Abstract
Differential repair of structurally distinct mutagenic lesions in critical genes may influence the cellular risk of malignant conversion. We have investigated rat mammary tumorigenesis induced by N-ethyl-N-nitrosourea (EtNU) versus N-methyl-N-nitrosourea (MeNU) with respect to tumor incidence, ras gene mutation, and gene-specific repair. Both carcinogens induced mammary adenocarcinomas at high yield. In mammary epithelia (very low expression of O6-alkylguanine-DNA alkyltransferase, MGMT), O6-methylguanine (O6-MeGua) was eliminated from transcribed (H-ras and β-actin) and inactive genes (IgE heavy chain) at the same slow rate as determined for bulk genomic DNA. The persistence of O6-MeGua in DNA correlated with a high frequency of G:C → A:T transition mutations at codon 12 of the H-ras gene in MeNU-induced tumors. Repair of O6-ethylguanine (O6-EtGua), too, was slow in the IgE heavy chain gene as in bulk DNA. Contrasting with O6-MeGua, however, O6-EtGua was removed ≈20 times faster from the active H-ras and β-actin genes via MGMT-independent mechanism(s). Accordingly, no H-ras codon 12 mutations were found in EtNU-induced tumors, and 5- to 8-fold surplus alkyltransferase activity of the mammary epithelia—via a bacterial ada transgene—did not significantly counteract tumorigenesis in EtNU-exposed contrary to MeNU-treated animals. Neither MeNU- nor EtNU-induced tumors exhibited mutations at codons 13 and 61 of H-ras or codons 12, 13, and 61 of K-ras. Fast repair of O6-EtGua, but not O6-MeGua, in transcribed genes thus prevents mutational activation of H-ras when rat mammary carcinogenesis is initiated by EtNU in place of MeNU.
Keywords: overall and gene-specific DNA repair, O6-alkylguanine-DNA alkyltransferase, ada-transgenic rats, mammary cancer
In the process of carcinogenesis, point mutations may activate, functionally alter, or silence critical genes in a cell type-specific, differentiation stage-specific, and carcinogen-specific manner. Carcinogenic risk caused by the conversion of DNA damage into mutations is inversely correlated with the capacity of target cells for DNA repair, which thus exerts a “tumor suppressor” function (1–4).
The formation and repair of specific DNA lesions do not occur randomly throughout the eukaryotic genome (5–10). Preferential nucleotide excision repair in the transcribed strand of active genes first was shown for UV-induced cyclobutane pyrimidine dimers (transcription-coupled repair; refs. 5–6 and 10). Repair rates may be heterogeneous at the level of individual nucleotides (7, 9). For example, mutational hot spots of the p53 gene in human skin tumors coincide with the slow repair of photoproducts at these very positions in human fibroblasts exposed to UV in vitro (7). Little information is available, however, connecting repair of carcinogen-specific DNA lesions and mutation frequency in transformation-associated genes with tumor incidence in established animal models of carcinogenesis.
For the present study, we have chosen rat mammary carcinogenesis by single-dose exposure to N-methyl-N-nitrosourea (MeNU) or N-ethyl-N-nitrosourea (EtNU) as a model, because: (i) alkylating N-alkyl-N-nitrosoureas are mutagens with high carcinogenic potential (11); (ii) the reaction products of MeNU and EtNU with cellular DNA, including the miscoding O6-alkylguanines and O2- and O4-alkylthymines, have been well characterized (12); (iii) pulse exposure of pubescent female rats to MeNU or EtNU induces mammary adenocarcinomas at high yield in a dose-dependent manner (13–14), the putative critical target cells being terminal end bud epithelia (15); (iv) an activating G:C → A:T transition mutation at the second position of codon 12 (exon 1) of the H-ras gene has been detected in a high proportion of MeNU-induced mammary tumors (MTs) and is considered a major initiating event in rat mammary epithelial cells (MEC) transformed by this carcinogen (16–17). This mutation can result from miscoding by O6-alkylguanine persisting unrepaired through DNA replication. O6-alkylguanines in DNA can be efficiently repaired by the “suicide” repair protein O6-alkylguanine-DNA alkyltransferase (MGMT) (18), and the capacity of cells for repair of O6-alkylguanines by MGMT depends on both the size of their MGMT pool and rate of MGMT synthesis; (v) because of their very low MGMT level (19), rat MEC are particularly suited for the modulation of DNA repair capacity by either functional inhibition or transgenic overexpression of MGMT; and (vi) suboptimal DNA repair may be a potential predisposing factor in human breast cancer (20). The rat model of mammary tumorigenesis could be informative in this regard, as it permits us to investigate carcinogenic risk as a function of the formation and repair of defined miscoding DNA lesions in specific genes.
MATERIALS AND METHODS
Animals.
Pathogen-free Sprague–Dawley rats (Zentralinstitut für Versuchstierzucht, Hannover, Germany) were used throughout this study.
O6-Alkylguanines in Genomic DNA.
High molecular weight DNA was isolated from tissues of rats sacrificed at different times after carcinogen exposure (21). After restriction of DNA by EcoRI, O6-alkylguanines were quantified by immuno-slot-blot (ISB; ref. 22) by using mAbs. Briefly, heat-denatured, single-stranded DNA was slot-blotted onto nylon membranes (NY13N; Schleicher & Schuell) and reacted with anti-(O6-alkyl-2′-deoxyguanosine) mAbs EM-21 or EM-2–3 (23), respectively, for O6-ethylguanine (O6-EtGua) or O6-methylguanine (O6-MeGua). DNA-bound mAbs were detected by enzyme-coupled antibodies [alkaline phosphatase/anti-mouse IgG (H+L); Schleicher & Schuell] and chemoluminescence substrate sheets (Schleicher & Schuell). Signals were recorded on x-ray films and evaluated densitometrically.
O6-Alkylguanines in Specific Genes.
O6-EtGua or O6-MeGua contents in specific gene sequences were determined by combined immunoaffinity/quantitative PCR (22, 24). Briefly, 5 μg of EcoRI restricted genomic DNA were spiked with internal standard DNA (100 fg of alkylated, linearized plasmid pHMOX; ref. 25), containing, on average, 1 O6-alkylguanine residue per plasmid molecule. DNA samples were incubated with purified anti-(O6-ethyl-2′-deoxyguanosine) mAb ER-6 (26) or mAb EM-2–3 in STE buffer (50 mM Tris⋅HCl, 100 mM NaCl, 1 mM EDTA; pH 7.5 for mAb ER-6 and pH 8.5 for mAb EM-2–3). DNA fragments containing O6-alkylguanine were trapped on nitrocellulose membranes (BA85; Schleicher & Schuell) as DNA-mAb complexes and thereafter re-eluted. Specific gene sequences in this fraction were coamplified with the internal standard sequence by quantitative multiplex PCR. PCR products were slot-blotted onto NY13N membranes and hybridized with 32P-labeled, internal sequence-specific oligonucleotides separately for each gene. Autoradiograms were evaluated densitometrically. Signals were normalized to those of the internal standard and converted into absolute gene copy numbers by using an external standard curve for calibration (22).
The PCR primers used for amplification of the cytoplasmic rat β-actin and IgE heavy chain genes, and the hybridization oligonucleotides for the respective PCR products, have been described (24). Primers hras1 and hras2 (see below) were used for amplification of H-ras exon 1. Primers for the yeast mox gene (27) were: primer mox-1 (bases 1,267–1,291), 5′-dTTCTGATGTACACCAGAGCCTCTGC-3′; primer mox-2 (complementary to bases 1,441–1,465), 5′-dAGGAAGTCC- TGGCACGTAGGATACG-3′. Internal hybridization oligonucleotides for PCR-amplified H-ras and mox sequences were: oligonucleotides hras-hyb (bases 1,151–1,170), 5′-dCTGTAGAAGCGATGACAGAA-3′; and mox-hyb (bases 1,366–1,385), 5′-dCTTACCAGCGTCCTTGCAAC-3′.
O6-EtGua in Individual Mammary Cells.
Mammary glands were isolated from 50-day-old females at different times after EtNU exposure. Cryosections (8 μm) immunostained with mAb EM-2–3 were counterstained with 4′,6-diamidino-2-phenylindole. O6-EtGua in nuclear DNA was visualized and quantified immunocytologically by using electronically intensified fluorescence and digital image analysis (22).
ada-Transgenic Rats.
The pSVmtv-ada vector (ref. 28; a generous gift from E. Waldstein, Tel Aviv), containing the bacterial O6-alkylguanine-DNA-alkyltransferase gene ada under the control of the simian virus 40 promoter and the mouse mammary tumor virus enhancer/promoter region, was linearized and microinjected into fertilized eggs, and one-cell embryos were transferred to the oviducts of pseudopregnant females according to published procedures (29, 30). The offspring were screened for the transgene by PCR amplification of ada-specific sequences in lymphocyte DNA. Primers for ada amplification (31) were: primer ada-1 (bases 645–669), 5′-dGGGCGATGATGACGCCACATTAATC-3′; primer ada-2 (complementary to bases 798–822), 5′-dTTGTTGCTGAAAAGCAGTGCCGCGA-3′. For differential display of endogenous MGMT and the Ada protein, lymphocytes were isolated from heparinized blood of 50-day-old normal or ada-transgenic animals by Ficoll-Hypaque centrifugation, and maintained for 18 hr at 37°C in RPMI 1640 medium with or without dexamethasone (1 mM). Cellular protein extracts were incubated with calf thymus DNA containing O6-[3H]MeGua (2). Proteins were separated by SDS/PAGE, and 3H-labeled alkyltransferase was located in 2-mm gel slices with protosol N (NEN) by liquid scintillation spectrometry. In addition to repair-proficient endogenous MGMT (apparent molecular mass, 25 kDa), hormone-stimulated transgenic lymphocytes contained a second 40-kDa protein with methyl group acceptor activity, comigrating with the authentic Ada protein (data not shown).
The ada-specific mRNA detected in the mammary gland of 50-day-old ada-transgenic females by semiquantitative reverse transcription–PCR using primers ada-1 and ada-2, was constitutive and independent of hormone treatment. Alkyltransferase activity in mammary gland cell extracts was determined by an mAb-based repair assay with double-stranded substrate oligonucleotides (32 mers) containing a single O6-MeGua or O6-EtGua residue (32). Briefly, protein extracts were prepared by sonication (3 × 5 s; 0°C) of minced tissue in a buffer containing 50 mM Tris⋅HCl (pH 7.8), 100 mM NaCl, 1 mM DTT, 1 mM EDTA, and 5% glycerol. Cell debris were removed by centrifugation (12,000 × g; 10 min). Varying amounts of protein were incubated with 2 fmol of 35S-labeled substrate for 30 min at 37°C in the same buffer without NaCl. The fractions of unrepaired substrate molecules were determined by a filter-binding assay using mAbs EM-2–3 or ER-6. Compared with nontransgenic littermates, alkyltransferase activity in mammary gland extracts of ada-transgenics was enhanced ≈6.5-fold: 11.4 ± 1.3 or 4.7 ± 0.2 fmol, respectively, of O6-MeGua or O6-EtGua repaired per mg of protein in transgenics versus 1.5 ± 0.5 and 0.9 ± 0.7 fmol/mg in nontransgenics (triplicate analyses in two independent experiments). The reference value was 58.0 ± 8.0 fmol of O6-MeGua repaired per mg protein by liver extracts from nontransgenics, as determined according to ref. 33.
Induction of Mammary Tumors.
Stock solutions (100 mg/ml of H2O-free dimethyl sulfoxide) of MeNU or EtNU (Serva), recrystallized twice from methanol, were stored at −20°C, and diluted with phosphate/citrate buffered saline (pH 4.2) immediately before i.p. injection. A single dose of MeNU (50 μg/g body weight) or EtNU (100 μg/g) was applied to 50-day-old normal or ada-transgenic females (15–26 animals per group). Animals were examined by palpation twice weekly. The time until MTs became first palpable was termed “induction period,” and MTs were resected when reaching a volume of ≈1 cm3. Portions of tumor tissue were stored in liquid nitrogen for cryosectioning and isolation of DNA or RNA. Hematoxylin/eosin-stained 5-μm sections of formalin-fixed material were used for histopathology.
Mutation Analyses.
Bulk DNA was isolated from MTs (21), and from selected areas of ≈50 tumor cells in 10-μm cryosections. After amplification by PCR, H-ras and K-ras sequences were analyzed for mutations by direct sequencing. H-ras alleles containing a G:C → A:T transition at the second position of codon 12 were enriched by digestion of DNA with Bpml (Biolabs), which recognizes the codon 12 wild-type sequence and cuts 16 bp downstream. By this procedure, one mutant allele was detectable among 50 wild-type alleles, as determined by serial dilution of homozygously mutant with wild-type DNA.
PCR (35 cycles; profile: 95°C, 0.5 min; 65°C, 1 min; 72°C, 2 min; final extension at 72°C, 10 min) was performed in 100 μl of PCR buffer (50 mM KCl/10 mM Tris⋅HCl, pH 8.3/1.5 mM MgCl2) containing 0.25 mM dNTP mixture, 0.15 μM each of upstream and 5′-biotinylated downstream primers, 2 units of Taq polymerase (Ampli Taq; Perkin–Elmer), and 500 ng of template DNA. Amplification of pseudogene sequences was avoided by using exon-/intron-specific primer pairs.
PCR primers for exon 1 (codons 12 and 13) of H-ras gene (34) were: primer hras-1 (bases 94–118) 5′-dCTGGCTAAGTGTGATTCTCATTGGC-3′; primer hras-2 (complementary to bases 206–230, 5′-biotinylated) 5′-dCAGCTGGATGGTCAGGGCACTCTTT-3′. Primers for H-ras exon 2 (codon 61) were: primer hras-4 (bases 418–442) 5′-dTCTCTGTCTAAGAAGAGGTAGGACC-3′; primer hras-5 (complementary to bases 619–643, 5′-biotinylated), 5′-dCTGTACTGATGGATGTCTTCAAAGG-3′. Primers for K-ras (EMBL database accession no. X74502) exon 1 (codons 12 and 13) were: primer kras-1 (bases 79–103), 5′-dACTTGATAATCTTGTGTGGAACATG-3′; primer kras-2 (complementary to bases 238–262, 5′-biotinylated) 5′-dCTCTATCGTAGGATCATATTCATCC-3′. Primers for K-ras exon 2 (codon 61; ref. 35) were: primer kras-3 (36) 5′-dATCCAGACTGTGTTTCTACC-3′; primer kras-4 (complementary to bases 99–123, 5′-biotinylated) 5′-dAAAGCCCTCCCCAGTTCTCATGTAC-3′. Streptavidin-coated magnetobeads (Dynal) were used for isolation of single-stranded PCR products for semi-automated sequencing with T7-DNA polymerase (A.L.F. DNA sequencer; Pharmacia). The neu/erbB-2 gene was analyzed for a T:A → A:T transversion at nucleotide 2,012 by using PCR/restriction fragment-length polymorphism methodology (37).
RESULTS
G:C → A:T transitions in DNA can result from unrepaired O6-alkylguanines and, in codon 12 of H-ras, have been invoked as critical initial genetic alterations in MeNU-induced rat mammary tumorigenesis (16–17). We therefore have investigated the overall and gene-specific repair of O6-alkylguanines in rat MEC after exposure to MeNU compared with EtNU.
Overall Repair of O6-Alkylguanines in Mammary Gland DNA.
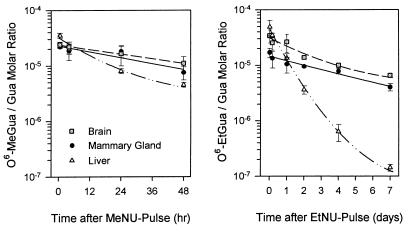
The kinetics of O6-EtGua versus O6-MeGua repair in bulk mammary gland DNA were determined in comparison to liver and brain DNA, after pulse-exposure of 50-day-old females to EtNU or MeNU under the conditions used for MT induction (see below). O6-alkylguanines in DNA were quantified by ISB at different times after carcinogen exposure. The initial rate of O6-EtGua elimination from DNA was 8- to 9-fold higher in the liver (t1/2 ≈ 6 hr) than in the mammary gland (t1/2 ≈ 48 hr) or brain (t1/2 ≈ 52 hr). At 4 days after exposure to EtNU, ≈40% of the input O6-ethylguanines still persisted in mammary gland and brain DNA, whereas nearly all had been repaired in liver DNA. Similar repair kinetics were obtained for O6-MeGua (liver, t1/2 ≈ 9 hr; mammary gland, t1/2 ≈ 40 hr; brain, t1/2 ≈ 42 hr) (Fig. 1; Table 1). These overall repair rates were in good agreement with the pool size of active MGMT determined in crude extracts of the respective tissues (see Materials and Methods).
Figure 1.
O6-alkylguanine elimination from bulk DNA of different tissues after exposure to MeNU or EtNU in vivo. DNA was isolated from mammary gland, liver, and brain, at different times after carcinogen exposure. O6-alkylguanine contents in DNA were quantified by ISB. Mean values of duplicate analyses from two (O6-MeGua) or four animals (O6-EtGua) per time point. Error bars: Range of measured values.
Table 1.
Overall and gene-specific repair of O6-alkylguanines in the DNA of mammary gland after exposure to MeNU or EtNU in vivo, and effect of MGMT inhibition by O6-BeGua
| O6-BeGua |
t1/2 (hr)
|
|||
|---|---|---|---|---|
| O6-MeGua
|
O6-EtGua
|
|||
| − | + | − | + | |
| Genomic DNA | 40 | >48* | 48 | >48† |
| IgE | 48 | >48* | 50 | >48‡ |
| H-ras | 36 | >48* | 2.5 | 2.6 |
| β-actin | 45 | >48* | 2.4 | 2.4 |
t1/2 values indicate the time periods required for removal of 50% of input O6-alkylguanines in DNA.
<10%,
26%, and
20% of input O6-alkylguanines repaired in the presence of O6-BeGua for 48 hr.
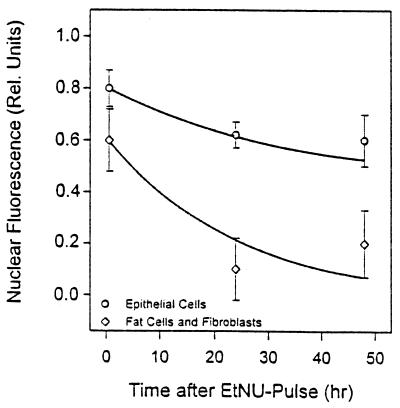
To determine whether the slow overall repair of O6-EtGua in mammary gland DNA was representative of the target MEC (as opposed to fat cells and fibroblasts), the elimination of O6-EtGua from nuclear DNA was measured immunocytologically at the level of individual cells at 1, 24, and 48 hr after EtNU exposure. O6-EtGua was removed from the DNA of MEC more slowly than from the DNA of the other cell types (Fig. 2), at a rate corresponding to that of bulk mammary gland DNA (Fig. 1, Table 1).
Figure 2.
Repair of O6-EtGua measured in individual mammary gland cells after exposure to EtNU in vivo. O6-EtGua in nuclear DNA was quantified immunocytologically. Values are means of ≥50 cells.
O6-EtGua, But Not O6-MeGua, Is Rapidly Repaired in Both Strands of Transcribed Genes as Opposed to Inactive Genes.
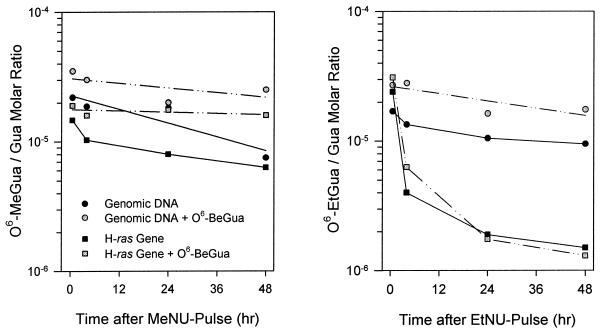
Semiquantitative reverse transcription–PCR analyses showed active transcription of H-ras in MEC of 50-day-old females (data not shown), in agreement with published data (38). The kinetics of O6-alkylguanine repair in the H-ras gene of the mammary gland were determined in comparison to repair in the active β-actin gene and the constant domains of the silent IgE heavy chain gene. Contrasting with the slow removal of both O6-EtGua and O6-MeGua from bulk DNA, very fast repair of O6-EtGua was observed in both the H-ras and β-actin gene. During the initial repair phase (Fig. 3; Table 1), the half-lives of O6-EtGua in H-ras (t1/2 = 2.5 hr) and β-actin (t1/2 = 2.4 hr) were shorter by a factor of ≈20 compared with the inactive IgE gene (t1/2 ≈ 50 hr) or bulk genomic DNA (t1/2 ≈ 48 hr). At 4 hr after EtNU exposure, ≈85% of O6-EtGua residues initially formed in the H-ras and β-actin genes had been eliminated, indicating fast removal of O6-EtGua from both strands of H-ras and β-actin. Contrary to O6-EtGua, O6-MeGua was repaired at a much slower rate in all gene sequences analyzed (t1/2 ≈ 36–48 hr; Fig. 3; Table 1).
Figure 3.
O6-alkylguanine elimination from bulk DNA and from the transcribed H-ras gene of the mammary gland after exposure to MeNU or EtNU in vivo, and effect of MGMT inhibition by O6-BeGua. DNA was analyzed for O6-MeGua or O6-EtGua by ISB or immunoaffinity/quantitative PCR. See Results for O6-BeGua exposure conditions. Values are means of duplicate analyses from two (O6-MeGua) or four animals (O6-EtGua) per time point.
Rapid Repair of O6-EtGua in Transcribed Genes Is Not Caused by MGMT.
The contribution of MGMT to the overall and gene-specific repair of O6-alkylguanines in the mammary gland was evaluated by inhibition of MGMT with O6-benzylguanine (O6-BeGua; ref. 39). Rats were injected i.p. with O6-BeGua (10 μg/g) 2 hr before carcinogen exposure, and the administration of O6-BeGua was repeated at 12-hr intervals. As shown in Table 1 and Fig. 3, repair of O6-EtGua in the silent IgE heavy chain gene (as in bulk DNA) was predominantly effected by MGMT (<26% of input O6-EtGua repaired in the presence of O6-BeGua for 48 hr), whereas the fast removal of O6-EtGua from H-ras and β-actin was not. However, neither from bulk DNA nor from the specific gene sequences analyzed was O6-MeGua removed to a significant extent under MGMT blocking conditions (<10% of input O6-MeGua repaired within 48 hr). O6-MeGua thus is removed from DNA predominantly by MGMT, and repair by MGMT appears to be independent of the transcriptional status of genes.
Induction of Mammary Tumors by MeNU or EtNU.
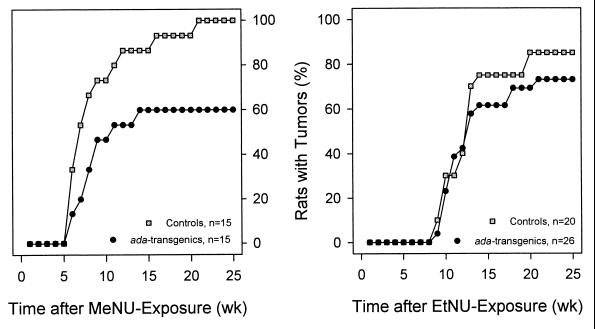
Pulse exposure to MeNU resulted in a 100% incidence of MT after a mean induction period of 49 days. The corresponding values for EtNU were ≈85% and 88 days (Fig. 4). All MTs were histopathologically classified as moderately differentiated papillary adenocarcinomas. No other malignancies were detected during the observation period of 175 days.
Figure 4.
Incidence of MeNU- or EtNU-induced mammary adenocarcinomas in normal or ada-transgenic rats. Percentages of animals with palpable tumors were determined at the time points indicated by • and □.
Surplus Alkyltransferase Expression Counteracted MeNU-Induced, But Not EtNU-Induced, Mammary Tumorigenesis.
Compared with nontransgenic littermates (100%), ada-transgenic rats exhibited a significantly (P < 0.01; Fisher’s exact test) reduced incidence of 60% and a prolonged mean induction period (from 49 to 74 days) of MTs after exposure to MeNU (Fig. 4). In contrast, EtNU-exposed ada-transgenic and normal rats did not exhibit significant differences regarding the induction period (97 versus 88 days) and the fraction of tumor-bearing animals (73% versus 85%). MTs of all four groups were neither distinguishable by histopathological criteria nor with respect to tumor progression.
G:C → A:T Mutations at H-ras Codon 12 Are Frequent in MeNU-Induced but Absent in EtNU-Induced MTs.
Direct sequencing of PCR-amplified H-ras exon 1 revealed G:C → A:T transitions at the second nucleotide of codon 12 (GGA → GAA) in ≈75% of MeNU-induced MTs (Table 2). No mutations were found at other positions of exon 1, including “hot spot” codon 13. H-ras codon 12 mutation frequencies were similar in MTs of normal and ada-transgenic animals (Table 2). In EtNU-induced MTs, mutations were neither detectable at codon 12 (Table 2) nor at any other position of H-ras exon 1. These findings were confirmed by analyses of tumor cells selected from cryosections.
Table 2.
Mutations at codon 12 of H-ras and K-ras in mammary tumors induced by MeNU or EtNU in normal and ada-transgenic rats
| MeNU
|
EtNU
|
|||
|---|---|---|---|---|
| Alkyltransferase status*
| ||||
| Normal | Surplus (adatransgenic) | Normal | Surplus (adatransgenic) | |
| H-ras codon 12 | 6/8† | 6/10 | 0/12 | 0/7 |
| K-ras codon 12 | 0/15 | ND | 0/12 | 0/3 |
ND, not determined.
Mammary epithelia at carcinogen exposure.
Number of tumors with mutant gene/number of tumors.
Analyses of K-ras exon 1 in 15 MeNU-induced MTs and in 12 EtNU-induced MTs showed wild-type sequences in all cases (Table 2).
No mutations at A:T base pairs of H-ras and K-ras codon 61 (CAA) were detected by direct sequencing of PCR-amplified exon 2 in MeNU-induced MTs (0/8 and 0/15 tumors, respectively). One of 20 EtNU-induced MTs harbored a transversion at H-ras codon 61 (CAA → CTA), whereas K-ras codon 61 was wild type in 12 of 12 EtNU-induced MTs. Similarly, no T:A → A:T transversions were detected at codon 664 (GTG) of neu/erbB-2 in a total of 13 MeNU- and 13 EtNU-induced MTs (including 10 MTs previously analyzed; S. N. Prokopenko, A. Yū. Nikitin and M.F.R., unpublished work).
DISCUSSION
The persistence of unrepaired O6-alkylguanines in bulk genomic DNA of different types of proliferation-competent cells correlates with enhanced carcinogenic risk (1–4). Surplus expression of either a bacterial ada or human MGMT transgene in mice results in accelerated removal of O6-alkylguanines from target cell DNA and reduced oncogenicity of N-nitroso carcinogens (3–4, 40–41), underscoring the importance of the repair of O6-alkylguanines as a risk determinant in malignant transformation. In rat hepatoma cells, we previously had observed that O6-EtGua is removed much more rapidly from the transcribed β-actin gene compared with the silent IgE heavy chain gene or bulk genomic DNA (24). We now have analyzed ras gene mutation frequencies and the repair of O6-MeGua versus O6-EtGua in the active H-ras and β-actin genes, as opposed to the nontranscribed IgE gene and bulk genomic DNA, in the rat model of mammary carcinogenesis induced by MeNU or EtNU.
Absence of H-ras Mutations in MTs Induced by EtNU in Place of MeNU.
In accordance with the literature (13–14, 16), pubescent female rats developed phenotypically indistinguishable mammary adenocarcinomas at high yield after pulse exposure to MeNU or EtNU (Fig. 4). Activating G:C → A:T transitions in H-ras codon 12 (but not in codons 13 or 61) were found in ≈75% of MTs induced by MeNU, but were absent in all of the EtNU-induced MTs (Table 2). No K-ras codon 12, 13, or 61 mutations were found in any of the MTs analyzed (Table 2), at variance with MTs resulting from exposure to MeNU on postnatal day 2 (17). MTs arising in pituitary-isografted mice exposed to MeNU have been reported to exhibit G:C → A:T transitions at codon 12 of K-ras, but not H-ras, whereas EtNU-induced MTs lacked this mutation in both ras genes (42–43).
Slow Overall Repair of O6-Alkylguanines in Genomic DNA.
The repair kinetics for O6-MeGua and O6-EtGua in bulk DNA were determined in mammary gland in comparison to brain and liver (Fig. 1), confirming earlier data for O6-MeGua (44). Consistent with the relative MGMT levels in these tissues (19, 45), both O6-MeGua and O6-EtGua were rapidly removed from bulk DNA of the liver, but highly persistent in mammary gland and brain DNA (Figs. 1 and 2). Because terminal end bud MEC of sexually mature rats have a mean cell cycle time of ≈15 hr and assuming ≤10% of mammary gland cells to be in cell cycle (15, 46), replicative bypass of O6-methylated guanines persisting in DNA through S-phase may lead to G:C → A:T transitions. Although the slow overall repair of O6-MeGua translates well into the high incidence of H-ras codon 12 transitions in MeNU-induced MTs, the similarly long persistence of O6-EtGua in the DNA of MEC appears to disagree with the absence of this mutation in MTs induced by EtNU.
Preferential Repair of O6-EtGua in Active Genes.
By using mAb-based immunoaffinity combined with quantitative PCR (22, 24), O6-alkylguanines can be quantified in single-copy genes at levels ≥1 O6-alkylguanine in 107 guanine molecules (≥250 O6-alkylguanines/diploid genome). The observed slow repair of O6-EtGua in the IgE heavy chain gene of the mammary gland corresponds to the slow repair of O6-EtGua in a silent lacZ transgene in mice (47). This slow repair in nontranscribed genes is contrasted by the ≈20 times more rapid removal of O6-EtGua from both strands of the active H-ras and β-actin genes (Table 1; Fig. 3). Mutation via unrepaired O6-alkylguanines in transcribed genes of the mammary gland thus is much less likely for O6-EtGua than for O6-MeGua, which was found to be equally persistent in both H-ras and β-actin and the inactive IgE heavy chain gene (Table 1; Fig. 3). The slow removal of O6-MeGua from the transcribed H-ras gene is in agreement with the observed high frequency of G:C → A:T transitions at codon 12 of H-ras in MeNU-induced MTs.
Functional inhibition of endogenous MGMT by O6-BeGua (39) in the mammary gland resulted in reduced overall repair of O6-MeGua in both transcribed and silent genes (as in bulk genomic DNA). However, the rapid removal of O6-EtGua from H-ras or β-actin remained almost unaffected, indicating efficient repair of this lesion in both strands of active genes by (an) as-yet-unidentified repair mechanism(s), e.g., nucleotide excision repair (Table 1; Fig. 3). Mutation analyses in rat fibroblasts and T lymphocytes have suggested efficient removal of O6-EtGua from both strands of the transcribed hprt gene (48). Functional repair analyses in different types of human cells suggest O6-EtGua elimination from DNA to be effected by both excision repair and MGMT (49–51). The present results underscore the dual character of O6-EtGua repair; i.e., slow overall repair predominantly by MGMT, in parallel to rapid removal from transcribed genes by other mechanism(s). To further characterize this fast repair process, cells with distinct DNA repair gene knockouts must be analyzed.
Surplus Transgenic Alkyltransferase Activity Counteracts MeNU-Induced, But Not EtNU-Induced, Mammary Tumorigenesis.
The expression of functional Ada protein controlled by the hormone-driven mouse MT virus promoter in ada-transgenic rats corroborates earlier findings in mice (52). Protection from DNA damage (O6-MeGua and O6-chloroethylguanine) by the PSVmtv-ada vector previously has been observed in hormone-stimulated HeLa cells (28). The reduced incidence and prolonged induction period of MeNU-induced MTs in ada-transgenic rats correlates with their surplus alkyltransferase activity compared with the low MGMT level in MEC of nontransgenic littermates. In a murine thymic lymphoma model, Dumenco et al. (3) have demonstrated pronounced protection from the oncogenic effect of MeNU by more drastically enhanced repair of O6-MeGua via targeted high-level expression of a human MGMT transgene. Other studies with methylating carcinogens support these findings (40–41).
Contrary to the situation with MeNU, and despite the efficient repair of O6-EtGua by the Ada protein (see Materials and Methods and ref. 53–55), there were no significant differences between normal rats and ada-transgenics regarding the incidence and induction period of EtNU-induced MTs. This finding is consistent with protection from O6-EtGua-derived mutations by efficient repair of O6-EtGua in transcribed genes of MEC in both groups of animals. DNA ethylation products other than O6-EtGua, which might be less efficiently repaired in active genes (e.g., O2- or O4-ethylthymine, O2-ethylcytosine; ref. 48) or, alternatively, O6-EtGua formed in temporarily silent genes, thus may be critical elements in EtNU-induced rat mammary tumorigenesis.
Because H-ras and K-ras are apparently not critical target genes in EtNU-induced rat mammary tumorigenesis, we have, in addition, analyzed the neu/erbB-2 gene for mutational activation. EtNU-induced rat schwannomas exhibit a diagnostic T:A → A:T transversion at codon 664 of the neu/erbB-2 gene (37, 56–57), and overexpression of either a wild-type or mutant neu/erbB-2 transgene induces malignant conversion of MEC in mice (58). However, all of the EtNU-induced rat MTs tested were neu/erbB-2 wild type.
From experiments with a different rat strain (Fischer 344), Cha et al. (59) have hypothesized that MTs induced by MeNU could have originated from MEC harboring a pre-existent mutant H-ras allele. If pre-existing H-ras mutations were critical to the induction of rat mammary cancer by N-alkyl-N-nitrosoureas, these mutations might have been detectable in both EtNU- and MeNU-initiated MTs; however, this was not the case in the present study.
Aside from the molecular basis and the biological implications of the puzzling disregard of O6-MeGua by the fast repair system for active genes, the contributions of different modes of repair remain to be investigated for other DNA lesions and in distinct types of target cells and stages of differentiation, with particular attention to the expression status of critical genes. Differential repair mechanisms, as described here for two nearly identical DNA alkylation products, may be important determinants that need to be factored into cancer risk estimates.
Acknowledgments
We thank E. Waldstein (Tel Aviv) and R. Roggenkamp (Düsseldorf) for providing the vectors pSVmtv-ada and pHMOX, respectively, W. Drosdziok for the synthesis of O6-benzylguanine, our Monoclonal Antibody Unit for its cooperation, B. Opgenoorth for expert technical assistance, K. Kamino (Hannover) for histopathological analyses, and K. Hochleitner (Dassel) for helpful discussions. Support by the Deutsche Forschungsgemeinschaft, Dr. Mildred Scheel Stiftung für Krebsforschung, and the National Foundation for Cancer Research is gratefully acknowledged.
ABBREVIATIONS
- MGMT
O6-alkylguanine-DNA alkyltransferase
- Ada
bacterial O6-alkylguanine-DNA alkyltransferase
- EtNU
N-ethyl-N-nitrosourea
- ISB
immuno-slot-blot
- MEC
mammary epithelial cells
- MeNU
N-methyl-N-nitrosourea
- MT
mammary tumor
- O6-BeGua
O6-benzylguanine
- O6-EtGua
O6-ethylguanine
- O6-MeGua
O6-methylguanine
References
- 1.Goth R, Rajewsky M F. Proc Natl Acad Sci USA. 1974;74:639–643. doi: 10.1073/pnas.71.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomale J, Huh N, Nehls P, Eberle G, Rajewsky M F. Proc Natl Acad Sci USA. 1990;87:9883–9887. doi: 10.1073/pnas.87.24.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dumenco L L, Allay E, Norton K, Gerson S L. Science. 1993;259:219–222. doi: 10.1126/science.8421782. [DOI] [PubMed] [Google Scholar]
- 4.Nakatsuru Y, Matsukuma S, Nemoto N, Sugano H, Sekiguchi M, Ishikawa T. Proc Natl Acad Sci USA. 1993;90:6468–6472. doi: 10.1073/pnas.90.14.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohr V A, Smith C A, Okumoto D S, Hanawalt P C. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 6.Mellon I, Spivak G, Hanawalt P C. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 7.Holmquist G P, Gao S. Mutat Res. 1997;386:69–101. doi: 10.1016/s1383-5742(96)00045-2. [DOI] [PubMed] [Google Scholar]
- 8.Denissenko M F, Pao A, Tang M, Pfeifer G P. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 9.Tu Y, Tornaletti S, Pfeifer G P. EMBO J. 1996;15:675–683. [PMC free article] [PubMed] [Google Scholar]
- 10.Verhage R A, van Gool A J, de Groot N, Hoeijmakers J H J, van de Putte P, Brouwer J. Mol Cell Biol. 1996;16:496–502. doi: 10.1128/mcb.16.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montesano R, Hall J, Wild C P. In: Relevance of Animal Studies to the Evaluation of Human Cancer Risk. d’Amato R, Slaga T J, Farland W H, Henry C, editors. New York: Wiley-Liss; 1992. pp. 175–196. [Google Scholar]
- 12.Beranek D T. Mutat Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 13.Gullino P M, Pettigrew H M, Grantham F H. J Natl Cancer Inst. 1975;54:401–414. [PubMed] [Google Scholar]
- 14.Stoica G, Koestner A, Capen C C. Am J Pathol. 1983;110:161–169. [PMC free article] [PubMed] [Google Scholar]
- 15.Russo J, Russo I H. Cancer Res. 1980;40:2677–2687. [PubMed] [Google Scholar]
- 16.Zarbl H, Sukumar S, Arthur A V, Martin-Zanca D, Barbacid M. Nature (London) 1985;315:382–385. doi: 10.1038/315382a0. [DOI] [PubMed] [Google Scholar]
- 17.Kumar R, Sukumar S, Barbacid M. Science. 1990;248:1101–1104. doi: 10.1126/science.2188364. [DOI] [PubMed] [Google Scholar]
- 18.Lindahl T, Sedgwick B, Sekiguchi M, Nakabeppu Y. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- 19.Fong L Y Y, Jensen D A, Magee P N. Carcinogenesis. 1990;11:411–417. doi: 10.1093/carcin/11.3.411. [DOI] [PubMed] [Google Scholar]
- 20.Parshad R, Price F M, Bohr V A, Cowans K H, Zujewski J A, Sanford K K. Br J Cancer. 1996;74:1–5. doi: 10.1038/bjc.1996.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Thomale J, Engelbergs J, Seiler F, Rajewsky M F. In: Technologies for Detection of DNA Damage and Mutations. Pfeifer G P, editor. New York: Plenum; 1996. pp. 87–101. [Google Scholar]
- 23.Eberle G. Ph.D. dissertation. Germany: University of Essen; 1989. [Google Scholar]
- 24.Thomale J, Hochleitner K, Rajewsky M F. J Biol Chem. 1994;269:1681–1686. [PubMed] [Google Scholar]
- 25.Eckart M. Ph.D. dissertation. Germany: University of Düsseldorf; 1988. [Google Scholar]
- 26.Rajewsky M F, Müller R, Adamkiewicz J, Drosdziok W. In: Carcinogenesis: Fundamental Mechanisms and Environmental Effects. Pullman B, Ts’o P O P, Gelboin H, editors. Dordrecht, The Netherlands: Reidel; 1980. pp. 207–218. [Google Scholar]
- 27.Ledeboer A M, Edens L, Maat J, Visser C, Bos J W, Verrips C T, Janowicz Z, Eckart M, Roggenkamp R, Hollenberg C P. Nucleic Acids Res. 1985;13:3063–3082. doi: 10.1093/nar/13.9.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waldstein E A. Carcinogenesis. 1990;11:21–26. doi: 10.1093/carcin/11.1.21. [DOI] [PubMed] [Google Scholar]
- 29.Hogan B, Constantini F, Lacy A. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 30.Nikitin A Y, Jin J J, Papewalis J, Prokopenko S N, Pozharisski K M, Winterhager E, Flesken-Nikitin A, Rajewsky M F. Oncogene. 1996;12:1309–1317. [PubMed] [Google Scholar]
- 31.Demple B, Sedgwick B, Robins P, Totty N, Waterfield M D, Lindahl T. Proc Natl Acad Sci USA. 1985;82:2688–2692. doi: 10.1073/pnas.82.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bender K, Federwisch M, Loggen U, Nehls P, Rajewsky M F. Nucleic Acids Res. 1996;24:2078–2094. doi: 10.1093/nar/24.11.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pegg A E, Roberfroid M, von Bahr C, Foote S R, Mitra S, Brésil H, Likhachev A, Montesano R. Proc Natl Acad Sci USA. 1982;79:3862–3165. doi: 10.1073/pnas.79.17.5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruta M, Wolford R, Dhar R, DeFeo-Jones D, Ellis R W, Scolnick E M. Mol Cell Biol. 1986;6:1706–1710. doi: 10.1128/mcb.6.5.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tahira T, Hayashi K, Ochiai M, Tsuchida N, Nagao M, Sugimura T. Mol Cell Biol. 1986;6:1349–1351. doi: 10.1128/mcb.6.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzui M, Yoshimi N, Tanaka T, Mori H. Mol Carcinogen. 1995;14:294–298. doi: 10.1002/mc.2940140410. [DOI] [PubMed] [Google Scholar]
- 37.Nikitin A Y, Ballering L A P, Lyons J, Rajewsky M F. Proc Natl Acad Sci USA. 1991;88:9939–9943. doi: 10.1073/pnas.88.22.9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu S J, Milligan J R, Archer M C. Mol Carcinogen. 1991;4:261–264. doi: 10.1002/mc.2940040403. [DOI] [PubMed] [Google Scholar]
- 39.Pegg A E, Boosalis M, Samson L, Moschel R C, Byers T L, Swenn K, Dolan M E. Biochemistry. 1993;32:11998–12006. doi: 10.1021/bi00096a009. [DOI] [PubMed] [Google Scholar]
- 40.Zaidi N H, Pretlow T P, O’Riordan M A, Dumenco L L, Allay E, Gerson S L. Carcinogenesis. 1995;16:451–456. doi: 10.1093/carcin/16.3.451. [DOI] [PubMed] [Google Scholar]
- 41.Becker K, Dosch J, Gregel C M, Martin B A, Kaina B. Cancer Res. 1996;56:3244–3249. [PubMed] [Google Scholar]
- 42.Guzman R C, Osborn R C, Swanson S M, Sakthivel R, Hwang S I, Miyamoto S, Nandi S. Cancer Res. 1992;52:5732–5737. [PubMed] [Google Scholar]
- 43.Swanson S M, Guzman R C, Tsukamoto T, Huang T T, Dougherty C D, Nandi S. Cancer Lett. 1996;102:159–165. doi: 10.1016/0304-3835(96)04175-4. [DOI] [PubMed] [Google Scholar]
- 44.Lu S J, Chaulk E J, Archer M C. Carcinogenesis. 1992;13:857–861. doi: 10.1093/carcin/13.5.857. [DOI] [PubMed] [Google Scholar]
- 45.Gerson S L, Trey J E, Miller K, Berger N A. Carcinogenesis. 1986;7:745–749. doi: 10.1093/carcin/7.5.745. [DOI] [PubMed] [Google Scholar]
- 46.Anderson C H, Beattie C W. Cancer Res. 1992;52:5076–5081. [PubMed] [Google Scholar]
- 47.Mientjes E J, Hochleitner K, Luiten-Schuite A, van Delft J H M, Thomale J, Berends F, Rajewsky M F, Lohman P H M, Baan R A. Carcinogenesis. 1996;17:2449–2454. doi: 10.1093/carcin/17.11.2449. [DOI] [PubMed] [Google Scholar]
- 48.Jansen J G, Mohn G R, Vrieling H, van Teijlingen C M M, Lohman P H M, van Zeeland A A. Cancer Res. 1994;54:2478–2485. [PubMed] [Google Scholar]
- 49.Maher V M, Domoradzki J, Bhattacharyya N P, Tsujimura T, Corner R C, McCormick J J. Mutat Res. 1990;233:235–245. doi: 10.1016/0027-5107(90)90166-2. [DOI] [PubMed] [Google Scholar]
- 50.Bronstein S M, Skopek T R, Swenberg J A. Cancer Res. 1992;52:2008–2011. [PubMed] [Google Scholar]
- 51.Buschfort C, Müller M R, Seeber S, Rajewsky M F, Thomale J. Cancer Res. 1997;57:651–658. [PubMed] [Google Scholar]
- 52.Krane I M, Leder P. Oncogene. 1996;12:1781–1788. [PubMed] [Google Scholar]
- 53.Samson L, Thomale J, Rajewsky M F. EMBO J. 1988;7:2261–2267. doi: 10.1002/j.1460-2075.1988.tb03066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morimoto K, Dolan M E, Scicchitano D, Pegg A E. Carcinogenesis. 1985;6:1027–1031. doi: 10.1093/carcin/6.7.1027. [DOI] [PubMed] [Google Scholar]
- 55.Liem L-K, Lim A, Li B F L. Nucleic Acids Res. 1994;22:1613–1619. doi: 10.1093/nar/22.9.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bargmann C I, Huang M-C, Weinberg R A. Cell. 1986;45:649–657. doi: 10.1016/0092-8674(86)90779-8. [DOI] [PubMed] [Google Scholar]
- 57.Perantoni A O, Rice J M, Reed C D, Watatani M, Wenk M L. Proc Natl Acad Sci USA. 1987;84:6317–6321. doi: 10.1073/pnas.84.17.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guy C T, Webster M A, Schaller M, Parsons T J, Cardiff R D, Muller W J. Proc Natl Acad Sci USA. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cha R S, Thilly W G, Zarbl H. Proc Natl Acad Sci USA. 1994;91:3749–3753. doi: 10.1073/pnas.91.9.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]