Abstract
Information on the interaction strength between small interfering RNA (siRNA) and chitosan can contribute to the understanding of the formation and stability of chitosan/siRNA nanoparticles used as siRNA delivery systems for gene silencing. In this study, we utilize atomic force microscopy to obtain force spectroscopy results of the interaction strengths between siRNA and chitosan measured in physiological phosphate buffered saline buffer at different pH. The force measurements revealed that the adhesive interactions decreased in force strength and force frequency as the pH was increased from 4.1 to 6.1, 7.4, and 9.5, exhibiting distinct multimodal distributions of the interaction forces between siRNA and chitosan molecules at acidic pH and only negligible adhesive forces were observed at neutral or high pH. The strong pH dependence of siRNA-chitosan interactions can provide a convincing rationale for siRNA/chitosan complex formation and nanoparticle stability under low acidic conditions. These findings demonstrate that the use of force spectroscopy for the adhesive force measurements allows an evaluation of the complexing ability between siRNA and chitosan that can be utilized to predict nanoparticle stability.
INTRODUCTION
Self-assembly of polycations and polyanions yielding polyelectrolyte nanoparticles has been widely used in gene delivery systems (1). In comparison to conventional DNA-based gene therapy, the use of small interfering RNA (siRNA, 21–30 nucleotides in length) for gene silencing by the method of RNA interference has recently attracted much attention (2,3) as a therapy potential for cancer and infectious diseases. Among nonviral gene delivery systems, chitosan has been demonstrated as a promising carrier for plasmid DNA (4–6) and plasmid-encoded siRNA targeting the respiratory syncytial virus NS1 gene (7). Previous studies of physiochemical characterization and cell transfection of chitosan/DNA nanoparticles have shown that the binding affinity of chitosan to DNA and the formation and stability of the chitosan/DNA nanoparticles can be influenced by structural parameters of the chitosan molecules (such as molecular weight, charge density, and degree of deacetylation (DD) of chitosan) (8,9) and buffer conditions (e.g., pH) (9,10). Electrostatic interaction between the polycation and DNA drives the formation and compaction of chitosan/DNA nanoparticles under acidic conditions. In addition, gel electrophoresis studies have shown that secondary (nonelectrostatic) interactions, such as hydrogen bonding and hydrophobic interactions, could be responsible for the binding between chitosan and DNA under neutral or alkaline conditions (11,12). The pH effect on the intermolecular interactions, affecting the extent of complexation between chitosan and nucleic acid (DNA or siRNA) and the stability and dissociation of these nanoparticles, can be studied in solution by using sensitive force-probing spectroscopy at the single molecular level.
Force spectroscopy studies obtained by means of atomic force microscopy (AFM) has proven to be a very interesting and versatile analytical approach to explore a wide range of intermolecular interactions forces down to the single molecular level (13–17). Force spectroscopy for electrostatic interaction measurements with a Si3N4 tip contacting a functionalized surface was first performed by Butt et al. (18). A wide variety of force spectroscopy studies has subsequently been carried out, ranging from the measurement of coordination interaction (19) and discrimination of chemical and chiral surface structures (20) to the study of complex biospecific interactions such as antibody-antigen interactions (21), protein-ligand interactions (22), DNA basepairing (23,24), and cell-protein interactions (25). In these measurements, a typical experimental design for using force spectroscopy to investigate the bimolecular interactions, such as protein-ligand interaction, involves ligand attachment to a modified AFM tip and the target protein onto a surface via flexible linkers (such as polyethylene glycol (21) and glutaraldehyde (26,27)) or vice versa. Such experiments make it possible to determine the binding affinity, rate constants, and structural data of the binding pocket and can also provide insights into the molecular dynamics of the biological recognition process by varying the loading rate of force appliance. So far, however, no studies have been carried out in which force spectroscopy has been applied to investigate the interaction of polycations, such as chitosan and nucleic acids in gene delivery system, to obtain a quantitative understanding of the strength of chitosan/DNA (or siRNA) interactions that affect the stability and dissociation of the nanoparticles relevant to biological applications.
In this work, based on a novel siRNA/chitosan nanoparticle system developed within our group (28), we investigate the molecular interactions between siRNA and chitosan in a phosphate-buffered saline (PBS) buffer using force spectroscopy to help understand the formation, stability, and dissociation of siRNA/chitosan nanoparticles. To prepare a feasible model system for the chitosan-siRNA interaction, siRNA was immobilized to the AFM tip, and chitosan was grafted onto a gold-coated surface via a linker of glutaraldehyde (29–31). The use of a glutaraldehyde spacer provides sufficient distance between grafted chitosan and the siRNA-modified tip, maximizing the spatial accessibility of siRNA to chitosan and at the same time minimizing undesirable nonspecific adhesions. The interaction strengths between siRNA and chitosan were measured in PBS buffer under different pH conditions relevant to siRNA/chitosan nanoparticle formation (low pH) and physiological application (neutral pH). Control experiments were performed by detecting the interactions between siRNA-modified AFM tips and hydroxyl- or SO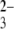 -terminated gold surfaces' terminated self-assembled monolayer (SAM) and also by blocking the interactions with the addition of excess of free siRNA to the buffer solution. The dynamics force measurements of the siRNA-chitosan interactions were performed to investigate the effect of loading rate on the interactions.
-terminated gold surfaces' terminated self-assembled monolayer (SAM) and also by blocking the interactions with the addition of excess of free siRNA to the buffer solution. The dynamics force measurements of the siRNA-chitosan interactions were performed to investigate the effect of loading rate on the interactions.
MATERIALS AND METHODS
Materials
siRNA duplex against the expression of enhanced green fluorescent protein (21 bp, sense sequence: GAC GUA AAC GGC CAC AAG UUC, antisense sequence: ACU UGU GGC CGU UUA CGU CGC) was from Dharmacon (Lafayette, CO). Once ssRNA nucleotides were modified with a thiol group at their 5′ end (DNA technology, Aarhus, Denmark), high pressure liquid chromatography (HPLC) was used to purify the oligos, and mass spectrometry was carried out to verify the mass. The thiol-modified siRNA was generated with two complementary ssRNA. The as modified siRNA was kept at −50°C for long-term storage or at 4°C under nitrogen until use. Chitosan (molecular mass 12 kDa, DD 77%) was obtained from Bioneer (Hørsholm, Denmark). siRNA/chitosan nanoparticles were prepared under acidic conditions as described elsewhere (28). All the chemicals were purchased from Sigma-Aldrich (Deisenhofen, Germany). All solutions used were adjusted to a different pH—4.1, 6.1, 7.4, and 9.5—using PBS buffer (138 mM NaCl, 2.7 mM KCl, pH 7.4 at 25°C, Biotechnology Performance Certified (Sigma-Aldrich)) with 1 M HCl or 1 M NaOH.
AFM tip and substrate preparation
Gold-coated AFM contact mode cantilevers (triangular, OMCL-TR400PSAHW, Olympus) were cleaned for 10 min by ultraviolet/ozone treatment (UVO-Cleaner, Jelight, CA), rinsed with deionized water, and dried under nitrogen flow. The clean cantilever tips were immediately immersed in a 4 μM aqueous solution of thiol-modified siRNA for 20 min, followed by insertion into a 1.0 μM aqueous solution of HS(CH2)2OH for 16 h and then rinsing with MilliQ water (Millipore, Bedford, MA). The uncovered gold surfaces of the AFM tips were eventually passivated by a SAM of HS(CH2)2OH, which can reduce the nonspecific adhesion of molecules (23,32). The as prepared tips were stored under a MilliQ water saturated atmosphere in a closed vessel at 4°C until use.
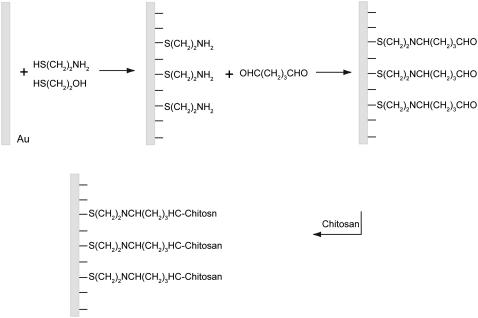
The chitosan-grafted surfaces were prepared according to a typical procedure described elsewhere (29,31), as shown in Scheme 1. Briefly, gold-coated wafers (Arrandee Supplies, Germany) were cleaned by Piranha solution (H2SO4/H2O2 7:3, v/v) at 75°C for 30 min (safety note: the Piranha solution should be handled with extreme caution). The freshly cleaned wafers were immediately immersed into the mixed ethanol solution of 1.3 μM HS(CH2)2NH2 and 1.0 μM HS(CH2)2OH (1:9 molar ratio final solution prepared by mixture of a two stock solution with different volumes) for 16 h. Unbonded alkanethiol aggregates on thiol-modified gold surfaces were briefly removed by gentle sonication in ethanol and then extensive rinsing with ethanol. The NH2-terminated wafers were then activated by incubation in 0.1% (v/v) glutaraldehyde solution in pure water for 1 h at room temperature and then rinsed with pure water and subsequently incubated for 1 h with 0.1 mg/mL of aqueous solution of chitosan at pH 5.7. The glutaraldehyde reaction with amine groups of chitosan is rapid in aqueous solution at pH 5.7 at room temperature, since this reaction takes place over a fairly wide pH range from 5.0 to 9.0 (29). The resulting surfaces were extensively rinsed three times with MilliQ water to remove loosely adsorbed aggregates. The as prepared surface was immediately used or kept at 4°C until use in PBS at pH 4.1 to render chitosan soluble.
SCHEME 1.
Schematic illustration of chitosan grafting on a gold surface, showing steps in chitosan surface treatment: animation, cross-linking with glutaraldehyde, and covalent binding of chitosan molecules.
AFM imaging and force spectroscopy
AFM images and force-distance curves were obtained using a commercial Digital Instruments Nanoscope IV MultiMode SPM (Veeco Instruments, Santa Barbara, CA) under ambient conditions. The system was equipped with a PicoForce controller and a low noise head. The images of unmodified and chitosan-grafted surfaces were recorded with silicon nitride probes (triangular, OMCL-TR400PSA-1; Olympus, Tokyo, Japan) in air and in PBS buffer at pH 7.4. The scan rate ranges within 1.0–2.0 Hz. Three images were obtained from separate locations on each sample surface for reproducibility.
The force spectroscopy measurements were performed using siRNA-modified cantilevers in fluid. Fluid with different pH was gently injected into a closed O-ring and subsequently allowed for 15 min of equilibrium. Spring constants were independently calibrated by a thermal tune procedure embedded in the PicoForce software (Version 6.12). The spring constant of the cantilever in force measurements was determined to be 0.03 ± 0.004 nN/m. Force-distance curves were recorded between siRNA-modified AFM tips and chitosan-terminated surfaces in PBS medium at pH 4.1, 6.1, 7.4, and 9.5. A relative trigger of 30 nm deflection of the cantilever was used to control the maximal force (<1 nN) of the tip against the surface, and the tip was allowed to rest on the plate for up to 0.5 s before the retrace at velocities varying from 100 to 1000 nm/s. For each experimental condition, force curves were recorded at 10 different spots, and at each spot 100 force curves were captured continuously. After each force spectroscopy experiment, AFM images were obtained to ensure the unbinding events on chitosan-terminated surfaces. Control experiments were carried out using the siRNA-modified tip and the pH-insensitive SAM surfaces of HS(CH2)2OH and also by blocking the interactions via adding excess of free siRNA to the buffer solution. The HS(CH2)2OH-terminated surface was prepared by immersing cleaned gold-coated glass wafers for 16 h in ethanol solution of HS(CH2)2OH (1.0 μM). The force-distance curves were analyzed using commercial PicoForce software (Version 6.12, Veeco). Data of unbinding forces were plotted in histograms, which were subsequently subjected to Gaussian filtering for analysis (25).
RESULTS AND DISCUSSION
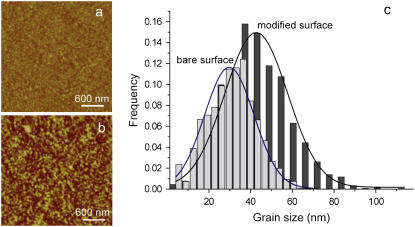
A feasibly simplified system of chitosan-siRNA interaction was molded by thiol-modified siRNA firmly anchored to gold-coated AFM tips and chitosan-grafted gold substrates via the cross-linking of cytosine and glutaraldehyde, as shown in Scheme 1. The quality of the chitosan-grafted gold substrates can be evaluated by AFM under liquid environments (25,33). Fig. 1 shows representative AFM images of the bare gold surface and the chitosan-grafted surface, which were recorded in PBS buffer at pH 7.4. The clean gold surface displays a polycrystalline surface with a root mean-square (RMS) roughness value of 2.5 nm over the image range of 3 × 3 μm2 (Fig. 1 a). With modification of chitosan, the chitosan-grafted surface displays many bright protrusions (grains) (Fig. 1 b), and the RMS roughness was determined to be 2.1 nm. This attenuation in RMS roughness of the chitosan-grafted surface is tentatively explained as being due to the uniform immobilization of spacers and chitosan molecules, which was observed for the functionalized surfaces with the lectin concanavalin A molecules reported previously (33). Furthermore, there was an apparent difference in grain size between chitosan-grafted surface and the clean gold surface (Fig. 1 c). The measured grains of the chitosan-grafted surface have a mean size of 43.2 nm, much larger than the grains with a mean size of 30.5 nm on the bare gold surface. The different heights and nanoparticle sizes of chitosan-modified surfaces can be attributed to the grafted chitosan molecules observed in PBS at pH 7.4, due to the aggregations of <10% amine groups protonated in the poor solution media at this pH (34). Compared to the bare gold and thiol-modified gold surfaces (not shown), the bright protrusions on the chitosan-modified surface clearly support that the spacer of glutaraldehyde was able to allow chitosan molecules distance from the thiol-modified surfaces and thus to afford the chitosan molecules suitable spatial freedom to contact siRNA in the following force measurements. Additionally, the grafting of chitosan to the surfaces was confirmed by scanning small areas of 1 × 1 μm2 at a larger force (5 nN) and subsequently a 3 × 3 μm2 image of the same area under a normal load (25,33). The 3 × 3 μm2 AFM image shows the underlying Au substrate (not shown), confirming the presence of the grafted chitosan molecules.
FIGURE 1.
Typical AFM images of (a) bare gold surface and (b) chitosan-grafted surfaces obtained in PBS solution (pH 7.4). (c) The comparison of grain size distribution between bare gold surface and chitosan-grafted surfaces, as determined by Gaussian fit.
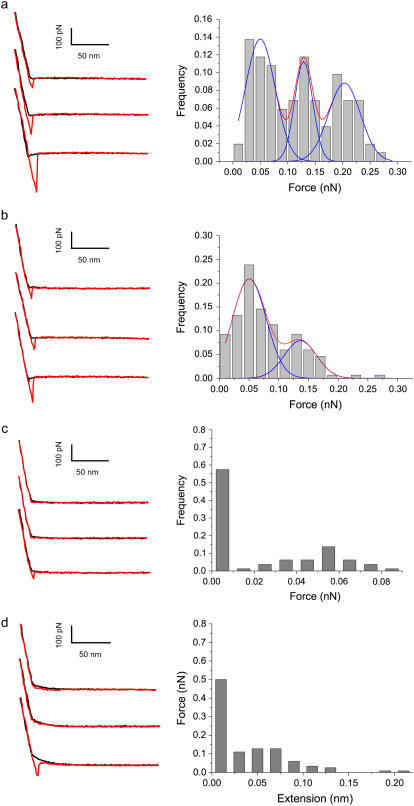
After AFM imaging, the bare AFM tip was replaced with siRNA-functionalized tips for the force spectroscopy experiments carried out in PBS buffer at different pH conditions. Fig. 2 shows typical force-distance curves and adhesive force distributions obtained using the same tip at the same rate from 1000 subsequent force curves at 10 different positions on the chitosan-grafted surface. All the left panels of Fig. 2 show that the adhesive forces follow linear retrace force slope. This observation means that no nonlinear stretched contour length was observed in the initial portion of the retraction curves. The characteristic of small siRNA tethered is dramatically different from the nonlinear stretching of flexible polyethylene glycol (PEG) linkers reported previously (17,25). Analysis of the force distributions and force-distance curves under varying pH conditions clearly shows that unbinding events were strongly affected by the pH of the buffer solution. Most of the curves at pH 4.1 and 6.1 show the pronounced unbinding events of adhesive forces (Fig. 2, a and b), whereas the unbinding event frequency was clearly reduced at neutral and alkaline pH (Fig. 2, c and d). The observed unbinding events with low probability (∼10%) exhibit the characteristic point of separation of the tip from the surfaces and adhesion events. The detailed analysis of the data from multiple force-distance curves revealed virtual overlap of the force-distance traces and different unbinding forces. At pH 4.1, the adhesive force histograms, determined by fitting the data to Gaussian distributions, revealed multimodal distributions of adhesive forces with maxima at 49 ± 19 pN, 128 ± 16 pN, and 203 ± 29 pN for pH 4.1 (Fig. 2 a). We attribute the peak of 49 pN to a single unbinding event and 128 pN and 203 pN, or two and four unbinding events, respectively (23,25,35). At pH 6.1 maxima were observed at 53 ± 21 pN and 136 ± 27 pN, corresponding to one and two unbinding events, respectively (Fig. 2 b) (23,25,35). The interactions can also be confirmed by plotting unbinding force peak values against peak number (23), and from the slope of a fitted line going through zero a single unbinding force of 51.7 pN is determined (23). These values suggest multiple-bridge cooperative interactions of the electrostatic interactions, with 51.7 pN being the adhesive strength quantum of single adhesion force, which is indeed in quantitative agreement with the ranges of electrostatic interaction reported in previous studies (25,36). For example, it has been shown that single unbinding forces are observed in the range of 40–70 pN for poly(acrylic acid) from SAMs with amine endgroups (36) and for poly(vinylamine) from silica substrates (37). In contrast to acidic pH, the adhesive forces observed at high pH—7.4 (Fig. 2 c) and 9.5 (Fig. 2 d)—showed dramatically low force frequency. The significant reductions of adhesive force and adhesion possibility observed reflect the decrease of the sites of the protonated amine groups complexing with siRNA.
FIGURE 2.
Zoomed typical force-distance curves of siRNA-functionalized tip interacting with a chitosan-grafted surface. The black and red traces indicate approach and retraction signals, respectively, in which siRNA-chitosan adhesive (interaction) force was observed during retraction. The three different force curves produced from the same siRNA-functionalized tip were displayed vertically for clarity. Histogram of unbinding force values for siRNA-chitosan obtained with the same AFM tip in PBS at different pH at a loading rate of 500 nm/s, showing the increase of adhesion frequency with increasing solution pH. (a) 4.1, (b) 6.1, (c) 7.4, and (d) 9.5.
A tentative explanation of the reduction in siRNA-chitosan interaction strength observed when increasing pH could be addressed in terms of the decreasing charge density of the amino groups and the molecular conformation transitions of chitosan. At a low pH 4.1, chitosan molecules are highly charged due to the fully protonated amine groups (6), favoring extendedly flexible-like structure and thus affording many sites contacting siRNA. At an intermediate pH of 6.1, very close to the apparent pKa (6.3) of chitosan, the deprotonation of the amino groups ( ) and the resulting formation of increasing amine (−NH2) groups in chitosan—allowing the chitosan molecule to undergo a less extended conformation transition and thus partially shielding the electrostatic contacting sites—reduced the frequency of the electrostatic attraction forces. Upon injection of solution with neutral pH, <10% of the amine groups of the chitosan are protonated (34). A significant decrease in unbinding events, as a result of the suppression of the electrostatic attraction force between siRNA and chitosan suppressed, was observed. At a high pH of 9.5, almost all amino groups' NH
) and the resulting formation of increasing amine (−NH2) groups in chitosan—allowing the chitosan molecule to undergo a less extended conformation transition and thus partially shielding the electrostatic contacting sites—reduced the frequency of the electrostatic attraction forces. Upon injection of solution with neutral pH, <10% of the amine groups of the chitosan are protonated (34). A significant decrease in unbinding events, as a result of the suppression of the electrostatic attraction force between siRNA and chitosan suppressed, was observed. At a high pH of 9.5, almost all amino groups' NH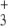 deprotonated to amine groups NH2, and the majority of chitosan molecules could be in conformations resembling three-dimensional hairy globular structure due to the collapsed network of intra- and intermolecular hydrogen bondings in chitosan. The existence of the weak siRNA-chitosan interactions that were still appreciably visible (Fig. 2 d) may be due to hydrogen bonding of siRNA with the primary amines of chitosan and also to hydrophobic interactions with N-acetyl groups or the neutralized segments involved in ion pairs (11).
deprotonated to amine groups NH2, and the majority of chitosan molecules could be in conformations resembling three-dimensional hairy globular structure due to the collapsed network of intra- and intermolecular hydrogen bondings in chitosan. The existence of the weak siRNA-chitosan interactions that were still appreciably visible (Fig. 2 d) may be due to hydrogen bonding of siRNA with the primary amines of chitosan and also to hydrophobic interactions with N-acetyl groups or the neutralized segments involved in ion pairs (11).
To confirm the reproducibility of the experimental findings, the force measurements were repeated twice using different AFM tips and samples to rule out any effects from tip shape and spring constants. In all the experiments carried out, the average value of the single unbinding force value was determined to be 55.8 pN at a retraction velocity of 500 nm/s in PBS (pH 4.1), suggesting reproducibility of our results.
Control experiments were conducted in PBS buffer within the involved pH range between an siRNA-modified tip and pH-insensitive hydroxyl- or SO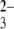 -terminated gold surfaces. The histograms and typical curves indicate negligible adhesive forces for hydroxyl- and SO
-terminated gold surfaces. The histograms and typical curves indicate negligible adhesive forces for hydroxyl- and SO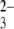 -terminated SAMs at pH 4.1, as shown in Fig. 3, a and b, respectively. In the absence of positively charged chitosan-terminated surfaces, siRNA-tethered AFM tips showed essentially no hysteresis between approach and pull-off curves in the experiments conduced under identical conditions, indicating that siRNA itself interacts very weakly with the modified surfaces. The quite low frequency interaction forces of the terminated gold surface clearly demonstrate the role in electrostatic interaction of contacting sites of the amino groups in chitosan polymers staying from the surfaces. For the SO
-terminated SAMs at pH 4.1, as shown in Fig. 3, a and b, respectively. In the absence of positively charged chitosan-terminated surfaces, siRNA-tethered AFM tips showed essentially no hysteresis between approach and pull-off curves in the experiments conduced under identical conditions, indicating that siRNA itself interacts very weakly with the modified surfaces. The quite low frequency interaction forces of the terminated gold surface clearly demonstrate the role in electrostatic interaction of contacting sites of the amino groups in chitosan polymers staying from the surfaces. For the SO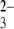 -terminated gold surface, our observation of no significant distribution interactions with anionic siRNA, very consistent with a previous study showing no dramatic interactions between ssDNA-modified tips and SO
-terminated gold surface, our observation of no significant distribution interactions with anionic siRNA, very consistent with a previous study showing no dramatic interactions between ssDNA-modified tips and SO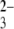 -terminated surfaces (23), essentially excludes that purely charge effects are responsible for the forces detected. Additionally, we also blocked the interactions by adding an excess of free siRNA to the buffer solution. The neglectable interactions measured (not shown) confirm our assignment of the electrostatic attraction forces between the siRNA-modified tip and chitosan-functionalized surfaces occurring at acidic conditions.
-terminated surfaces (23), essentially excludes that purely charge effects are responsible for the forces detected. Additionally, we also blocked the interactions by adding an excess of free siRNA to the buffer solution. The neglectable interactions measured (not shown) confirm our assignment of the electrostatic attraction forces between the siRNA-modified tip and chitosan-functionalized surfaces occurring at acidic conditions.
FIGURE 3.
The specificity of the siRNA-chitosan unbinding events is shown in control experiments. The distributions of adhesive forces in control experiments between siRNA-modified AFM tip and (a) SO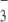 and (b) HO-terminated surfaces. Representative force-distance curves show that the specific interaction signals disappear, and the trace follows the retrace very well.
and (b) HO-terminated surfaces. Representative force-distance curves show that the specific interaction signals disappear, and the trace follows the retrace very well.
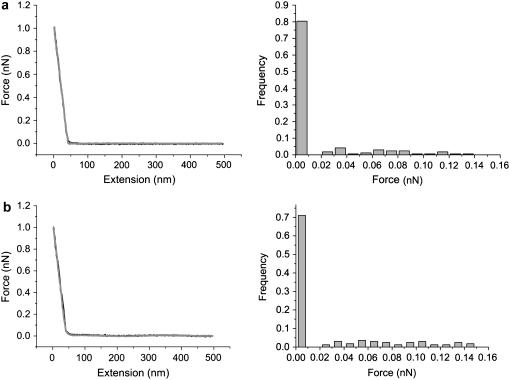
The dynamics force measurements of the siRNA-chitosan interactions were performed to investigate the effect of loading rate on the interactions. Fig. 4 shows the slight dependence of unbinding forces at several retraction rates at pH 6.1, indicating a nonequilibrium process of the force measurements. This result is different from a desorption plateau observed in most cases for the desorption of molecules interacting mainly by an electrostatic mechanism. In this study, siRNA molecules were directly immobilized on the tip, without the linking of PEG spacer. Due to the rather small siRNA tethered, it is very challenging to identify the desorption plateau generally characteristic of unbinding of specific interactions between molecular pairs immobilized to the AFM tip and substrate by polymeric spacers (such as PEG) in a nonequilibrium process (17).
FIGURE 4.
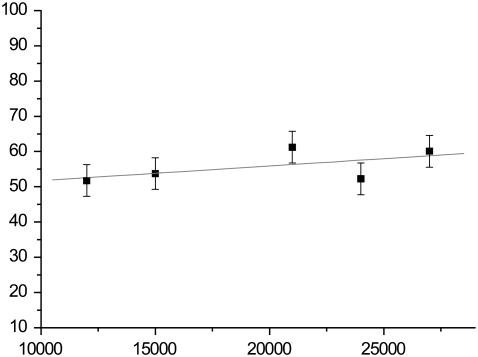
Dynamic force spectroscopy of single unbinding events on loading rates at pH 6.1 reflects the pH dependence of unbinding force.
Our result of the differences in unbinding events at variable pH conditions can be used to predict nanoparticle stability and siRNA intracellular release. The stability is also a very important parameter for gene silencing protocols to control the intracellular release of the DNA vector for gene expression (38–40). At low pH, under formulation conditions strong interactions between chitosan and siRNA can result in more compact nanoparticles. At neutral or at alkaline pH conditions, weak binding affinities can lead to poor compaction and low stability of siRNA/chitosan nanoparticles and the possible fast release of siRNA from siRNA/chitosan nanoparticles at these pH conditions.
CONCLUSION
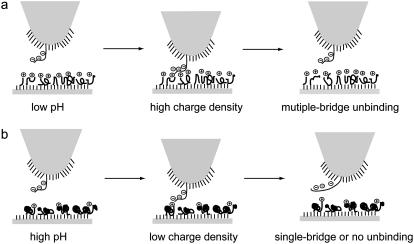
The interaction between siRNA and chitosan has been investigated by direct force measurements in PBS solution using AFM-based force spectroscopy with the aim of obtaining a quantitative understanding of the interaction strength within siRNA/chitosan nanoparticles for prediction of nucleic acid delivery behaviors. The effect of media pH on the interaction between chitosan and siRNA was observed, which is relevant to the formation and stability of nanoparticles and the dissociation of the nanoparticles for silencing within the cell. At low pH conditions, the strong siRNA-chitosan interaction can be ascribed to the multiple contacting sites of siRNA with high charge density and freely extended coil of chitosan (Fig. 5 a), reflecting good compaction and stability of siRNA/chitosan nanoparticles. At neutral or alkaline pH conditions, no clear interaction between chitosan and siRNA molecules occurs, due to the less contacting pairs resulting from the decreasing charge density and aggregates of chitosan molecules (Fig. 5 b), suggesting low stability of chitosan/siRNA and possibly a fast release of siRNA. This quantitative investigation of the siRNA-chitosan interactions demonstrates a novel use of force spectroscopy for sensitive studies of siRNA-carrier interactions for applications in drug delivery (41).
FIGURE 5.
Schematic illustration of the interaction between an siRNA-modified tip and a chitosan-immobilized surface under variable pH conditions. (a) At low pH conditions, due to high charge density of chitosan molecule, the strong chitosan-siRNA interactions show multiple bridge unbinding events. (b) At high pH conditions, the weak chitosan-siRNA interactions result in no or few unbinding events.
Acknowledgments
We thank Prof. Yves F. Dufrêne and Prof. Cees Dekker for fruitful suggestions.
We greatly acknowledge financial support from the Danish Ministry for Science, Technology and Innovation through the iNANO Center, from the Danish Research Councils, and from the Carlsberg Foundation.
Editor: Robert Callender.
References
- 1.Kabanov, A. V., P. L. Felgner, and L. W. Seymour. 1998. Self-Assembling Complexes for Gene Delivery: From Laboratory to Clinical Trial. Wiley, Chichester, UK.
- 2.Morris, K. V., S. W. L. Chan, S. E. Jacobsen, and D. J. Looney. 2004. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 305:1289–1292. [DOI] [PubMed] [Google Scholar]
- 3.Dorsett, Y., and T. Tuschl. 2004. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 3:318–329. [DOI] [PubMed] [Google Scholar]
- 4.Mansouri, S., P. Lavigne, K. Corsi, M. Benderdour, E. Beaumont, and J. C. Fernandes. 2004. Chitosan-DNA nanoparticles as non-viral vectors in gene therapy: strategies to improve transfection efficacy. Eur. J. Pharm. Biopharm. 57:1–8. [DOI] [PubMed] [Google Scholar]
- 5.Borchard, G. 2001. Chitosans for gene delivery. Adv. Drug Deliv. Rev. 52:145–150. [DOI] [PubMed] [Google Scholar]
- 6.Il'ina, A. V., and V. P. Varlamov. 2005. Chitosan-based polyelectrolyte complexes: a review. Appl. Biochem. Microbiol. 41:5–11. [PubMed] [Google Scholar]
- 7.Zhang, W. D., H. Yang, X. Y. Kong, S. Mohapatra, H. San Juan-Vergara, G. Hellermann, S. Behera, R. Singam, R. F. Lockey, and S. S. Mohapatra. 2005. Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nat. Med. 11:56–62. [DOI] [PubMed] [Google Scholar]
- 8.Danielsen, S., S. Strand, C. de Lange Davies, and B. T. Stokke. 2005. Glycosaminoglycan destabilization of DNA-chitosan polyplexes for gene delivery depends on chitosan chain length and GAG properties. Biochim. Biophys. Acta. 1721:44–54. [DOI] [PubMed] [Google Scholar]
- 9.Strand, S. P., S. Danielsen, B. E. Christensen, and K. M. Varum. 2005. Influence of chitosan structure on the formation and stability of DNA-chitosan polyelectrolyte complexes. Biomacromolecules. 6:3357–3366. [DOI] [PubMed] [Google Scholar]
- 10.Liu, W. G., S. J. Sun, Z. Q. Cao, Z. Xin, K. D. Yao, W. W. Lu, and K. D. K. Luk. 2005. An investigation on the physicochemical properties of chitosan/DNA polyelectrolyte complexes. Biomaterials. 26:2705–2711. [DOI] [PubMed] [Google Scholar]
- 11.Messai, I., D. Lamalle, S. Munier, B. Verrier, Y. Ataman-Onal, and T. Delair. 2005. Poly(D,L-lactic acid) and chitosan complexes: interactions with plasmid DNA. Colloids Surf. A Physicochem. Eng. Asp. 255:65–72. [Google Scholar]
- 12.Schatz, C., C. Pichot, T. Delair, C. Viton, and A. Domard. 2003. Static light scattering studies on chitosan solutions: from macromolecular chains to colloidal dispersions. Langmuir. 19:9896–9903. [Google Scholar]
- 13.Dufrêne, Y. F. 2004. Using nanotechniques to explore microbial surfaces. Nat. Rev. Microbiol. 2:451–460. [DOI] [PubMed] [Google Scholar]
- 14.Evans, E. 2001. Probing the relation between force—lifetime—and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 30:105–128. [DOI] [PubMed] [Google Scholar]
- 15.Janshoff, A., M. Neitzert, Y. Oberdorfer, and H. Fuchs. 2000. Force spectroscopy of molecular systems—single molecule spectroscopy of polymers and biomolecules. Angew. Chem. Int. Ed. Engl. 39:3213–3237. [DOI] [PubMed] [Google Scholar]
- 16.Butt, H. J., B. Cappella, and M. Kappl. 2005. Force measurements with the atomic force microscope: technique, interpretation and applications. Surf. Sci. Rep. 59:1–6. [Google Scholar]
- 17.Kienberger, F., A. Ebner, H. J. Gruber, and P. Hinterdorfer. 2006. Molecular recognition imaging and force spectroscopy of single biomolecules. Acc. Chem. Res. 39:29–36. [DOI] [PubMed] [Google Scholar]
- 18.Butt, H.-J. 1991. Electrostatic interaction in atomic force microscopy. Biophys. J. 60:777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auletta, T., M. R. De Jong, A. Mulder, F. Van Veggel, J. Huskens, D. N. Reinhoudt, S. Zou, S. Zapotoczny, H. Schonherr, G. J. Vancso, and L. Kuipers. 2004. Beta-cyclodextrin host-guest complexes probed under thermodynamic equilibrium: thermodynamics and AFM force spectroscopy. J. Am. Chem. Soc. 126:1577–1584. [DOI] [PubMed] [Google Scholar]
- 20.Mckendry, R., M. E. Theoclitou, T. Rayment, and C. Abell. 1998. Chiral discrimination by chemical force microscopy. Nature. 391:566–568. [Google Scholar]
- 21.Hinterdorfer, P., W. Baumgartner, H. J. Gruber, K. Schilcher, and H. Schindler. 1996. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 93:3477–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merkel, R., P. Nassoy, A. Leung, K. Ritchie, and E. Evans. 1999. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 397:50–53. [DOI] [PubMed] [Google Scholar]
- 23.Ling, L. S., H. J. Butt, and R. Berger. 2004. Rupture force between the third strand and the double strand within a triplex DNA. J. Am. Chem. Soc. 126:13992–13997. [DOI] [PubMed] [Google Scholar]
- 24.Sattin, B. D., A. E. Pelling, and M. C. Coh. 2004. DNA base pair resolution by single molecule force spectroscopy. Nucleic Acids Res. 32:4876–4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dupres, V., F. D. Menozzi, C. Locht, B. H. Clare, N. L. Abbott, S. Cuenot, C. Bompard, D. Raze, and Y. F. Dufrêne. 2005. Nanoscale mapping and functional analysis of individual adhesins on living bacteria. Nat. Met. 2:515–520. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, Y. X., F. Qin, Y. Q. Li, X. H. Fang, and C. L. Bai. 2004. Measuring specific interaction of transcription factor ZmDREB1A with its DNA responsive element at the molecular level. Nucleic Acids Res. 32:e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang, Y. X., C. F. Zhu, L. S. Ling, L. J. Wan, X. H. Fang, and C. Bai. 2003. Specific aptamer-protein interaction studied by atomic force microscopy. Anal. Chem. 75:2112–2116. [DOI] [PubMed] [Google Scholar]
- 28.Howard, K. A., U. L. Rahbek, X. D. Liu, C. K. Damgaard, S. Z. Glud, M. Ø. Andersen, M. B. Hovgaard, A. Schmitz, J. R. Nyengaard, F. Besenbacher, and J. Kjems. 2006. RNA interference in vitro and in vivo using a chitosan/siRNA nanoparticle system. Mol. Ther. 14:476–484. [DOI] [PubMed] [Google Scholar]
- 29.Carrara, C. R., and A. C. Rubiolo. 1994. Immobilization of beta-galactosidase on chitosan. Biotechnol. Prog. 10:220–224. [Google Scholar]
- 30.Jameela, S. R., and A. Jayakrishnan. 1995. Glutaraldehyde crosslinked chitosan microspheres as a long acting biodegradable drug delivery vehicle: studies on the in vitro release of mitoxantrone and in vivo degradation of microsheres in rat muscle. Biomaterials. 16:769–775. [DOI] [PubMed] [Google Scholar]
- 31.Liu, Y. J., Y. L. Li, S. C. Liu, J. Li, and S. Z. Yao. 2004. Monitoring the self-assembly of chitosan/glutaraldehyde/cysteamine/Au-colloid and the binding of human serum albumin with hesperidin. Biomaterials. 25:5725–5733. [DOI] [PubMed] [Google Scholar]
- 32.Krautbauer, R., M. Rief, and H. E. Gaub. 2003. Unzipping DNA oligomers. Nano Lett. 3:493–496. [Google Scholar]
- 33.Touhami, A., B. Hoffmann, A. Vasella, F. A. Denis, and Y. F. Dufrene. 2003. Probing specific lectin-carbohydrate interactions using atomic force microscopy imaging and force measurements. Langmuir. 19:1745–1751. [Google Scholar]
- 34.Schatz, C., C. Viton, T. Delair, C. Pichot, and A. Domard. 2003. Typical physicochemical behaviors of chitosan in aqueous solution. Biomacromolecules. 4:641–648. [DOI] [PubMed] [Google Scholar]
- 35.Radtchenko, I. L., G. Papastavrou, and M. Borkovec. 2005. Direct force measurements between cellulose surfaces and colloidal silica particles. Biomacromolecules. 6:3057–3066. [DOI] [PubMed] [Google Scholar]
- 36.Friedsam, C., A. D. Becares, U. Jonas, M. Seitz, and H. E. Gaub. 2004. Adsorption of polyacrylic acid on self-assembled monolayers investigated by single-molecule force spectroscopy. N. J. Phys. 6:1–16. [Google Scholar]
- 37.Hugel, T., M. Grosholz, H. Clausen-Schaumann, A. Pfau, H. Gaub, and M. Seitz. 2001. Elasticity of single polyelectrolyte chains and their desorption from solid supports studied by AFM based single molecule force spectroscopy. Macromolecules. 34:1039–1047. [Google Scholar]
- 38.Köping-Höggård, M., Y. S. Mel'nikova, K. M. Varum, B. Lindman, and P. Artursson. 2003. Relationship between the physical shape and the efficiency of oligomeric chitosan as a gene delivery system in vitro and in vivo. J. Gene Med. 5:130–141. [DOI] [PubMed] [Google Scholar]
- 39.Köping-Höggård, M., I. Tubulekas, H. Guan, K. Edwards, M. Nilsson, K. M. Varum, and P. Artursson. 2001. Chitosan as a nonviral gene delivery system. Structure-property relationships and characteristics compared with polyethylenimine in vitro and after lung administration in vivo. Gene Ther. 8:1108–1121. [DOI] [PubMed] [Google Scholar]
- 40.Schaffer, D. V., N. A. Fidelman, N. Dan, and D. A. Lauffenburger. 1999. Vector unpackaging as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol. Bioeng. 67:598–606. [DOI] [PubMed] [Google Scholar]
- 41.Javier, A. M., O. Kreft, A. P. Alberola, C. Kirchner, B. Zebli, A. S. Susha, E. Horn, S. Kempter, A. G. Skirtach, A. L. Rogach, J. R. Ädler, G. B. Sukhorukov, M. Benoit, and W. J. Parak. 2006. Combined atomic force microscopy and optical microscopy measurements as a method to investigate particle uptake by cells. Small. 2:394–400. [DOI] [PubMed] [Google Scholar]