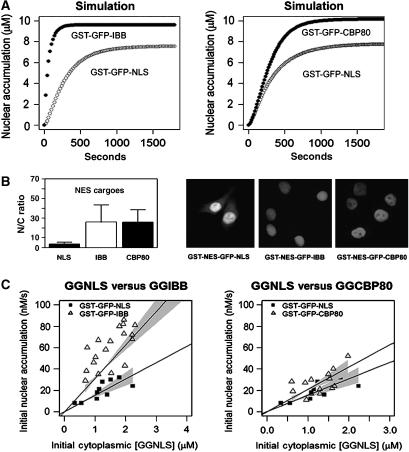
Figure 2.
Comparison of import rates and steady-state nuclear accumulation for NLS, CBP80, and IBB protein cargoes. (A) Left panel: simulation results predict that cargo imported directly by Impβ has greater initial rate and steady-state level of nuclear accumulation than cargo imported via the Impα adapter. Right panel: the bipartite NLS, CPB80, is predicted to accumulate more efficiently than the monopartite NLS. (B) A total of 20 μM GST-NES-GFP-NLS, GST-GFP-NES-CBP80, or GST-NES-GFP-IBB was injected into HeLa cell cytoplasm. Confocal images were collected 30 min post-injection. N/C ratios were calculated from mean pixel intensities for the nuclear and cytoplasmic compartments. Values are means of 15 cells±1 s.d. (C) Experimental validation of the computer predictions. A portion of 20 μM cargo was injected into the cytoplasm of HeLa cells. Images were collected every 20 s. Pixel intensity was converted to concentration by use of an external series of standards. Initial rate was defined as the rate of change of nuclear concentration during the first 30 s. Initial concentration for each cell was plotted against initial rate and the slope was fit by linear regression. Each data point represents an individual cell. Gray areas indicate the 99% confidence interval of the regression line.
To evaluate steady-state accumulation of Impα/Impβ cargo in our experimental system, we built GST-NES-GFP-NLS, which contains both an import and an export signal. The export signal is an NES from protein kinase inhibitor (PKI) that is recognized by CRM1 (Henderson and Eleftheriou, 2000). To compare Impα adapter-mediated import with direct import, we prepared a second cargo protein, GST-NES-GFP-IBB, which contains an IBB motif (Gorlich et al, 1996a). The IBB domain is a 41 amino acid arginine-rich fragment from Impα that is representative of a class of NLSs that bind directly to Impβ (Palmeri and Malim, 1999). To describe these shuttling cargoes in the model, we added pathways for export through CRM1 and cofactor RanBP3 (Figure 1B).
We injected each cargo into the cytoplasm of HeLa cells and recorded the nuclear/cytoplasmic ratio (N/C ratio) after 30 min (Materials and methods). The direct Impβ (IBB) cargo achieved a significantly larger nuclear/cytoplasmic gradient than the Impβ/Impα (NLS) cargo (Figure 2B). To compare initial rates of import, we used a GST-GFP-IBB cargo. After microinjecting either GST-GFP-IBB or GST-GFP-NLS into the cytoplasm of HeLa cells, initial import rates were recorded as described previously (Riddick and Macara, 2005). Import of GST-GFP-IBB was significantly faster than that of GST-GFP-NLS (Figure 2C).
Next, we sought to determine if the results for the monopartite NLS were generalizable to a bipartite NLS. Bipartite NLSs, like that found in nuclear CAP-binding protein subunit p80 (CBP80), are known to bind to Impα with a greater affinity than monoparite NLSs (Robbins et al, 1991; Fontes et al, 2000). Simulation of bipartite NLS cargo import predicted a similar initial rate, but increased N/C ratio as compared to monopartite NLS cargo (Figure 2A). To test this prediction, we measured initial import rates and steady-state accumulation for GST-GFP-CBP80 and GST-NES-GFP-CBP80 in HeLa cells. GST-GFP-CBP80 shows a similar initial rate to the monopartite cargo and GST-NES-GFP-CBP80 a much higher N/C ratio at steady state, in agreement with the model (Figure 2B and C).
Steady-state sensitivity analysis
If adapter proteins do not allow the production of a greater cargo gradient, what other advantage might they offer? Previously, we used sensitivity analysis to explore the coupling of reactant concentrations to cargo import rates (Riddick and Macara, 2005). Impα had the largest dynamic range of control over initial rate, although Ran and NTF2 also functioned as limiting reactants in the system. Surprisingly, Impβ, CAS, and the guanine exchange factor RCC1 inhibited import at higher levels of concentrations. As steady-state cargo concentrations are likely to have greater cellular consequences than initial rates, we performed a sensitivity analysis to correlate reactant concentrations with steady-state cargo accumulation. Initial reactant concentrations were varied individually from 0.0001- to 10 times that of their original values. After allowing the system to reach steady state, cytoplasmic injection of a shuttling cargo was simulated by increasing concentration of the cargo in silico instantaneously from 0 to 4 μM. After a return to steady state, the ratio of nuclear/cytoplasmic concentration was calculated (Supplementary Figure 1). Steady-state nuclear/cytoplasmic cargo ratio followed the same general trends as the initial import rates (Riddick and Macara, 2005); Impα, Ran, and NTF2 act as limiting reactants, whereas high concentrations of RCC1 and Impβ reduce the N/C ratio. The inhibitory effect of Impβ has two primary sources. First, excess Impβ can travel through the NPC without cargo and bind with RanGTP in the nucleus before returning to the cytoplasm. This process, called ‘futile cycling', depletes the RanGTP gradient. Second, Impβ must react with RanGTP on the nuclear side of the NPC to be released from a binding site on nucleoporins in the nuclear basket. Without sufficient RanGTP, Impβ, and cargo complexes arrest within the nuclear pore, blocking traffic in both directions.
Impα increases dynamic range of control over import
To test these predictions, we perturbed reactant concentrations in intact cells. The shuttling cargo was injected together with recombinant proteins in a known molar ratio. By quantifying the amount of the fluorescent cargo in the cytoplasm, we could then reliably calculate the concentration of the co-injected protein. To test for decreased levels of Impβ and Impα1, we transfected siRNAs targeted against these karyopherins. The shuttling cargo was then injected and the N/C ratio was recorded at steady state. To evaluate the level of knockdown in the injected cells, they were fixed and immunostained using anti-Impβ or anti-Impα1 and secondary antibodies conjugated to Texas Red. Pixel intensities of the Texas Red signal recorded by confocal microscopy could then be used to estimate whole-cell concentration of the karyopherin, relative to control cells.
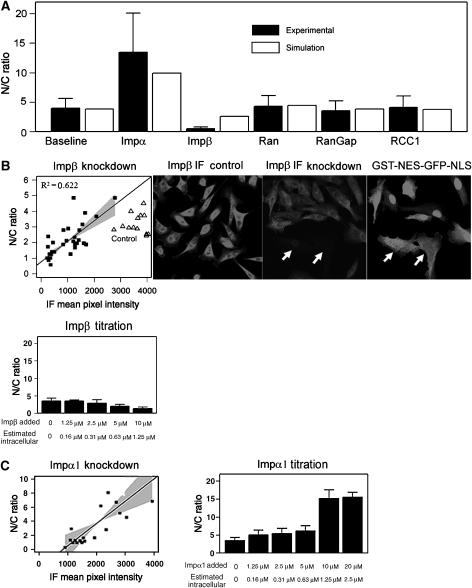
Knockdown of Impβ reduced nuclear accumulation in agreement with the sensitivity analysis (Figure 3B). Knockdown of Impα1 shows that changing Impα1 levels exert larger corresponding changes in nuclear cargo accumulation (Figure 3C), indicating a larger dynamic range of control.
Figure 3.
Effect of transport protein levels on the steady-state accumulation of the shuttling GST-NES-GFP-NLS cargo. (A) HeLa cells were injected with a mixture of 20 μM GST-NES-GFP-NLS and 4 μM recombinant transport protein. Cells were kept at 37°C in physiological saline for the duration of the experiment. N/C ratios were measured 30 min post-injection. Simulation results using identical conditions (white) are displayed next to experimental results (black). (B) Upper panel: HeLa cells were transfected with siRNA for Impβ. After incubation in DMEM (5% CS, 5% FCS, 1% PS) for 72 h, cells were injected with the GST-NES-GFP-NLS cargo. After further incubation at 37°C for 30 min, cells were fixed with formalin and immunostained with anti-Impβ and an Alexa-546 conjugated secondary antibody. N/C ratio of the cargo was defined by the ratio of mean pixel intensity in the nucleus divided by that of the cytoplasm. IF intensity was measured as the mean pixel value of the entire cell. IF intensity, which reflects the relative abundance of Impβ, was plotted against N/C ratio of GST-NES-GFP-NLS. The slope was fit by linear regression. Gray areas represent 99% CI of the regression line. (B) Lower panel: HeLa cells were co-injected with 20 μM GST-NES-GFP-NLS and various concentrations of recombinant Impβ (1.25, 2.5, 5 and 10 μM). Intracellular concentration of Impβ was estimated using the approximate average cargo concentration (3 μM) from fluorescence as described previously and the known molar ratio of cargo to recombinant protein. (C) Left panel: HeLa cells were transfected with siRNA for Impα1, microinjected with GST-NES-GFP-NLS, and immunostained with anti-Impα1 as previously described. (C) Right panel: HeLa cells were co-injected with 20 μM GST-NES-GFP-NLS and various concentrations of recombinant Impα1 (1.25, 2.5, 5, 10, and 20 μM). Intracellular concentration of Impα1 was estimated using the approximate average cargo concentration (3 μM) from fluorescence as described previously and the known molar ratio of cargo to recombinant protein.
Co-injected recombinant Impα1 strongly upregulated the nuclear accumulation of the shuttling cargo (Figure 3A), whereas co-injected Impβ suppressed nuclear accumulation of the cargo, as predicted. Titration of co-injected Impα1 and Impβ levels shows how increases in Impα1 concentration have a greater effect on the range of cargo accumulation (Figure 3B and C). Although our current model includes competition for nucleoporins, it does not include a three-dimensional spatial representation of the NPC that could capture the blocking effect of Impβ in the confined space of the nuclear basket. This difference may explain the somewhat greater inhibitory effect of excess Impβ seen in vivo.
Discussion
Using a combined modeling/experimental approach, we have shown that adapter-mediated import offers no advantage in driving a cargo gradient, despite using twice as much energy as that of direct import. However, Impα shows a large dynamic range of control over cargo accumulation. In contrast, Impβ shows an ‘inverted U' response curve in which either increasing or decreasing Impβ inhibits import. This behavior results from the high-affinity binding of Impβ both to RanGTP and to the nuclear basket. Excess of Impβ can cause futile cycling and deplete the RanGTP gradient. Diminished nuclear RanGTP can cause accumulation of Impβ at the nuclear basket to the point at which it begins to occlude the pore, severely restricting transport of cargo complexes.
Adapter proteins may, therefore, have evolved as a means to more flexibly control cargo gradients under different cellular conditions. For example, recent work has shown how the modulation of nuclear transport rates maintains the fidelity of wave propagation in cell cycle progression (Becskei et al, 2004). Flexibility in biological systems describes an organism's ability to adjust to changing environments. This control comes at the expense of efficiency and requires an additional expenditure of energy. But the advantage that comes with this flexibility is increased robustness, the ability of a system to remain stable in the face of external perturbations (Csete and Doyle, 2002; Stelling et al, 2002). This trade-off between regulation and efficiency is fundamental to all control systems and is likely to be widespread in cell biology.
Another group has recently looked at the dynamics of cargo import in yeast cells (Timney et al, 2006). They found a simple linear relationship between initial cytoplasmic concentration and initial import rate similar to that previously found in mammalian cells. This relationship remained constant even up to 100 μM initial concentration of cargo, demonstrating the remarkable capacity of the NPC to handle large amounts of cargo transport. Import rates were found to be 0.07–1.2 cargo molecules/NPC/μM, comparable to that found previously in our study of import in mammalian cells.
Timney et al (2006) also found that the initial rate of an NLS-GFP cargo increased with higher Karyopherin concentrations up to about 15 μM, at which point import rates began to show saturation kinetics. Although the Timney et al (2006) contrast this result with our finding that Impβ inhibits import at higher concentrations, Kap95p is most closely related to Impβ in mammalian cells. Kap95p shows a high affinity to the nucleoporins in the nuclear basket in a similar way to Impβ, so a test of Kap95p abundance in yeast cells would be a more definitive comparison. We suggest that the high-affinity binding of Impβ and Kap95p to the nuclear side of the NPC make these Karyopherins especially sensitive to limitations in the Ran gradient as well as competition for binding sites that can cause the inhibition of transport we have observed at high concentrations of these receptors.
Import rates were successfully fit using a simple model of cargo-binding kinetics, passive import, and passive leak kinetics. Timney et al (2006) interpreted these findings as evidence that nuclear transport follows a simple ‘pump-leak' model in which import rates are largely determined by the number of Karyopherin–Cargo complexes that form and the rate at which cargo passively diffuses back to the cytoplasm. This model is consistent with our findings that the Karyopherin Impα acts as a limiting reactant for import and the bipartite NLS (which binds Impα more tightly) leads to greater nuclear accumulation of cargo. Both these factors would act to increase the amount of effective Karyopherin–cargo complexes in vivo. Timney et al (2006) also use their model to predict that the Ran gradient is not limiting for import. However, we have shown previously (Riddick and Macara, 2005) that both Ran and NTF2 are limiting for import in whole HeLa cells. Our results do agree with Timney et al (2006) in showing that the capacity of the NPC is enormous and not likely to be limiting for cargo import rates.
Recent work from another group has shown that the rate of cargo transport through the NPC could be modulated ∼10-fold by Impβ in permeabilized mammalian cells (Yang and Musser, 2006). However, permeabilized cells have an NPC depleted of native Karyopherins and would not show competitive inhibition seen in whole cells. This is reflected by experimental evidence that import rates in whole cells are more than an order of magnitude less than those of permeabilized cells (Timney et al, 2006). Therefore, any quantitative result from permeabilized cells should be interpreted with caution. As total Karyopherin concentration in whole cells has been estimated to be 15 μM (Smith et al, 2002), Impβ might be expected to start inhibiting transport above these levels in permeabilized cells.
Our nuclear transport model is the first to include a detailed reconstruction both of nuclear import through Impα/Impβ and export through CRM1. It will allow future work to explore how the regulation of nuclear import couples to signal transduction pathways such as PKA/CREB and Jak-Stat in which shuttling factors may increase responsiveness of the system to changes in the state of the receptor at the cell surface.
Materials and methods
Computer model
Our original computer model for nucleo-cytoplasmic transport considered only the receptor-mediated import of cargo. In addition, it ignored the complexities of translocation through the NPC as a complex, and described nucleo-cytoplasmic movements by single permeability constant (Riddick and Macara, 2005). Experimental work has shown that this simple linear model can successfully describe the translocation process to a first approximation, and has the advantage that the permeability constant can be derived experimentally. However, more complex behaviors, such as competition between transport receptors at the NPC, cannot be represented in this way.
To create a more realistic model of the translocation process, we added a third compartment that represents the entire volume of all nuclear pores embedded in the nuclear envelope. Compartmental models assume that all species diffuse completely within compartments. Therefore, nucleoporins were modeled as freely diffusing but effectively trapped within the nuclear pore. Although most nucleoporins do not freely diffuse in vivo, this approximation should be reasonably accurate, as all kinetic measurements of nucleoporins have been measured from dilute solutions. Nucleoporins show an increasing gradient of affinity for Impβ, with interior nucleoporins showing an affinity of approximately 100 nM (Ben-Efraim and Gerace, 2001). We chose this value as an average representational value for all nucleoporins in the NPC compartment of the model.
In the three-compartment model, the three permeability equations used to represent transport are as follows: (1) permeability between the cytoplasm and NPC; (2) binding to nucleoporins within the NPC; and (3) permeability between the NPC and nucleus. Under conditions in which nucleoporins are not saturated, karyopherins transit the pore with permeability that approximates that used in the two-compartment model.
We also added terms in the model to represent Crm1-mediated protein export, and included the interaction of Crm1 with RanBP3, which can function as a cofactor to enhance cargo binding to Crm1. These modifications permit the analysis of shuttling cargoes, which possess both an NLS and a nuclear export signal (NES). The NES acts as a constant load against which the import machinery must work to maintain nuclear accumulation of the cargo. Differences in the efficiency of import can then be assessed by measuring the N/C ratio at steady state (which in the absence of an NES would approach infinity).
The complete model was simulated using Jarnac, a Biochemical simulation package for Windows (Sauro et al, 2003). The reactions were converted internally by Jarnac to a series of coupled ODEs. Jarnac was then used to solve the ODEs based on a set of initial values. Cellular concentrations for Impα, CRM1, and RanBP3 were measured experimentally as described below. Rate constants for CRM1/RanBP3 export were taken from the literature wherever possible (Table I). The entire model will be available from the Biomodels database (http://www.ebi.ac.uk/biomodels/) in SBML format.
Table 1.
Kinetic constants used in the computer model
| Reaction | Kon M s−1 | Koff s−1 | Reference |
|---|---|---|---|
| Impβ+NUP | 1.00E+08 | 10 | Ben-Efraim and Gerace (2001) |
| NTF2+NUP | 1.00E+07 | 200 | Chaillan-Huntington et al (2000) |
| Cas+NUP | 1.00E+07 | 200 | Estimate from Chaillan-Huntington et al (2000) |
| CRM1+NUP | 1.00E+07 | 200 | Estimate from Chaillan-Huntington et al (2000) |
| RanGTP+RanBP3 | 1.00E+07 | 10 | Englmeier et al (2001) |
| CRM1+RanBP3 | 1.00E+07 | 1 | Englmeier et al (2001) |
| CRM1−RanBP3+NES | 1.00E+07 | 1 | Lindsay (2001) |
| CRM1−RanBP3+Rt | 1.00E+07 | 1 | Englmeier et al (2001) |
| Crm1−RanBP3−NES+RanGTP | 1.00E+07 | 0.3 | Englmeier et al (2001) |
| CRM1−RanBP3−RanGTP+NES | 1.00E+07 | 0.3 | Lindsay (2001) |
| CRM1−RanBP3−NES−RanGTP+RanBP1 | 3.00E+06 | 4.00E-10 | Estimate from Askjaer et al (1999) |
Constructs and cell culture
The GST-GFP-NLS construct has been described previously (Riddick and Macara, 2005). The IBB domain (Impα1 residues 1–88) was first subcloned from GFP-IBB into GST-GFP to produced GST-GFP-IBB. GST-GFP-CPB80 has been described previously (Tachibana et al, 1999). The PKI NES (LALKLAGLDI) was then subcloned into GST-GFP-NLS, GST-GFP-IBB, and GST-GFP-CBP80 to produce the shuttling constructs GST-NES-GFP-NLS and GST-NES-GFP-IBB, and GST-NES-GFP-CBP80.
Quantitative Western blotting
Concentrations of recombinant Ran, Impα, CRM1, and RanBP3 were first quantified by comparison to BSA standards on SDS–PAGE stained with Coomassie blue. Known concentrations of Ran and each transport receptor from 0.5 to 0.05 μg were then run on SDS–PAGE, together with 20 μl HeLa cell lysate. After blotting for both Ran and each transport receptor individually, concentration of proteins in the HeLa cell lysate was estimated by comparison to the recombinant standards. Estimated cellular concentration for transport receptor proteins (Impα 1 μM; CRM1 0.3 μM; RanBP3 0.05 μM) was then calculated based on the previously published concentration of Ran in HeLa cells (6 μM).
siRNA knockdown
HeLa cells were transfected with Impβ siRNA, Impα siRNA, or the control siRNA (Dharmacon random 21-mer) using SiPort (Ambion) (Impα), or Oligofectamine (Invitrogen) (Impβ) according to the manufacturers protocol. After 72 h, the media was changed to physiological saline and cells were cytoplasmically injected with the shuttling GST-NES-GFP-NLS protein. Media was then changed to DMEM (5% FCS, 5% CS, 1% PS) and the cells were incubated for 30 min at 37°C. Following incubation, cells were fixed and immunostained as described previously (Smith et al, 1998) for Impβ (ABS) or Impα (Transduction Laboratories).
Microinjection and confocal microscopy
Microinjection and microscopy were performed as described previously (Riddick and Macara, 2005). Nuclear to cytoplasmic ratio was defined by the ratio of mean nuclear pixel intensity to mean cytoplasmic pixel intensity 30 min after injection.
Supplementary Material
Supplementary Figure 1
Acknowledgments
We thank Ting Chen for the preparation of recombinant Ran. We also thank to Leslie Loew, Boris Slepchenko, and Jim Schaff of the Virtual Cell Group at the University of Connecticut for helpful discussions on compartmental simulation. This work was supported by grants GM50525 and U54 RR 022232 from the National Institutes of Health, DHHS.
References
- Askjaer P, Bachi A, Wilm M, Bischoff FR, Weeks DL, Ogniewski V, Ohno M, Niehrs C, Kjems J, Mattaj IW, Fornerod M (1999) RanGTP-regulated interactions of CRMI with nucleoporins ans a shuttling DEAD-box helicase. Mol Cell Biol 19: 6276–6285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becskei A, Boselli MG, van Oudenaarden A (2004) Amplitude control of cell-cycle waves by nuclear import. Nat Cell Biol 6: 451–457 [DOI] [PubMed] [Google Scholar]
- Ben-Efraim I, Gerace L (2001) Gradient of increasing affinity of importin ß for nucleoporins along the pathway of nuclear import. J Cell Biol 152: 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Gorlich D (1997) RanBP1 is crucial for the release of RanGTP from importin β-related nuclear transport factors. FEBS Lett 419: 249–254 [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Ponstingl H (1991) Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354: 80–82 [DOI] [PubMed] [Google Scholar]
- Chaillan-Huntington C, Braslavsky CV, Kuhlmann J, Stewart M (2000) Dissecting the interactions between NTF2, RanGDP, and the nucleoporin XFXFG repeats. J Biol Chem 275: 5874–5879 [DOI] [PubMed] [Google Scholar]
- Chook YM, Blobel G (2001) Karyopherins and nuclear import. Curr Opin Struct Biol 11: 703–715 [DOI] [PubMed] [Google Scholar]
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J (1998) Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94: 193–204 [DOI] [PubMed] [Google Scholar]
- Csete ME, Doyle JC (2002) Reverse engineering of biological complexity. Science 295: 1664–1669 [DOI] [PubMed] [Google Scholar]
- Englmeier L, Fornerod M, Bischoff FR, Petosa C, Mattaj IW, Kutay U (2001) RanBP3 influences interactions between CRM1 and its nuclear protein export substrates. EMBO Rep 2: 926–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog B, Aebi U (2003) The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol 4: 757–766 [DOI] [PubMed] [Google Scholar]
- Floer M, Blobel G, Rexach M (1997) Disassembly of RanGTP karyopherinβ complex, an intermediate in nuclear protein import. J Biol Chem 272: 19538–19546 [DOI] [PubMed] [Google Scholar]
- Fontes MR, Teh T, Kobe B (2000) Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J Mol Biol 297: 1183–1194 [DOI] [PubMed] [Google Scholar]
- Gorlich D, Henklein P, Laskey RA, Hartmann E (1996a) A 41 amino acid motif in importin-alpha confers binding to importin- beta and hence transit into the nucleus. EMBO J 15: 1810–1817 [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–660 [DOI] [PubMed] [Google Scholar]
- Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR (1996b) Identification of different roles for RanGDP and RanGTP In nuclear protein import. EMBO J 15: 5584–5594 [PMC free article] [PubMed] [Google Scholar]
- Henderson BR, Eleftheriou A (2000) A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp Cell Res 256: 213–224 [DOI] [PubMed] [Google Scholar]
- Herold A, Truant R, Wiegand H, Cullen BR (1998) Determination of the functional domain organization of the importin alpha nuclear import factor. J Cell Biol 143: 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Gorlich D (1997) Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell 90: 1061–1071 [DOI] [PubMed] [Google Scholar]
- Lindsay M (2001) RanBP3 is a cofactor for Crm1-mediated nuclear protein export. J Cell Biol. 153: 1391–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara IG (2001) Transport into and out of the Nucleus. Microbiol Mol Biol Rev 65: 570–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeri D, Malim MH (1999) Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol Cell Biol 19: 1218–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C, Orem N, Trueheart J, Thorner JW, Macara IG (2000) Random mutagenesis and functional analysis of the Ran-binding protein, RanBP1. J Biol Chem 275: 4081–4091 [DOI] [PubMed] [Google Scholar]
- Ribbeck K, Lipowsky G, Kent HM, Stewart M, Gorlich D (1998) NTF2 mediates nuclear import of Ran. EMBO J 17: 6587–6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddick G, Macara IG (2005) A systems analysis of importin-{alpha}-{beta} mediated nuclear protein import. J Cell Biol 168: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J, Dilworth SM, Laskey RA, Dingwall C (1991) Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64: 615–623 [DOI] [PubMed] [Google Scholar]
- Sauro HM, Hucka M, Finney A, Wellock C, Bolouri H, Doyle J, Kitano H (2003) Next generation simulation tools: the systems biology Workbench and BioSPICE integration. OMICS 7: 355–372 [DOI] [PubMed] [Google Scholar]
- Smith A, Brownawell A, Macara IG (1998) Nuclear import of Ran:GDP is mediated by NTF2. Curr Biol 8: 1403–1406 [DOI] [PubMed] [Google Scholar]
- Smith AE, Slepchenko BM, Schaff JC, Loew LM, Macara IG (2002) Systems analysis of Ran transport. Science 295: 488–491 [DOI] [PubMed] [Google Scholar]
- Stelling J, Klamt S, Bettenbrock K, Schuster S, Gilles ED (2002) Metabolic network structure determines key aspects of functionality and regulation. Nature 420: 190–193 [DOI] [PubMed] [Google Scholar]
- Suntharalingam M, Wente SR (2003) Peering through the pore: nuclear pore complex structure, assembly, and function. Dev Cell 4: 775–789 [DOI] [PubMed] [Google Scholar]
- Tachibana T, Hieda M, Yoneda Y (1999) Up-regulation of nuclear protein import by nuclear localization signal sequences in living cells. FEBS Lett 442: 235–240 [DOI] [PubMed] [Google Scholar]
- Timney BL, Tetenbaum-Novatt J, Agate DS, Williams R, Zhang W, Chait BT, Rout MP (2006) Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J Cell Biol 175: 579–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K (2002) Nucleocytoplasmic transport: cargo trafficking across the border. Curr Opin Cell Biol 14: 328–335 [DOI] [PubMed] [Google Scholar]
- Yang W, Musser SM (2006) Nuclear import time and transport efficiency depend on importin beta concentration. J Cell Biol 174: 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1