Abstract
Tomato (Solanum lycopersicum) is susceptible to grey mold (Botrytis cinerea). Partial resistance to this fungus has been identified in accessions of wild relatives of tomato such as Solanum habrochaites LYC4. In a previous F2 mapping study, three QTLs conferring resistance to B. cinerea (Rbcq1, Rbcq2 and Rbcq4a) were identified. As it was probable that this study had not identified all QTLs involved in resistance we developed an introgression line (IL) population (n = 30), each containing a S. habrochaites introgression in the S. lycopersicum cv. Moneymaker genetic background. On average each IL contained 5.2% of the S. habrochaites genome and together the lines provide an estimated coverage of 95%. The level of susceptibility to B. cinerea for each of the ILs was assessed in a greenhouse trial and compared to the susceptible parent S. lycopersicum cv. Moneymaker. The effect of the three previously identified loci could be confirmed and seven additional loci were detected. Some ILs contains multiple QTLs and the increased resistance to B. cinerea in these ILs is in line with a completely additive model. We conclude that this set of QTLs offers good perspectives for breeding of B. cinerea resistant cultivars and that screening an IL population is more sensitive for detection of QTLs conferring resistance to B. cinerea than the analysis in an F2 population.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-006-0500-2) contains supplementary material, which is available to authorized users.
Introduction
Cultivated tomato (Solanum lycopersicum) displays only a limited amount of genotypic and phenotypic variation. Wild relatives of tomato are a useful source of variation for improving tomato (Rick 1982). Several wild Solanum accessions are crossable with tomato but barriers like unilateral incompatibility, hybrid inviability and sterility sometimes have to be overcome to obtain viable progeny. After successful hybridization, the introgression of specific traits from wild Solanum is often laborious and time consuming. The practical feasibility of introgressing specific chromosomal regions can be hampered by reduced recombination and/or linkage drag. Despite these difficulties, many favorable traits such as disease resistances (Bai et al. 2003; Haanstra et al. 2000; Kabelka et al. 2002), tolerance to abiotic stresses (Foolad et al. 2003) and agronomic traits (Bernacchi et al. 1998; Eshed and Zamir 1995; Lecomte et al. 2004) have been successfully introduced into tomato.
The focus of our research was the identification of quantitative trait loci (QTL) for increased disease resistance to Botrytis cinerea Pers:Fr [teleomorph Botryotinia fuckeliana (de Bary) Whetzel], a necrotrophic pathogenic fungus with a wide host range (Elad et al. 2004; Jarvis 1977). To date, Arabidopsis thaliana is the principal model to study the interaction with B. cinerea. Glazebrook (2005) reviewed the complex interactions between B. cinerea and mutants of A. thaliana. Denby et al. (2004) performed QTL mapping to elucidate the genetics of the interactions between B. cinerea and A. thaliana and reported the presence of multiple QTLs controlling resistance to B. cinerea.
Heirloom and modern hybrid tomato cultivars are susceptible to B. cinerea although a number of cultivars show quantitative resistance. A high level of resistance to B. cinerea has been identified in the wild accession Solanum habrochaites LYC4 (ten Have et al. 2007; Urbasch 1986). To study the genetics of this resistance, an F2 mapping population (n = 174) of the cross between S. lycopersicum cv. Moneymaker and S. habrochaites LYC4 has been developed (Finkers et al. 2007). Initially, two QTLs for resistance to B. cinerea were identified in this F2 mapping study (denominated Rbcq1 and Rbcq2, located on Chromosomes 1 and 2, respectively). A third QTL (Rbcq4 on Chromosome 4) that showed an interaction with Rbcq2 was detected in segregating BC2S1 progenies (Finkers et al. 2007).
A limitation of QTL-mapping in interspecific segregating F2 populations is the wide variation in plant growth rate, morphology and physiology but also the presence of major QTLs that potentially disguise QTLs with minor effects. Furthermore, it is difficult to do repeated tests as each F2 plant is a unique genotype. Alternatively, a genetic library consisting of a set of introgression lines (IL) can be used for mapping purposes. Each IL ideally harbors a single, defined chromosome segment that originates from the donor species in an otherwise uniform genetic background (Zamir 2001). Such lines facilitate the identification of QTLs because phenotypic variation between the line and the control cultivar is directly associated with the introgressed segment. Each line typically contains more than 95% of the recurrent cultivated parent genome and minor quantitative effects can easily be identified by comparison with the recurrent parent. Minimizing negative epistatic effects caused by other regions of the wild genome may lead to identification of novel QTLs (Eshed and Zamir 1995). Each line is homozygous and immortal and thus allows multiple testing (in multiple environments). Finally, sterility problems are less prominent due to the fact that the genetic constitution of each line is largely identical to the cultivated variety, which is usually very fit and fertile.
The first IL population was developed in wheat (Wehrhahn and Allard 1965), but the majority of IL populations have been developed during the last decade by means of marker-assisted selection (MAS) using different marker systems and breeding approaches. In tomato, ILs have been developed from Solanum pennellii LA716 (Eshed and Zamir 1994), S. habrochaites LA1777 (Monforte and Tanksley 2000a), S. habrochaites LA407 (Francis et al. 2001) and S. lycopersicoides LA2951 (Canady et al. 2005). ILs in tomato has proved to be extremely helpful in the identification of QTLs (Eshed and Zamir 1995; Rousseaux et al. 2005), fine mapping of QTLs (Eshed and Zamir 1996; Ku et al. 2001; Monforte et al. 2001; Monforte and Tanksley 2000b) and cloning of QTLs (Frary et al. 2000; Fridman et al. 2000; Liu et al. 2002). Besides tomato, IL populations have been developed for barley (von Korff et al. 2004), cabbage (Ramsay et al. 1996), lettuce (Jeuken and Lindhout 2004), melon (Eduardo et al. 2005), rice (Lin et al. 1998) and wheat (Pestova et al. 2001).
In general, IL populations have been developed by MAS using co-dominant SSR and RFLP markers but this is labor intensive and results in a lower marker density (ranging from 62 to 350 markers) compared to using AFLP markers (757 markers for lettuce). The advantage of SSR and RFLP markers, however, is that their positions are usually known, allowing the selection of a core set of markers covering the complete genome. AFLPs are high throughput markers but often with an unknown position and unequal coverage of the genome. Clustering of AFLP markers is a potential pitfall for certain enzyme combinations (Haanstra et al. 1999), however, in other cases AFLP markers identified introgressions that remained undetected using RFLP markers (Bonnema et al. 2002).
Our research aimed at verification of the QTLs for resistance to B. cinerea that have previously been identified in an interspecific F2 population (Finkers et al. 2007) and the identification of additional QTLs. This paper describes the development of an IL population (BC5S2) of S. habrochaites LYC4 in the genetic background of an indeterminate growing tomato by using AFLP as the platform for MAS. Each of the developed ILs was screened for its level of susceptibility to B. cinerea.
Materials and methods
Plant material and development of the ILs
The parents of the present study were the B. cinerea susceptible, indeterminate growing S. lycopersicum cv. Moneymaker (hereafter referred to as SL) and the resistant S. habrochaites LYC4 (ten Have et al. 2007; Urbasch 1986; hereafter referred to as SH, syn. Lycopersiconhirsutum). Seeds of SL were in stock of Wageningen University and seeds of SH were obtained from the Institute for Plant Genetics and Crop Plant Research, Gatersleben, Germany. One F1 plant was self pollinated to obtain F2 seeds and also backcrossed to SL to obtain BC1 seeds. The F2 seeds were sown to grow an F2 population (n = 174), which has previously been used for the construction of a genetic linkage map (Finkers et al. 2007). The BC1 was used to develop the IL population (Fig. 1).
Fig. 1.
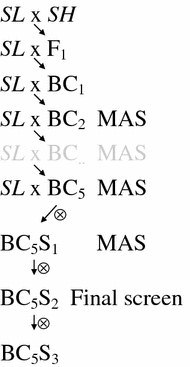
Backcross and selection strategy used to obtain the S. habrochaites LYC4 IL population introgressed in the S. lycopersicum cv. Moneymaker genetic background. Starting from BC2, marker assisted selection (MAS) was applied by using AFLP markers. The BC5S2 population was screened with a combination of AFLP and CAPS markers
Marker analysis
Genomic DNA was isolated from two young (rolled up) leaves using a CTAB based protocol according to Steward and Via (1993), adjusted for high throughput DNA isolation using 1 ml micronic tubes (Micronic BV, Lelystad, The Netherlands) and ground using a Retsch 300 mm shaker at maximum speed (Retsch BV, Ochten, The Netherlands).
AFLP™ analysis was done according to Vos et al. (1995). AFLP fragments were resolved on a LI-COR 4200 DNA sequencer, essentially following the method published by Myburg et al. (2001). The selective Pst primer was labeled with an IRD700 or IRD 800 fluorescent label. AFLP gel images were scored using AFLP-Quantar™ Pro software (Keygene, Wageningen, The Netherlands). Primer and adapter sequences have been described by Bai et al. (2003).
CAPS primers were obtained from the “Solanaceae Genomics Website” (http://www.sgn.cornell.edu) or designed on sequences of genomic or cDNA clones available from the same source. Polymorphisms between SL and SH were determined using the CAPS digestion approach described by Bai et al. (2004). Marker sequences, PCR conditions, and specific restriction endonucleases used for genotyping are presented in Supplementary Table 1. PCR products were separated using a 2.5% agarose gel.
Table 1.
Estimated mean disease incidence (DI), lesion size (LS) and lesion growth (LG) in the introgression lines (IL) and control lines. Means of each trait for each IL were compared to the mean of S. lycopersicum cv. Moneymaker (SL) using a Dunnett test and significant differences are marked with * P < 0.05 or ** P < 0.01
| IL | QTL | Na | Disease incidence (%) | Lesion size (mm) | Lesion growth rate (mm/day) |
|---|---|---|---|---|---|
| 1–1 | 41 | 65 ± 6.7 | 56 | 5.9 | |
| 1–2 | Rbcq1? | 40 | 47 ± 6.7 | 33 | 3.4 |
| 1–3 / 3–3 | Rbcq9b | 29 | 45 ± 9.1** | 30** | 1.7** |
| 1–4 | Rbcq9b | 44 | 37 ± 6.4** | 34 | 2.4* |
| 2–1 | Rbcq2 | 44 | 41 ± 6.4** | 30* | 3.0 |
| 2–2 | Rbcq2 | 44 | 37 ± 6.5** | 26* | 2.8 |
| 2–3 | 44 | 58 ± 6.5 | 44 | 3.5 | |
| 3–1 | Rbcq3 | 43 | 47 ± 6.6 | 41 | 2.8* |
| 3–2 | Rbcq3 | 44 | 46 ± 6.5* | 35 | 4.2 |
| 4–1 | Rbcq4a | 44 | 41 ± 6.4** | 26** | 2.5* |
| 4–2 | Rbcq4a | 42 | 45 ± 6.7* | 33 | 3.8 |
| 4–3 | Rbcq4b, Rbcq9a | 20 | 51 ± 9.6 | 29 | 2.8 |
| 5–1 | 44 | 61 ± 6.6 | 53 | 4.8 | |
| 5–2 | 43 | 69 ± 6.6 | 64 | 5.4 | |
| 6–1 | Rbcq6 | 44 | 49 ± 6.5* | 44 | 3.6 |
| 6–2 / 7–2 | 44 | 55 ± 6.3 | 39 | 3.7 | |
| 6–3 | 44 | 79 ± 6.5 | 49 | 4.6 | |
| 7–1 | 44 | 50 ± 6.4 | 35 | 3.1 | |
| 8–3 | 44 | 59 ± 6.5 | 43 | 3.9 | |
| 9–1 | Rbcq9a | 44 | 69 ± 6.5 | 34* | 3.0* |
| 9–2 | Rbcq9b | 44 | 49 ± 6.4* | 33 | 3.1 |
| 10–1 | 43 | 60 ± 6.6 | 47 | 4.3 | |
| 10–2 | 44 | 62 ± 6.5 | 49 | 4.4 | |
| 10–3 | 44 | 70 ± 6.4 | 53 | 4.7 | |
| 10–4 | 44 | 76 ± 6.6 | 47 | 4.8 | |
| 11–1 / 9–3 | Rbcq9b | 44 | 48 ± 6.5* | 36 | 4.3 |
| 11–2 | Rbcq11 | 44 | 34 ± 6.4** | 33* | 3.2 |
| 12–1 | Rbcq12 | 44 | 51 ± 6.4 | 35 | 4.7 |
| 12–2 | Rbcq12 | 43 | 52 ± 6.4 | 37 | 4.0 |
| 12–3 | Rbcq9b, Rbcq12 | 24 | 24 ± 8.6** | 21** | 2.3 |
| SL | 156 | 73 ± 4.0 | 46 | 4.6 | |
| SH | 44 | 3 ± 6.4** | NDb | ||
| BRC-5 | Rbcq1 andRbcq4b | 39 | 15 ± 6.9** | 20* | NDc |
| Durinta | 68 | 42 ± 5.5** | 29** | 2.3** |
aN Number of inoculation sites
bOnly one outgrowing lesion, observation excluded from the statistical analysis
cFirst observation of outgrowing lesions was after 12 days, therefore lesion growth could not be determined
Graphical genotyping
Graphical representation of the genotypes for each backcross population and the ILs were obtained using the software program GGT (van Berloo 1999). For the calculation of introgression size and genome percentages, the half-intervals flanking a marker locus were considered to be of the same introgression as implemented by the software. Missing marker data were estimated from the flanking markers; if these had identical genotypes then the missing marker was assumed to have the same genotype as the two flanking makers. If the two flanking makers had contrasting genotypes, the missing data were recorded as truly missing.
Experimental design and disease evaluations
To assess susceptibility, 16 incomplete randomized blocks were used in which in total 11 replications for each IL were tested. Each block contained at least two SL plants and one plant of S. lycopersicum cv. Durinta, a commercial cultivar producing truss tomatoes with a fair level of resistance to B. cinerea. Six weeks after sowing, plants were transplanted into the soil of two greenhouse compartments with a set day/night regime of 19/15°C and a photo period of 16 h light. After 11 weeks, two incisions of 15 mm were made into the stem of each plant at approximately 1.5 and 1.7 m height using a knife. Each wound was inoculated with a 1 cm2 agar plug containing a culture of B. cinerea B05.10 (Benito et al. 1998) and subsequently covered with tape. Two weeks later a third and fourth inoculation were performed on the same plants below the initial inoculation sites. Plants were watered at the beginning of the evening to maintain a humid climate during the night. Disease parameters were measured nine and 12 days after inoculation. The parameters were: disease incidence as percentage of growing lesions (DI), lesion size (using a caliper) after 12 days minus the size of the inoculation wound (15 mm, LS), and lesion growth rate expressed as the difference in lesion size between nine and 12 days post inoculation expressed in mm/day (LG).
Statistical analysis
Statistical analysis was performed using SPSS 12.0 software (SPSS Inc., Chicago, USA). Using a general linearized model (GLM) procedure, means for each IL/trait were estimated according to the following models: DI = constant + IL + block + inoculation position, LS = constant + IL + block + inoculation position and LG = constant + IL + block + inoculation position
Mean values of each trait were compared to the mean of the susceptible control genotype SL using a Dunnett test (Dunnett 1955) and probabilities smaller then 0.05 were considered as significant. To analyze LS and LG, a square root transformation was applied to normalize the data of both traits. Correlations between traits were calculated using the Pearson correlation coefficient.
Results
IL population
An introgression line (IL) population of S. habrochaites LYC4 (SH) in the genetic background of S. lycopersicum cv. Moneymaker (SL) was generated according to the procedure illustrated in Fig. 1. One F1 plant from the cross between SL and SH was backcrossed to SL. Subsequently a random set of 14 BC1 plants was backcrossed to SL to obtain a BC2 progeny (n = 59). All BC2 plants were genotyped and a selected set was backcrossed to SL. This set was chosen in such a way that the combined introgressions covered as much of the SH genome as possible and that each alien chromosome was represented by three ILs. The backcrossing and selection procedure was repeated until BC5 (Fig. 1). Eventually, 31 BC5 plants were selected, mainly containing one or two heterozygous introgressions. These 31 plants were self pollinated. Up to 12 plants of each of the 31 BC5S1 families were screened with AFLP markers to obtain a selected BC5S2 progeny (n = 44) homozygous for the (single) SH introgressions. The selected BC5S2 progeny was grown once more to allow a more detailed marker screen in which 457 AFLP markers (result of 10 AFLP enzyme/primer combinations) and 34 CAPS markers were analyzed. The total of 457 AFLP markers reported here is a twofold increase in marker density compared to the F2 population (Finkers et al. 2007). The marker screen resulted in the selection of a core set of 30 ILs with the highest possible coverage of the SH genome (Fig. 2) in as few lines as possible. Each selected line was self pollinated to obtain BC5S3 seeds.
Fig. 2.
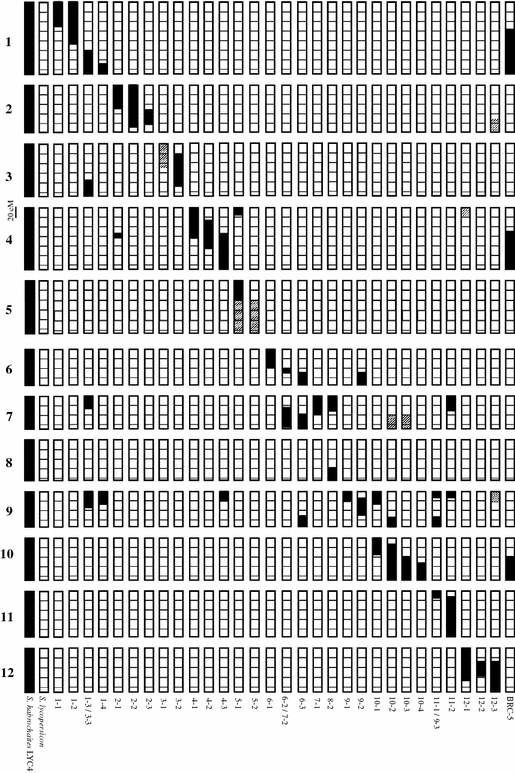
Graphical representation of the genotypes of the S. lycopersicum cv. Moneymaker × S. habrochaites LYC4 introgression line population. All chromosomes are drawn to scale in 20 cM segments according to the F2 genetic linkage map (Finkers et al. 2007) or estimated using the S. lycopersicum × S. pennellii linkage map (Tanksley et al. 1992; http://www.sgn.cornell.edu). Genetic length of the chromosome 9 introgressions was estimated from recombination frequencies of BC5S2 marker data. Homozygous introgressions from S. habrochaites are in black and heterozygous introgressions in gray
The IL library consists of 15 lines with a single introgression, ten lines with two introgressions, four lines with three introgressions while one IL contained four introgressions. The proportion of SH genome ranged from 20 (1.7%) to 122 cM (10.6%) with an average of 60 cM (5.2%). The IL library covers 95% of the F2 AFLP linkage map (Finkers et al. 2007), which however, does not cover the entire SH genome. Additional CAPS analysis on Chromosomes 3 (top of the short arm), 4 (top of the short arm), 5 (long arm) and 9 (top of the short arm) provided markers that were nearer to the telomeres than the most distal AFLP marker. The sizes of the latter introgressions were estimated, using the high density RFLP map (Tanksley et al. 1992; http://www.sgn.cornell.edu) and were estimated to be 20, 50 and 32 cM for Chromosome 4, 5 and 9, respectively. The size of the introgression at the top arm of Chromosome 3 could not be estimated.
No ILs homozygous SH for the top of Chromosome 3 (IL3-1) was obtained. Plants homozygous SH for IL5-1 and 5-2 failed to set seeds and these lines could only be maintained as heterozygotes for these introgressions. No ILs were obtained containing an introgression of Chromosome 1 harboring CAPS marker TG460, the distal end of the long arm of Chromosome 2 and the top of the short arm of Chromosome 8. Introgressions with the top of the short arm of Chromosome 7 and 9 were present in multiple ILs (Fig. 2). Selection against introgression on Chromosome 7 was hampered by an excess of SH alleles. Introgressions for the top of Chromosome 9 could only be detected during the more detailed CAPS marker screen of the BC5S2 ILs, as mentioned above. Introgressions homozygous SH in this region were present in 30% of the ILs.
Susceptibility of introgression lines to B. cinerea
The population of 30 ILs was grown in the greenhouse to the adult stage, inoculated with B. cinerea and evaluated for disease symptoms. On 9 and 12 days after inoculation the quantitative susceptibility to B. cinerea was quantified by determining the parameters disease incidence (DI), lesion size (LS) and lesion growth rate (LG), the results of which are shown in Table 1. The resistant parent SH hardly showed symptoms while 73% of the inoculation sites on the susceptible control SL developed into an expanding lesion with a mean LS of 46 mm and a LG of 4.6 mm/day. In total 14 ILs showed reduced susceptibility compared to SL and thus contain QTLs increasing resistance. Overall, 12 of these 14 ILs showed a significantly lower DI (24–49%), seven a significantly reduced LS (21–33 mm) and five a significantly lower LG (1.7–3.0 mm/day). Two lines, IL4-1 and IL1-3/3–3, showed a significant reduction of all three parameters (DI, LS and LG). The commercial cultivar S. lycopersicum cv. Durinta, with a fair level of resistance, also showed lower disease parameters compared to the susceptible control, SL.
Pearson correlation coefficients were calculated to asses the correlations between DI, LS and LG. Significant correlations (P < 0.01) were present between DI and LS (r = 0.82); DI and LG (r = 0.65) and LG and LS (r = 0.82) and are in agreement with the presence of lines significantly reduced for more than one disease parameter. The high correlations show that most of the QTLs alleles of SH result in the reduction of all three parameters (DI, LS and LG). We designated the identified QTLs as resistance to B. cinerea QTL (Rbcq; Table 1) followed by the number of the chromosome on which they are located, an analogous to the designation used for describing the QTLs in the F2 population (Finkers et al. 2007).
Three QTLs, Rbcq2, Rbcq4a (previously named Rbcq4; Finkers et al. 2007) and Rbcq6, unambiguously conferred increased resistance. The lower levels of susceptibility in IL3-1 and IL3-2, containing overlapping introgressions, are most probably the effect of one QTL, i.e., Rbcq3.
Others ILs those are less susceptible to B. cinerea than the reference SL contain multiple introgressions, thus complicating the allocation of resistance loci to specific chromosomes. In some cases, the most likely location can be deduced. For instance, IL9-2 contains introgressions of Chromosome 6 and 9. The introgression on Chromosome 6 is also present in IL6-3, which is equally susceptible to B. cinerea as SL. Thus, the increased resistance of IL9-2 is likely caused by a QTL on Chromosome 9 and there is no contribution from Chromosome 6. Furthermore, IL9-1 is partly overlapping with IL9-2 and has a similar LS as IL9-2, but a DI similar to SL. Therefore, we propose that two QTLs are present in IL9-2, Rbcq9a causing reduced LS in both IL9-1 and IL9-2 and Rbcq9b causing reduced DI only in IL9-2. By analogous deductions, the reduced disease symptoms in IL1-3/3-3, 1-4, 11–2 and 12–3 are likely to be caused by their Chromosome 9 introgressions which partly overlap the introgressions of IL9-1 and IL9-2. However, IL12-3 is significantly more resistant then IL9-2 suggesting an additional effect on Chromosome 12 (Rbcq12). The reduced susceptibility in IL12-1 and IL12-2, although statistically not significant, suggest that Rbcq12 might be located in the region overlapping between IL12-1, 12–2 and 12–3. IL11-2 has a significantly lower DI than IL9-1 and the introgression on Chromosome 9 is smaller then in IL9-1. Therefore the lower DI is probably conferred by another QTL: Rbcq11. IL1-3/3-3 and 1–4 are not significantly less susceptible then IL9-2 therefore the resistance in these lines is probably conferred by the presence of Rbcq9b.
Due to the absence of SH alleles for the CAPS marker TG460, which is linked to Rbcq1, it is unclear whether this locus is represented within the IL population. However, from previous experiments (Finkers et al. 2007) a highly resistant BC2S2 genotype was selected, denominated BRC-5, containing three homozygous introgressions on Chromosomes 1 (including Rbcq1 and homozygous SH for TG460), 4 and 10, representing in total 18% of the SH genome (Fig. 2). BRC-5 was the least susceptible line showing a DI of only 15%. No decreased susceptibility has been identified in any of the four Chromosome 10 ILs even though these ILs cover the entire length of Chromosome 10. It is therefore unlikely that the Chromosome 10 introgression contributes to the resistance of BRC-5. The Chromosome 4 introgression of BRC-5 does not overlap with the previously mapped position of Rbcq4a. IL4-3 showed a decreased susceptibility, although statistically not significant, suggesting the presence of a second QTL Rbcq4b. The combined effect of Rbcq1 and Rbcq4b is likely responsible for the increased resistance of BRC-5, however, this needs to be confirmed by experimental evidence.
In summary, ten QTLs were identified all increasing resistance to B. cinerea. The evidence for QTLs Rbcq4b, Rbcq11 and Rbcq12 is only circumstantial due to the presence of Rbcq9a/b, and their effects should be confirmed in the absence of the introgression on Chromosome 9.
Discussion
IL population
An IL population (n = 30) was developed of S. habrochaites in the genetic background of the cultivated tomato S. lycopersicum. During generation of the IL population, 239 additional AFLP markers were used which were not identified during analysis of the initial F2 population (Finkers et al. 2007) providing a nearly two-fold increase in marker density in spite of the use of the same AFLP primer combinations. A similar increase in marker density was observed during the development of a lettuce IL population (Jeuken and Lindhout 2004), mainly due to the reduced complexity of the AFLP-patterns of the ILs, thereby enabling the scoring of closely migrating or faint amplification products.
Although PstI/MseI markers are more equally distributed over the tomato chromosomes compared to EcoRI/MseI markers (Haanstra et al. 1999), ten randomly chosen primer combinations did not provide complete genome coverage. Compared to the high density RFLP reference map (Tanksley et al. 1992) about 32% of the genetic length of Chromosome 9 was not detected during the AFLP screenings. In an IL population of S. pennellii that was developed using 350 RFLP markers (Eshed and Zamir 1994), Chromosome 10 introgressions remained unnoticed using RFLP markers but could be detected using AFLP markers (Bonnema et al. 2002). A combination of AFLP with CAPS or SSR markers is a good strategy to develop introgression lines with a full genome coverage.
Reproductive behavior of the IL population
IL populations are generally believed to be less prone to reproductive problems, yet the development of some specific ILs has proven to be cumbersome. No ILs containing the top of Chromosome 8 were obtained. ILs generated with S. pennellii LA716 (Eshed and Zamir 1995) and S. habrochaites LA1777 (Monforte and Tanksley 2000a), as progenitors appeared to lack lines with introgressions for a small region on the short arm of Chromosome 8. Attempts to fixate this region of the wild relative remained unsuccessful, presumably due to the presence of a male sterility locus. Compared to both previously mentioned studies, a larger portion of Chromosome 8 is lacking in our population. Developing additional ILs covering at least part of this region should be feasible.
Plants those were homozygous SH for introgressions on the long arm of Chromosome 5 failed to set seeds. The S. habrochaites LA1777 IL population (Monforte and Tanksley 2000a) contains a QTL for reduced self seed set on Chromosome 5, possibly causing zygotic incompatibility resulting in early seed abortion (Moyle and Graham 2005). The occurrence of homozygous introgressions of Chromosome 7 and 9 was skewed towards homozygous SH resulting in the presence of these introgressions in multiple ILs (Fig. 2). Similar observations were made in other advanced interspecific tomato populations involving wild relatives such as S. galapagense (Paran et al. 1995), L. peruvianum (Fulton et al. 1997) and S. habrochaites (Monforte and Tanksley 2000a).
One of the advantages of an IL population is that each line has a high resemblance to SL and therefore sterility problems are expected to be minimal. For the development of a full set of homozygous ILs, however, extra effort must be undertaken to select lines containing introgressions flanking deleterious genes (e.g., on Chromosomes 5 and 8) or select against overrepresentation of introgressions on which an advantageous gene is located (e.g., on Chromosomes 7 and 9).
Confirmation of previously identified loci for resistance to B. cinerea
Two different disease assays were applied to quantify resistance to B. cinerea. Initially, QTLs were detected in an F2 population and confirmed in BC2S1 families (Finkers et al. 2007) using a bioassay on stem segments (ten Have et al. 2007) leading to the identification of three QTLs: Rbcq1 reducing LG, Rbcq2 and Rbcq4a both reducing DI (Finkers et al. 2007). Stem resistance in the IL population was quantified in whole adult plants; wound inoculated with a B. cinerea containing agar plug and disease progress was recorded during a longer period. In all tests, Rbcq2 and Rbcq4a conferred resistance to B. cinerea showing the robustness of each QTL. Rbcq1 could not be confirmed in the IL population, but it remains uncertain whether Rbcq1 was represented in the ILs tested. Marker analysis showed that the introgression in lines IL1-2 and IL1-3/3-3 do not overlap. A small SH introgression for Chromosome 1 is therefore not represented within the ILs (Fig. 2). Rbcq1 is however, certainly present in BRC-5, the most resistant introgression line tested. Thus, all three previously identified loci for increased resistance to B. cinerea were detected using both bioassays.
Identification of additional resistance loci using an IL population
The main purpose of developing an IL population and assessing its resistance to B. cinerea was to identify additional QTLs to the ones previously identified in the F2 population (Finkers et al. 2007). As resistance to B. cinerea is polygenic, multiple minor quantitative effects can easily be overlooked in an interspecific F2 population. Seven additional QTLs were detected in the IL population: Rbcq3, Rbcq4b, Rbcq6, Rbcq9a, Rbcq9b, Rbcq11 and Rbcq12. No indications for any of the seven new QTLs were found in the analysis of the F2 population, even when reexamining these data (data not shown). Most newly identified loci were less efficient in reducing symptoms compared to Rbcq2. These “minor loci” might have been identified in a larger F2 population, but they were readily detected in an IL population. Also in an Arabidopsis RIL population, a larger number of QTLs for resistance to B. cinerea were reported, some of which were considered to be specific for distinct pathogen isolates (Denby et al. 2004). This IL population provides an excellent tool to study the possible existence of isolate-specific QTLs in tomato and study resistance levels in different environments.
Potential of pyramiding QTLs
None of the QTLs was sufficient to confer the full level of resistance of the SH parent. To reach such a level QTLs have to be combined. Pyramiding of ten QTLs is practically impossible and probably not necessary. An example of an additive effect is illustrated by Rbcq9b and Rbcq12. IL12-1 and IL12-2 with only Rbcq12 had on average a 22% lower DI (although not significant) while IL9-2 with Rbcq9b had a DI significantly decreased by 24%, compared to SL. Assuming an additive model, the combination of Rbcq9b and Rbcq12 would lead to a decrease in DI by 46%. The observation that IL12-3, combining both Rbcq9b and Rbcq12, had a DI that was 49% lower is in agreement with this additive model. These results show the potential of pyramiding multiple QTLs in order to get an elevated resistance to B. cinerea but redundancy of QTLs for resistance is expected. The IL population is an excellent tool to combine multiple QTLs and study their interaction in order to select the best set of QTLs for exploitation in a commercial breeding program. More detailed metabolome, proteome and transcriptome analysis of each of the ILs may provide insight in the resistance mechanism conferred by each QTL.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1: Primer sequences, lengths of PCR products and enzymes revealing a polymorphism for CAPS/SCAR markers. (DOC 227 kb)
Acknowledgments
This project is sponsored by grants of the Dutch Technology Foundation (STW, grant no. WBI.4172) and De Ruiter Seeds R&D BV, The Netherlands. The authors would like to thank Petra van den Berg, Pham Thi Thu Huong and Roeland Voorrips for their contribution and valuable suggestions.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-006-0500-2) contains supplementary material, which is available to authorized users.
References
- Bai Y, Huang CC, van der Hulst R, Meijer Dekens F, Bonnema G, Lindhout P (2003) QTLs for tomato powdery mildew resistance (Oidium lycopersici) in Lycopersicon parviflorum G1.1601 co-localize with two qualitative powdery mildew resistance genes. Mol Plant Microbe Interact 16:169–176 [DOI] [PubMed]
- Bai Y, Feng XH, van der Hulst R, Lindhout P (2004) A set of simple PCR markers converted from sequence specific RFLP markers on tomato Chromosomes 9 to 12. Mol Breed 13:281–287 [DOI]
- Benito EP, ten Have A, van ‘t Klooster JW, van Kan JAL (1998) Fungal and plant gene expression during synchronized infection of tomato leaves by Botrytis cinerea. Eur J Plant Pathol 104:207–220 [DOI]
- Bernacchi D, Beck Bunn T, Eshed Y, Lopez J, Petiard V, Uhlig J, Zamir D, Tanksley S (1998) Advanced backcross QTL analysis in tomato. I. Identification of QTLs for traits of agronomic importance from Lycopersicon hirsutum. Theor Appl Genet 97:381–397 [DOI]
- Bonnema G, Berg P, Lindhout P (2002) AFLPs mark different genomic regions compared with RFLPs: a case study in tomato. Genome 45:217–221 [DOI] [PubMed]
- Canady MA, Meglic V, Chetelat RT (2005) A library of Solanum lycopersicoides introgression lines in cultivated tomato. Genome 48:685–697 [DOI] [PubMed]
- Denby KJ, Kumar P, Kliebenstein DJ (2004) Identification of Botrytis cinerea susceptibility loci in Arabidopsis thaliana. Plant J 38:473–486 [DOI] [PubMed]
- Dunnett CW (1955) A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc 50:1096–1121 [DOI]
- Eduardo I, Arus P, Monforte AJ (2005) Development of a genomic library of near isogenic lines (NILs) in melon (Cucumis melo L.) from the exotic accession PI161375. Theor Appl Genet 112:139–148 [DOI] [PubMed]
- Elad Y, Williamson B, Tudzynski P, Delen N (2004) Botrytis: biology, pathology and control. Kluwer, Dordrecht
- Eshed Y, Zamir D (1994) A genomic library of Lycopersicon pennellii in L. esculentum: a tool for fine mapping of genes. Euphytica 79:175–179 [DOI]
- Eshed Y, Zamir D (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141:1147–1162 [DOI] [PMC free article] [PubMed]
- Eshed Y, Zamir D (1996) Less-than-additive epistatic interactions of quantitative trait loci in tomato. Genetics 143:1807–1817 [DOI] [PMC free article] [PubMed]
- Finkers R, van den Berg P, van Berloo R, ten Have A, van Heusden AW, van Kan JAL, Lindhout P (2007) Three QTLs for Botrytis cinerea resistance in tomato. Theor Appl Genet. org/10.1007/s00122-006-0458-0 [DOI] [PubMed]
- Foolad MR, Zhang LP, Subbiah P (2003) Genetics of drought tolerance during seed germination in tomato: inheritance and QTL mapping. Genome 46:536–545 [DOI] [PubMed]
- Francis DM, Kabelka E, Bell J, Franchino B, St. Clair D (2001) Resistance to bacterial canker in tomato (Lycopersicon hirsutum LA407) and its progeny derived from crosses to L. esculentum. Plant Dis 85:1171–1176 [DOI] [PubMed]
- Frary A, Nesbitt TC, Grandillo S, Knaap Evd, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD (2000) fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289:85–88 [DOI] [PubMed]
- Fridman E, Pleban T, Zamir D (2000) A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc Natl Acad Sci USA 97:4718–4723 [DOI] [PMC free article] [PubMed]
- Fulton TM, Beck Bunn T, Emmatty D, Eshed Y, Lopez J, Petiard V, Uhlig J, Zamir D, Tanksley SD (1997) QTL analysis of an advanced backcross of Lycopersicon peruvianum to the cultivated tomato and comparisons with QTLs found in other wild species. Theor Appl Genet 95:881–894 [DOI]
- Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227 [DOI] [PubMed]
- Haanstra JPW, Wye C, Verbakel H, Meijer Dekens F, van den Berg P, Odinot P, van Heusden AW, Tanksley S, Lindhout P, Peleman J (1999) An integrated high density RFLP-AFLP map of tomato based on two Lycopersicon esculentum × L. pennellii F2 populations. Theor Appl Genet 99:254–271 [DOI]
- Haanstra JPW, Meijer Dekens F, Lauge R, Seetanah DC, Joosten M, de Wit P, Lindhout P (2000) Mapping strategy for resistance genes against Cladosporium fulvum on the short arm of chromosome 1 of tomato: Cf-ECP5 near the Hcr9 Milky Way cluster. Theor Appl Genet 101:661–668 [DOI]
- Jarvis WR (1977) Botryotinia and Botrytis species: taxonomy, physiology, and pathogenicity; a guide to the literature. Monograph 15
- Jeuken MJW, Lindhout P (2004) The development of lettuce backcross inbred lines (BILs) for exploitation of the Lactuca saligna (wild lettuce) germplasm. Theor Appl Genet 109:394–401 [DOI] [PubMed]
- Kabelka E, Franchino B, Francis DM (2002) Two loci from Lycopersicon hirsutum LA407 confer resistance to strains of Clavibacter michiganensis subsp. michiganensis. Phytopathology 92:504–510 [DOI] [PubMed]
- Ku HM, Liu J, Doganlar S, Tanksley SD (2001) Exploitation of Arabidopsis-tomato synteny to construct a high-resolution map of the ovate-containing region in tomato chromosome 2. Genome 44:470–475 [DOI] [PubMed]
- Lecomte L, Saliba Colombani V, Gautier A, Gomez Jimenez MC, Duffe P, Buret M, Causse M (2004) Fine mapping of QTLs of chromosome 2 affecting the fruit architecture and composition of tomato. Mol Breed 13:1–14 [DOI]
- Lin SY, Sasaki T, Yano M (1998) Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines. Theor Appl Genet 96:997–1003 [DOI]
- Liu JP, Van Eck J, Cong B, Tanksley SD (2002) A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc Natl Acad Sci USA 99:13302–13306 [DOI] [PMC free article] [PubMed]
- Monforte AJ, Tanksley SD (2000a) Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in a L. esculentum genetic background: a tool for gene mapping and gene discovery. Genome 43:803–813 [DOI] [PubMed]
- Monforte AJ, Tanksley SD (2000b) Fine mapping of a quantitative trait locus (QTL) from Lycopersicon hirsutum chromosome 1 affecting fruit characteristics and agronomic traits: breaking linkage among QTLs affecting different traits and dissection of heterosis for yield. Theor Appl Genet 100:471–479 [DOI]
- Monforte AJ, Friedman E, Zamir D, Tanksley SD (2001) Comparison of a set of allelic QTL-NILs for chromosome 4 of tomato: deductions about natural variation and implications for germplasm utilization. Theor Appl Genet 102:572–590 [DOI]
- Moyle LC, Graham EB (2005) Genetics of hybrid incompatibility between Lycopersicon esculentum and L. hirsutum. Genetics 169:355–373 [DOI] [PMC free article] [PubMed]
- Myburg AA, Remington DL, O’ Malley DM, Sederoff RR, Whetten RW (2001) High-throughput AFLP analysis using infrared dye-labeled primers and an automated DNA sequencer. Biotechniques 30:348–357 [DOI] [PubMed]
- Paran I, Goldman I, Tanksley SD, Zamir D (1995) Recombinant inbred lines for genetic mapping in tomato. Theor Appl Genet 90:542–548 [DOI] [PubMed]
- Pestsova EG, Borner A, Roder MS (2001) Development of a set of Triticum aestivum–Aegilops tauschii introgression lines. Hereditas 135:139–143 [DOI] [PubMed]
- Ramsay LD, Jennings DE, Bohuon EJR, Arthur AE, Lydiate DJ, Kearsey MJ, Marshall DF (1996) The construction of a substitution library of recombinant backcross lines in Brassica oleracea for the precision mapping of quantitative trait loci. Genome 39:558–567 [DOI] [PubMed]
- Rick CM (1982) The potential of exotic germplasm for tomato improvement. In: Vasil IK, Scowcroft WR, Frey KJ (eds) Plant improvement and somatic cell genetics. Academic, New York 1982:1–28
- Rousseaux MC, Jones CM, Adams D, Chetelat R, Bennett A, Powell A (2005) QTL analysis of fruit antioxidants in tomato using Lycopersicon pennellii introgression lines. Theor Appl Genet 111:1396–1408 [DOI] [PubMed]
- Steward CN, Via LE (1993) A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14:748–750 [PubMed]
- Tanksley SD, Ganal MW, Prince JP, de Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB (1992) High density molecular linkage maps of the tomato and potato genomes. Genetics 132:1141–1160 [DOI] [PMC free article] [PubMed]
- ten Have A, van Berloo R, Lindhout P, van Kan JAL (2007) Partial stem and leaf resistance against the fungal pathogen Botrytis cinerea in wild relatives of tomato. Eur J Plant Pathol 117:153–166 [DOI]
- Urbasch I (1986) Resistenz verschiedener Kultur- und Wildtomatenpflanzen (Lycopersicon spp.) gegenüber Botrytis cinerea Pers. J Phytopathol 116:344–351
- van Berloo R (1999) GGT: software for the display of graphical genotypes. J Hered 90:328–329 [DOI]
- von Korff M, Wang H, Leon J, Pillen K (2004) Development of candidate introgression lines using an exotic barley accession (Hordeum vulgare ssp. spontaneum) as donor. Theor Appl Genet 109:1736–1745 [DOI] [PubMed]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414 [DOI] [PMC free article] [PubMed]
- Wehrhahn C, Allard RW (1965) The detection and measurement of the effects of individual genes involved in inheritance of a quantitative character in wheat. Genetics 51:109–119 [DOI] [PMC free article] [PubMed]
- Zamir D (2001) Improving plant breeding with exotic genetic libraries. Nat Rev Genet 2:983–989 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
Table S1: Primer sequences, lengths of PCR products and enzymes revealing a polymorphism for CAPS/SCAR markers. (DOC 227 kb)


