Abstract
Paramyxoviruses comprise several major human pathogens. Although a live-attenuated vaccine protects against measles virus (MV), a member of the paramyxovirus family, the virus remains a principal cause of worldwide mortality and accounts for approximately 21 million cases and 300,000 to 400,000 deaths annually. The development of novel antivirals that allow improved case management of severe measles and silence viral outbreaks is thus highly desirable. We have previously described the development of novel MV fusion inhibitors. The potential for preexisting or emerging resistance in the field constitutes the rationale for the identification of additional MV inhibitors with a diverse target spectrum. Here, we report the development and implementation of a cell-based assay for high-throughput screening of MV antivirals, which has yielded several hit candidates. Following confirmation by secondary assays and chemical synthesis, the most potent hit was found to act as a target-specific inhibitor of MV replication with desirable drug-like properties. The compound proved highly active against multiple primary isolates of diverse MV genotypes currently circulating worldwide, showing active concentrations of 35 to 145 nM. Significantly, it does not interfere with viral entry and lacks cross-resistance with the MV fusion inhibitor class. Mechanistic characterization on a subinfection level revealed that the compound represents a first-in-class nonnucleoside inhibitor of MV RNA-dependent RNA polymerase complex activity. Singly or in combination with the fusion inhibitors, this novel compound class has high developmental potential as a potent therapeutic against MV and will likely further the mechanistic characterization of the viral polymerase complex.
Measles virus (MV), a member of the paramyxovirus family, remains a principal cause of worldwide morbidity and mortality, being responsible for approximately 300,000 to 400,000 deaths annually, despite the existence of a live-attenuated vaccine (8, 50). Globally, measles is the leading cause of childhood death from a vaccine-preventable disease (http://www.cdc.gov/programs/global06.htm) and remains among the 10 most lethal human pathogens. Transmitted via the respiratory route, the virus is highly communicable and one of the most infectious pathogens identified (19, 21, 47). Complications associated with MV infection include acute encephalitis in approximately 0.1% of cases and subacute sclerosing panencephalitis, a lethal late sequela that occurs years after the primary infection (19, 22).
Currently, no therapeutics are available for case management of severe measles or the rapid silencing of local outbreaks. Ribavirin, the only drug available for the treatment of some paramyxovirus infections (9, 41), has been used experimentally for the treatment of measles, but with limited efficacy (2). This makes desirable the development of cost-effective antivirals against MV that augment the existing vaccination program.
MV infection is initiated by pH-independent fusion of the viral envelope with the target cell plasma membrane (19). The hemagglutinin (H) envelope glycoprotein mediates particle attachment (13, 18, 32, 46), followed by membrane fusion orchestrated by the fusion (F) envelope protein (26). Viral-gene expression and subsequent genome replication then take place in the cytosol (19). Both processes are mediated by the viral RNA-dependent RNA polymerase (RdRp) complex, which consists minimally of a homotetramer of the viral phosphoprotein (P) and a single polymerase (L) protein (6, 25). The sole target for RdRp is a ribonucleoprotein complex of viral RNA encapsidated by the MV nucleocapsid (N) protein (6), minimizing the presence of naked genomic RNA in the host cell. Considering that human and animal tissues lack a known homologue of the RdRp or the fusogenic envelope proteins, the polymerase complex and components of the entry machinery constitute particularly attractive targets for virus-specific small-molecule inhibitors.
Despite its critical role in the viral life cycle, our mechanistic understanding of the MV RdRp is still limited and the structural characterization of its components is sparse. An abundance of structural disorder has been found in the MV N and P proteins (23, 27), and no paramyxovirus polymerase has been purified yet (6). In addition to their therapeutic potential, small-molecule compounds interfering with the function of the MV RdRp complex may constitute viable tools for a better molecular and structural characterization of the viral replication machinery.
In contrast to the RdRp, considerable structural information is available for the paramyxovirus attachment (12, 53) and fusion proteins, including structures of the latter in both the prefusion (52) and intermediate to postfusion (10, 51) conformations. In previous work, we identified a new class of MV fusion inhibitors, substituted anilides, in a structure-based drug design approach (36, 38). The lead compound of this inhibitor class, AS-48 (35, 44), shows activity in the low micromolar range (50% inhibitory concentration, 0.6 to 3.0 μM) against a panel of MV field isolates. A single sub-Saharan isolate is resistant to inhibition by AS-48, however, and in vitro adaptation has resulted in the appearance of characteristic escape mutants after four to seven passages (14), suggesting that resistance may emerge rapidly in the field. The identification of additional drug candidates against MV with diverse target characteristics is therefore imperative. In addition to counteracting preexisting resistance, combined administration of compounds with different target sites may reduce the rate of viral escape or result in impaired fitness of virions that develop multiple resistance.
Toward this goal, we report here the development of a robust, cell-based assay for high-throughput screening (HTS) of MV inhibitor candidates. Implementation of this assay has yielded several hit candidates, which were subsequently confirmed in manual secondary assays. The structure of the most potent candidate was confirmed by independent synthesis. It has desirable drug-like properties, does not block viral entry, and is not subject to cross-resistance with the AS-48 class of MV fusion inhibitors. Mechanistic characterization has revealed that the compound acts late in the viral life cycle, prompting us to address the question of whether it specifically interferes with the activity of the viral polymerase complex.
MATERIALS AND METHODS
Cell culture, transfection, and production of MV stocks.
All cell lines were maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin. Vero-SLAM cells, derived from Vero cells (African green monkey kidney epithelial cells; ATCC CCL-81) and stably expressing human SLAM/CD150w; Vero-dogSLAM cells (40) stably expressing dog SLAM; and BSR T7/5 cells (7) stably expressing T7 polymerase were incubated at every third passage in the additional presence of G-418 (Geneticin) at a concentration of 100 μg/ml. Lipofectamine 2000 (Invitrogen) was used for transient-transfection experiments. To prepare virus stocks, cells were infected at a multiplicity of infection (MOI) of 0.001 PFU/cell and incubated at 37°C. The cells were scraped in OPTIMEM (Invitrogen), virus was released by two freeze-thaw cycles, and titers were determined by 50% tissue culture infective dose (TCID50) titration according to the Spearman-Karber method (43) as previously described (37). MV-Edmonston (MV-Edm) stocks were grown and titered on Vero cells, while for MV field isolates, Vero-SLAM cells were used, and for canine distemper virus (CDV), Vero-dogSLAM cells were used. All MV field isolates were originally derived from peripheral blood mononuclear cell samples, and the viruses were isolated and minimally passaged on SLAM-positive B95-a cells or Vero-SLAM cells.
High-throughput compound screening.
For screening, Vero cells were seeded in 96-well microtiter plates at a density of 7,500 cells per well in 100 ml growth medium. After a 4-hour incubation period at 37°C and 5% CO2, compound was added in 1.0-μl/well doses (25 μM final concentration) with a Sciclone automated liquid handler system (Caliper, MA), followed by infection with a recombinant MV that harbors enhanced green fluorescent protein (eGFP) as an additional transcription unit (rMV-eGFP) (15, 17) at an MOI of 0.25 PFU/cell in 100 ml serum-free medium. Final solvent (dimethyl sulfoxide [DMSO]) concentrations were 0.5%, at which no adverse effect on cell viability or virus growth could be detected in control samples. All virus stocks used for screening were subjected to dialysis against phosphate-buffered saline (PBS) to remove contaminating eGFP that had been synthesized during virus growth. Following a 64-hour incubation period at 37°C, green fluorescence, indicating expression of viral proteins, was quantified using an Analyst HT microplate reader (Molecular Devices). To validate the assay, the MV fusion inhibitor AS-48 (35) was added in an otherwise identical setting, and z′ values were calculated according to the following formula: z′ = 1 − (3SDC + 3SDB)/(meanC − meanB), where C is the control and B is the background (54). As a first-pass test to exclude false-positive compounds, cytotoxicity was assessed microscopically for all wells that showed low fluorescence intensity and, for selected compounds, photodocumented at a magnification of ×200. The compound library used was a diversity set purchased by the Emory Chemical Biology Discovery Center from ChemDiv (San Diego, CA).
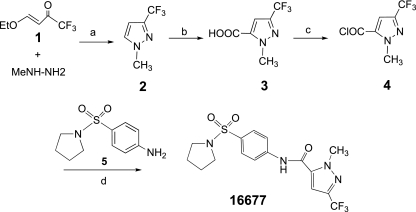
Chemical synthesis of compound 16677.
For synthesis of compound 16677, 1-methyl-3-trifluoromethyl-5-pyrazolecarboxylic acid (see Fig. 4, step 3) was prepared from commercially available compound 1 (see Fig. 4), as previously described (39). Compound 3 (see Fig. 4) (820 mg; 4.2 mmol) in dichloromethane (10 ml) was treated with oxalyl chloride (2.0 M in CH2Cl2; 8.5 mmol; 4.2 ml) and a catalytic amount of dimethylformamide. The reaction mixture was incubated at room temperature for 5 h. Evaporation of solvent delivered yellow acyl chloride (see Fig. 4, step 4) in a quantitative yield. A portion of the latter in dichloromethane (0.55 mmol) was added to a cold solution of 4-amino-prolidinyl sulfonamide (see Fig. 4, step 5) (113.1 mg; 0.5 mmol) and pyridine (48 μl; 0.6 mmol) in dichloromethane (2 ml). The reaction mixture was warmed to room temperature (18 h), poured into dilute hydrochloric acid (1 N), extracted with dichloromethane (three times 15 ml), and dried over anhydrous Na2SO4. The product was purified by chromatography using hexane-ethyl acetate (3:1) to obtain compound 16677 as a white powder (110.2 mg; 55% yield). 1H nuclear magnetic resonance (400 MHz, CDCl3) δ1.76-1.80 (4H, m), 3.24-3.27(4H, m), 4.28 (3H, s), 7.75-7.77 (2H, m), 7.84-7.87 (3H, m). High resolution mass spectrometry calculated for C16H17 F3N4O3S, 402.0974, found 403.1044 (M + 1). Analysis calculated for C16H17 F3N4O3S: C, 47.76; H, 4.26; N, 13.92, found C, 47.71; H, 4.23; N, 13.81. Subsequent to hit identification, the synthetic sample was used for all experiments.
FIG. 4.
Synthesis of 1-methyl-3-(trifluoromethyl)-N-[4-(pyrrolidinylsulfonyl)-phenyl]-1H-pyrazole-5-carboxamide (16677). Reagents: (a) MeOH, reflux; (b) n-BuLi/i-Pr2NH, then CO2; (c) (COCl)2, dimethylformamide(cat.), CH2Cl2, 0°C-rt; (d) py, CH2Cl2, rt.
Quantification of compound cytotoxicity.
Two independent assays, a nonradioactive cytotoxicity assay (CytoTox 96 Non-Radioactive Cytotoxicity Assay; Promega) and a trypan blue exclusion assay, were employed to determine the cytotoxicities of compounds. For the cytotoxicity assay, 12,000 cells per well in a 96-well plate format were incubated at 37°C for 24 h in four replicates per concentration tested in the presence of a range of compound concentrations in twofold dilutions (highest, 150 μM; 300 μM for compound 16677). Conversion of a tetrazolium salt into a colored formazan product by cellular lactate dehydrogenase released into the culture supernatants was then measured at 490 nm using a Bio-Rad plate reader. Values were calculated according to the following formula: percent viability = 100 − [(experimental − background)/(maximum − background) × 100]. For the trypan blue exclusion assay, 2 × 105 cells per well were seeded in a six-well plate format and incubated at 37°C for 30 h in three replicates per concentration tested in the presence of a range of compound concentrations in fivefold dilutions (highest, 500 μM). The cells were then detached from the culture dishes, aliquots were incubated with trypan blue solution for 15 min at room temperature, and the viable cells were counted using a hemacytometer.
Dose-response inhibition curves based on virus yields.
To generate virus yield-based dose-response curves, 4 × 105 cells per well were infected in a six-well plate format with rMV-Edm, MV field isolates, CDV, or human parainfluenza virus type 2 (hPIV2) as specified at an MOI of 0.1 PFU/cell in the presence of a range of compound concentrations in twofold dilutions (highest, 75 μM) or equivalent volumes of solvent (DMSO) only and incubated in the presence of compound at 37°C. For assessment of clinical MV isolates, compound was added in threefold dilutions (highest, 37.5 μM). Thirty-six hours postinfection, cell-associated viral particles were harvested and titered as described above. Plotting virus titers as a function of the compound concentration allowed the calculation of 50% effective concentrations (EC50s), at which virus yields are 50% of DMSO-treated controls.
Compound specificity.
To determine compound specificity, 4 × 105 cells in a six-well plate format were infected in serum-free growth medium at an MOI of 0.1 or 0.5 PFU/cell, as specified, in the presence of a range of 16677 concentrations in fourfold dilutions (highest, 12.5 μM). Bovine serum albumin (BSA) was added to some samples at a final concentration of 10 mg/ml. When virus-induced cytopathicity in DMSO control samples reached approximately 75%, the complete series was harvested, titers of cell-associated viral particles were determined by TCID50 titration, and EC50s were calculated for each series.
Compound stability.
To assess compound stability under physiological conditions, inhibitor 16677 was dissolved in growth medium (15 μM final concentration) and incubated at 37°C and the physiological pH for different time intervals (longest, 24 h). Control samples contained equal amounts of DMSO and were likewise incubated for 24 h. Subsequent to preincubation, MV was added to the compound aliquots, and the mixtures were transferred to 4 × 105 target cells seeded in a six-well plate format (resulting MOI, 0.1 PFU/cell). Thirty-six hours postinfection, cell-associated viral particles were harvested and virus titers were determined by TCID50 titration.
Transient fusion-inhibition assays.
To assess the ability of compound 16677 to inhibit cell-to-cell fusion induced by transiently expressed MV glycoproteins, a previously established assay was employed (35). Briefly, 6 × 105 cells per well were transfected in a 6-well plate format with 4 μg plasmid DNA each encoding MV H and F genes, and the cells were transferred 4 h posttransfection to 96-well plates containing compound 16677 or AS-48 in a range of concentrations in twofold dilutions (highest, 150 μM). Fusion activity was assessed microscopically 48 h posttransfection, and the extent of cytotoxicity as a consequence of extensive syncytium formation was quantified according to the following formula, using the cytotoxicity assay described above: percent cytotoxicity = (experimental − background)/(maximum − background) × 100. For some experiments, the cells were photodocumented 24 h posttransfection.
Dissociation assays.
Viral particles (8 × 104 PFU, equivalent to an MOI of 0.1 PFU/cell) were mixed with compound 16677 (final concentration, 15 μM) dissolved in 3 ml PBS. After a 10-minute incubation at 37°C to allow compound binding, the sample was divided into two aliquots (1.5 ml each) that were either subjected to dialysis against PBS (cutoff, 75 kDa; dilution factor, 100,000×; 4°C; 10 h) or incubated for 10 h at 4°C without dialysis. A control sample was treated with DMSO only and subjected to dialysis. All samples were then transferred to 4 × 105 target cells seeded in a six-well plate, cell-associated viral particles were harvested 36 h postinfection, and viral titers were determined by TCID50 titration.
Virus entry assays.
For entry experiments, viral particles (MOI = 0.5 PFU/cell) were adsorbed to 4 × 105 target cells in a six-well plate format at 4°C in the presence of 10 μM 16677 or an equal amount of DMSO for 1 hour. The cells were then shifted to 37°C for 30 min, followed by inactivation of adsorbed extracellular virions by a 2-minute acid treatment (40 mM sodium citrate, 10 mM KCl, 135 mM NaCl, pH 3.0) at 25°C as previously described (33, 35, 36). Subsequent incubation for 30 h at 37°C in the presence or absence of 10 μM 16677, as specified, was followed by determination of cell-associated virus titers by TCID50 titration.
Time of compound addition assays.
Cells (3 × 105/well in a 12-well plate format) were infected with MV at an MOI of 1.0 PFU/cell, and compound 16677 (final concentration, 15 μM) or AS-48 (final concentration, 75 μM) was added at the indicated time points. Control cells were infected in the presence of equal amounts of DMSO. Twenty hours postinfection, when virus-induced cytopathicity exceeded 90%, cell-associated viral particles were harvested and subjected to TCID50 titration.
Minireplicon assays.
BSR T7/5 cells (5 × 105 per well in a six-well plate format) were transfected with plasmid DNA encoding MV L (0.24 μg), MV N (0.94 μg), or MV P (0.29 μg) and 2 μg of the MV chloramphenicol acetyltransferase (CAT) minigenome reporter plasmid (42). For analysis of Nipah virus polymerase activity, cells were transfected with plasmid DNAs encoding Nipah virus L (0.4 μg), N (1.25 μg), or P (0.8 μg) protein and 3.5 μg of the Nipah CAT reporter plasmid as previously described (20). Control wells included identical amounts of reporter and helper plasmids but lacked the plasmids harboring the respective L gene. Two hours posttransfection, compound 16677 was added in a range of concentrations in threefold dilutions (highest, 30 μM), while some wells received compound AS-48 or equal amounts of DMSO for comparison. Thirty-eight hours posttransfection, the cells were lysed and CAT concentrations in the lysates were determined using a CAT-enzyme-linked immunosorbent assay system (Roche).
In vitro protein transcription/translation.
Rabbit reticulocyte lysates were mixed with 0.5 μg plasmid DNA encoding MV F under the control of the T7 promoter (pT7-MV F), 20 μCi [35S]methionine, and compound 16677 (final concentration, 50 μM) or an equal volume of DMSO. Samples were incubated at 30°C for 90 min, mixed with urea buffer (200 mM Tris, pH 6.8, 8 M urea, 5% sodium dodecyl sulfate, 0.1 mM EDTA, 0.03% bromphenol blue, 1.5% dithiothreitol), and fractionated on 12% polyacrylamide gels. The dried gels were exposed to XAR films.
RESULTS
Development of a primary assay suitable for automated screening of MV antivirals.
To identify novel MV inhibitor candidates, we have developed a protocol for the automated screening of compound libraries for their activities against live MV. The assay relies on rMV-eGFP (15, 17) and detects eGFP-mediated fluorescence as an indicator for expression of the viral genome. Due to the positioning of the eGFP-encoding transcription unit in the viral genome, every infectious cycle results in eGFP synthesis prior to expression of the viral proteins. Consequently, compounds that interfere with vial entry or viral-gene expression should predominantly be identified. To exclude compounds with substantial cytotoxicity prior to secondary assays, cell viability was determined microscopically in a second step for all wells in which the automated system detected low fluorescence intensities.
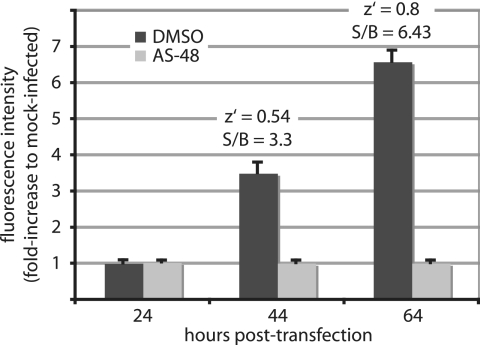
When employing the MV entry inhibitor AS-48 to evaluate this assay, we observed a time-dependent increase in signal intensity, reflecting virus growth and genome expression. At 64 h postinfection, the signal-to-background ratio reached 6.43 and the overall z′ value was 0.8 (Fig. 1). Robust screening protocols typically yield z′ values from 0.5 to 1, indicating the broad suitability of an assay for automated screening (54). These results therefore recommended our protocol as a suitable strategy for HTS-based hit identification.
FIG. 1.
Development of a cell-based assay for the automated identification of MV inhibitors. Cells seeded in microtiter plates were infected at an MOI of 0.25 PFU/cell with rMV-eGFP, and the fluorescence intensity was determined at the indicated times postinfection. For a control, cells were infected with rMV-eGFP in the presence of 37.5 μM AS-48, an MV fusion inhibitor. Average values of four replicates are shown; the error bars represent standard deviations. z′ = 1 − (3SDC + 3SDB)/(meanC − meanB), where C is the control and B is the background; S/B, signal to background ratio.
Library screening yields four sulfonamide hit candidates.
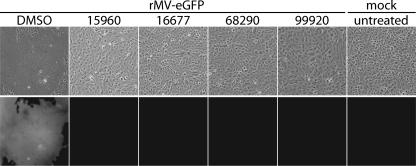
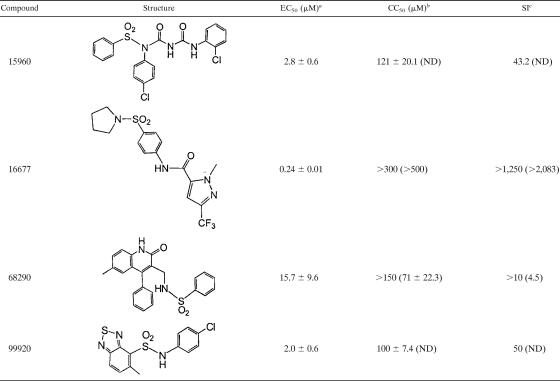
We applied this assay to the automated screening of 34,000 compounds. The process yielded 12 potential hits that reduced GFP fluorescence to background levels and showed low cytotoxicity when wells were microscopically assessed. Retesting of these molecules in the same assay reduced the number of reproducible hits to four compounds (Fig. 2), equivalent to a hit identification rate of approximately 0.01%. The chemical structures of the four hit compounds are shown in Table 1. While they represent different structural classes overall, it is remarkable that each of these compounds incorporates a sulfonamide unit. The compounds were not found in searches within SciFinder Scholar (http://www.cas.org/SCIFINDER/SCHOLAR) and PubChem (http://pubchem.ncbi.nlm.nih.gov), however, revealing that this is likely the first report of biological activity associated with them.
FIG. 2.
HTS identifies four hit candidates with potent anti-MV properties. Shown are phase-contrast and fluorescence micrographs of cells infected with rMV-eGFP in the presence of 25 μM compound. Controls included cells infected with rMV-eGFP in the presence of equal volumes of DMSO and mock-infected cells. Representative fields of view are shown.
TABLE 1.
Structures of confirmed HTS hits and summary of secondary-assay results
a Concentration at which a 50% reduction of virus yields (MV-Edm strain) was observed. Averages and standard deviations (SDs) are based on three experiments.
b Determined for Vero cells using a CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega). Averages and SDs are based on four experiments. The highest concentrations examined were 150 μM for compounds 15960, 68290, and 99920 and 300 μM for compound 16677. The values in parentheses reflect a 50% reduction in the number of viable cells after a 30-hour incubation in the presence of compound, determined using a trypan blue exclusion assay. Averages and SDs are based on three experiments. ND, not determined.
c SI, selectivity index (CC50/EC50). The values in parentheses are based on CC50s obtained using the trypan blue exclusion assay.
Secondary confirmatory assays identified compound 16677 as the most potent hit.
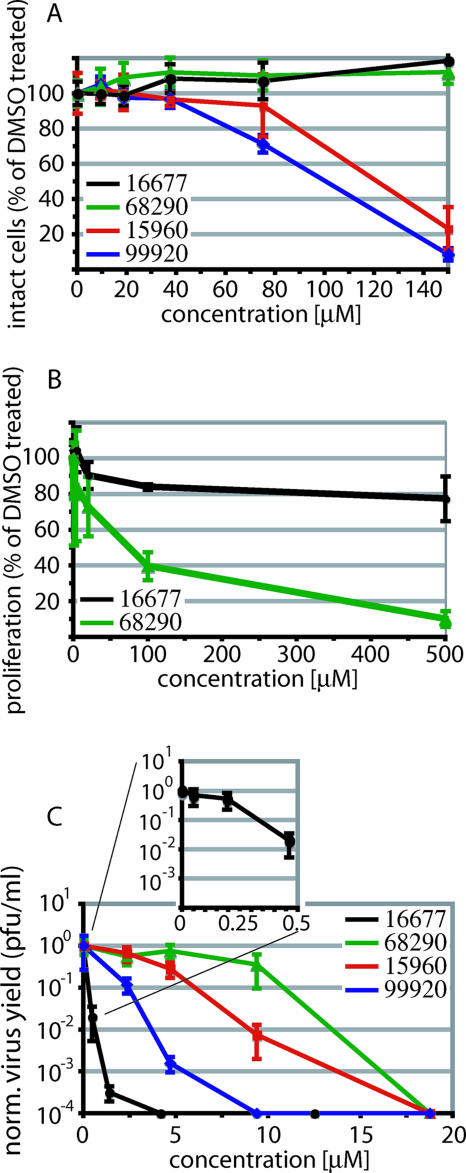
To compare antiviral activities and assess whether these compounds represent true hits that warrant hit-to-lead development, each was subjected to independent secondary assays. We first quantified the inherent cytotoxicities of the four compounds using a colorimetric cytotoxicity assay (1). Candidates 99920 and 15960 revealed 50% cytotoxic concentrations (CC50s) of 100 and 121 μM, respectively, while compounds 68290 and 16677 showed no detectable toxicity at 150 μM and 300 μM, respectively, the highest concentrations examined (Fig. 3A and Table 1).
FIG. 3.
Manual secondary assays confirm anti-MV activities of the four hit candidates. (A) Quantification of the extent of chemical lysis of cells incubated in the presence of compound. The values reflect the percentages of signal intensity compared with cells incubated in the presence of DMSO. Averages of four replicates are shown, and the error bars represent standard deviations (SDs). (B) Quantification of proliferation activities of cells incubated in the presence of compound. The number of live cells was determined 30 h after compound addition. The values indicate the percentages of live cells compared with DMSO-treated controls. Averages of three experiments are shown, and the error bars represent SDs. (C) Virus yield assay to determine the reduction of virus loads. Cells were infected with MV-Edm in the presence of different compound concentrations, and titers of cell-associated viral particles were determined by TCID50 titration 36 h postinfection. Titers were normalized for DMSO-treated control infections. Average values of two experiments are shown, and the error bars represent standard errors of the mean.
These experiments were complemented for compounds 68290 and 16677 by an independent trypan blue exclusion assay that determined the effects of the inhibitor candidates on cell proliferation rates and viabilities. For compound 16677, the number of live cells was reduced by only 20% after incubation in the presence of compound concentrations as high as 500 μM, while for compound 68290, a 50% reduction was observed at approximately 71 μM (Fig. 3B and Table 1).
Since the CC50s of all candidate compounds substantially exceeded the concentrations used in the primary HTS assay (25 μM), all four were subjected to a confirmatory assay that measured the reduction of virus yields by the inhibitor. Using as the viral target rMV-Edm, which was also employed for the original HTS screen, compound 16677 was found to have the greatest antiviral potency, with an EC50 of approximately 240 nM (Fig. 3C and Table 1). The EC50s for the other three candidates are summarized in Table 1.
Taken together, these secondary assays recommended compound 16677 as a particularly promising candidate MV inhibitor. Since compounds incorporated in HTS libraries, particularly those dissolved in DMSO, are known to degrade and/or rearrange upon prolonged storage (45), the integrity of 16677 was confirmed by chemical synthesis. The synthesis strategy is summarized in Fig. 4. Pure synthetic 16677 proved highly effective against MV and was used for all experiments subsequent to hit identification.
Compound 16677 is a target-specific MV inhibitor.
To examine the protein binding and viral specificity of candidate 16677, we first subjected the compound to two assays that were reportedly suitable to determine nonspecific protein binding (29, 30). For these and all following experiments, the MV field isolate Alaska (MV-Alaska) was employed, since this strain was found to be highly sensitive to 16677 (see below).
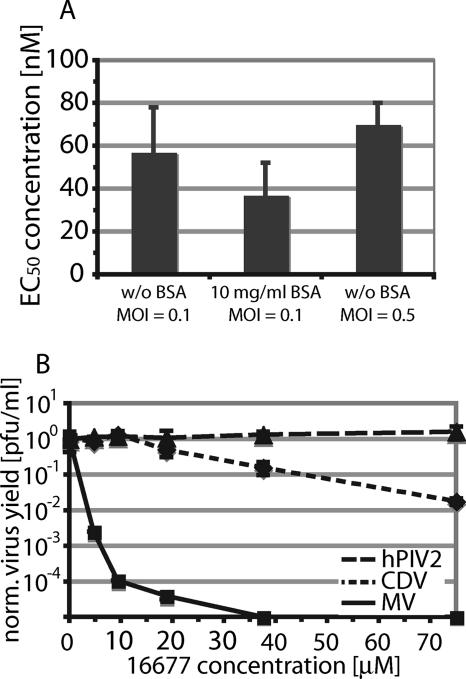
The first assay measured the effect of additional inert protein (BSA) on compound activity, and the second monitored the outcome of an increase in the amount of available target. BSA is thought to compete with the target protein for nonspecific binding to compound, while an increase in the number of target molecules would quickly saturate the absorption capacity of compound aggregates. In either case, a sharp drop in activity reflected by increased EC50s would result, while the EC50s of well-behaved inhibitors would be essentially unaffected. When EC50s were determined for 16677 after incubation of infected cells in the presence and absence of 10 mg/ml BSA or after infection with a fivefold-higher virus inoculum, no significant differences were observed (Fig. 5A), indicating little nonspecific protein binding.
FIG. 5.
Compound 16677 is a well-behaved, target-specific MV inhibitor. (A) Unrelated protein material or variation of the amount of target molecules does not affect 16677 activity. Virus yield-based dose-response curves as described in the legend to Fig. 4B were generated for 16677 upon infection of cells with MV-Alaska in the absence of BSA, in the presence of BSA, and after infection with a fivefold-higher virus inoculum. The values represent average EC50s of two experiments, and the error bars represent standard errors of the mean (SEMs). (B) 16677 is highly MV specific. Virus yield-based dose-response curves were generated for MV, the closely related CDV, and the more distantly related hPIV2. The titers were normalized for DMSO-treated control infections to facilitate comparison of different experiments. Average values of two experiments are shown, and the error bars represent SEMs.
Assessment of activities of 16677 against two related members of the paramyxovirus family, CDV and hPIV2, corroborated these findings. Like MV, CDV belongs to the genus Morbillivirus, and the two viruses share approximately 61% protein identity, while hPIV2 is more distantly related and shows only 21% protein identity with MV. When dose-response curves were generated for these viruses, compound 16677 yielded an EC50 of 28 μM against CDV and was not inhibitory against hPIV2 at 75 μM, the highest concentration examined (Fig. 5B). These data thus demonstrate high specificity of compound 16677 for MV and to a lesser degree for the intimately related CDV and argue against a nonspecific mechanism of activity.
Inhibitory activity is stable under physiological conditions, and 16677 binding is reversible.
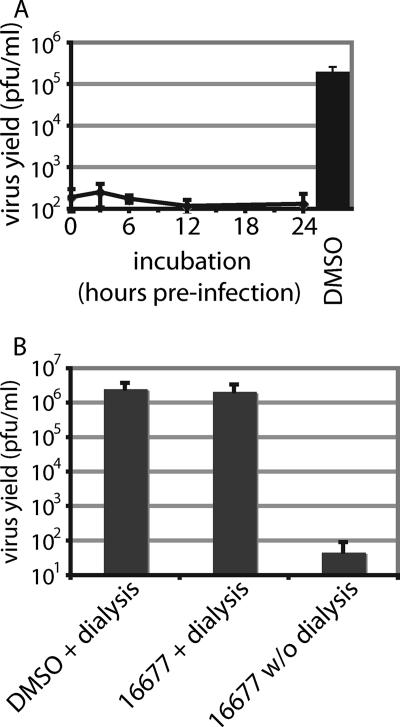
A long half-life of antiviral activity under physiological conditions and absence of chemical reactivity are desirable properties of inhibitor candidates. To assess its stability under physiological conditions, 16677 was preincubated in cell culture medium at 37°C for different periods ranging from 0 to 24 h, followed by mixing with MV-Alaska and infection of target cells. While virus grew efficiently in solvent-only-treated control infections, no loss in antiviral activity of 16677 could be detected even after preincubation for 24 h (Fig. 6A), indicating a favorable stability profile for the compound.
FIG. 6.
Compound 16677 is stable under physiological conditions, and inhibition is reversible. (A) Compound dilutions in growth medium (final concentration, 15 μM) were preincubated at 37°C for the indicated periods, followed by transfer to cells and infection with MV-Alaska. For the control (untreated), equal dilutions of DMSO in growth medium were preincubated for 24 h. The values represent titers of cell-associated viral particles determined 36 h postinfection through TCID50 titration. Averages of two experiments are shown, and the error bars represent standard errors of the mean (SEMs). (B) Removal of 16677 through dialysis fully restores viral replication. Virus dilutions (equivalents of 0.1 MOI) were mixed with 16677 (final concentration, 15 μM) and subjected to dialysis against PBS. The cells were subsequently infected with the mixtures, and viral titers were determined 36 h postinfection by TCID50 titration. Controls include DMSO-treated virions and compound-treated samples maintained under the same conditions without (w/o) dialysis. Averages of two experiments are shown, and the error bars represent SEMs.
To explore whether 16677 chemically reacts with its target or whether compound docking is reversible, MV aliquots were incubated with the compound in the absence of target cells for 10 min at 37°C, followed by dialysis at 4°C with a molecular mass cutoff of 75 kDa, which ensured free diffusion of the compound but not the viral particles (total dilution factor of the compound, 100,000-fold). Controls included solvent-only-treated particles that were similarly subjected to dialysis and 16677-treated virus samples that were, instead of being dialyzed, held at 4°C for the same period. Subsequent infection of cells with the different virus samples and titration of infectious particles produced 30 h postinfection revealed that the inhibitory activity of 16677 is ablated by dialysis prior to infection (Fig. 6B), indicating that compound docking is reversible and is not based on a chemical reaction of the inhibitor with the target.
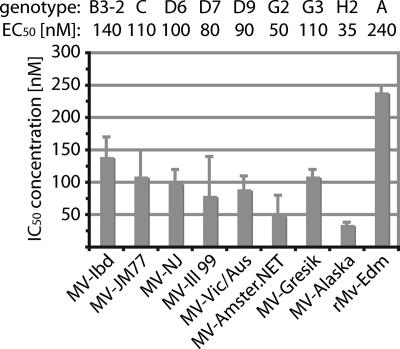
Potent activity of 16677 against primary MV isolates, including an isolate resistant to the MV entry inhibitor AS-48.
To explore the potential value of candidate 16677 as a clinically relevant inhibitor of MV, we assessed its activities against a panel of primary MV isolates that represented several genotypes circulating worldwide (see Table S2 in the supplemental material). This panel included the sub-Saharan isolate of genotype B3, MV-Ibd, which we previously found to be resistant to our series of MV entry inhibitors (14). Depending on the genotype of the MV strain analyzed, we calculated EC50s ranging from 35 to 140 nM for these isolates (Fig. 7). MV-Ibd was likewise inhibited by 16677, indicating that no cross-resistance exists between this compound class and the previously characterized MV entry inhibitors. All primary isolates tested were also more sensitive to compound 16677 than was the recombinant MV-Edm virus (genotype A), on which the original hit discovery was based. Viruses of genotype A have not been endemic in several decades, underscoring the therapeutic potential of the inhibitor class represented by 16677.
FIG. 7.
16677 is active against a panel of clinical MV isolates. Dose-response curves were generated on the basis of virus yields determined by TCID50 titration, and EC50s were calculated. Viral genotypes are given above the graph. Averages of two experiments are shown, and the error bars represent standard errors of the mean (SEMs). For viruses MV-Alaska and rMV-Edm, the EC50s are based on three experiments and the error bars represent standard deviations.
Compound 16677 targets a postentry step in the viral life cycle.
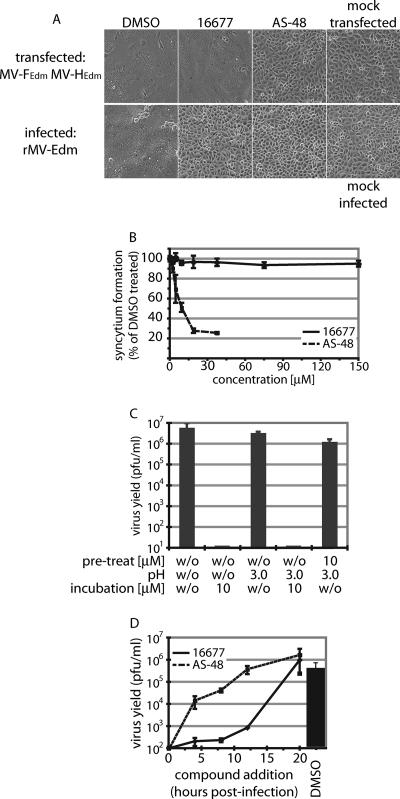
To gain insight into the mechanism of 16677 antiviral activity, we subjected the compound to an initial mechanistic characterization. We first addressed the question of whether membrane fusion, and hence viral entry, is inhibited by the compound. In the presence and absence of 16677, cells were transiently transfected with expression plasmids encoding the MV H and F envelope glycoproteins, and membrane fusion activity was examined microscopically. Controls included transfected cells treated with the lead entry inhibitor AS-48 and 16677-treated cells that were infected with MV rather than plasmid transfected. In contrast to the strong inhibitory effect of AS-48, cell-to-cell fusion mediated by transiently expressed MV H and F proteins was uninhibited by 16677 (Fig. 8A). However, virus-mediated cytopathicity was fully suppressed by the compound, confirming the specificity of the assay. Quantification of the envelope glycoprotein-induced cytopathicity and generation of dose-response curves for both 16677 and AS-48 confirmed these microscopic observations, since MV H and F protein-mediated cell-to-cell fusion was uninhibited even at 16677 concentrations of 150 μM (Fig. 8B).
FIG. 8.
16677 does not interfere with viral entry and is active late in the replicative cycle. (A) Cell-to-cell fusion induced by transiently expressed MV glycoproteins is not inhibited by the compound. Transfected cells, treated with 15 μM 16677 or DMSO, were photographed after a 24-hour incubation period. For comparison, cells treated with the fusion inhibitor AS-48, cells infected with MV, and mock-transfected/infected cells are shown. (B) Quantification of syncytium formation of transfected cells treated as described for panel A. The values represent averages of four replicates and are expressed as percentages of the syncytium formation observed for DMSO-treated samples. The error bars represent standard deviations. (C) Presence of 16677 during adsorption of virions to target cells does not render particles sensitive to neutralization by a pH 3.0 wash, indicating successful viral entry in the presence of compound. Virions were adsorbed to target cells in the presence of DMSO or 16677, followed by pH 3.0 treatment to neutralize all particles that were in a premembrane fusion state and incubation in the presence of DMSO or 16677 as indicated. Yields of cell-associated viral particles were determined by TCID50 titration. Average values of two experiments are shown, and the error bars represent standard errors of the mean (SEMs). (D) Antiviral activity of 16677 persists to late phases of a replicative cycle. The compound (final concentration, 15 μM) was added at the indicated times postinfection to a one-step replicative cycle. Cell-associated viral particles of compound-treated samples and DMSO-treated controls were harvested 20 h postinfection, and virus yields were determined by TCID50 titration. For comparison, infected cells were treated with the fusion inhibitor AS-48. Average titers of two experiments are shown, and the error bars represent SEMs.
A viral-entry assay was employed to assess whether the transient-expression assay accurately reflected the conditions of virus infection. As demonstrated for several enveloped viruses (24), the infectivity of MV particles is completely ablated by a brief pH 3.0 wash (35, 36). Capitalizing on this, we adsorbed MV particles (equivalent to an MOI of 0.5 PFU/cell) to target cells at 4°C in the presence or absence of 16677, followed by removal of unbound virus and a 30-minute incubation period at 37°C to allow viral entry to proceed. Subsequently, particles that were in a premembrane fusion state were neutralized through a pH 3.0 wash, followed by incubation with or without compound and determination of virus yields. If 16677 interfered with viral entry, particle absorption in the presence of compound followed by low-pH treatment should result in a sharp drop in virus yields, as exemplified by the previous analysis of members of the AS-48 entry inhibitor class in this assay (35, 36). However, virus yields were only slightly affected by the pH 3.0 wash when 16677-treated samples were compared to DMSO-treated cells (Fig. 8C). Substantial reduction of virus yields by 16677 was observed only when the compound was present during the incubation period subsequent to the wash step, confirming its effectiveness (Fig. 8C).
To specify the time interval postentry in which the virus remained sensitive to inhibition by 16677, the effects of compound administration on replication efficiency at different stages of the viral life cycle were assessed. For this assay, viral growth was synchronized by infection of cells with MV at an MOI of 1.0 PFU/cell, and all samples were harvested 20 h postinfection, thus essentially excluding secondary infections. Even when added 12 h postinfection, 16677 caused an approximately 99.9% reduction in virus yields, thus demonstrating high antiviral effectiveness (Fig. 8D). This was in contrast to the entry inhibitor AS-48, which, as expected, had to be present at the time of infection to achieve full inhibition. Taken together, these findings demonstrate that 16677 does not interfere with the viral entry machinery and point to a postentry step as its mechanism of action.
Compound 16677 inhibits RdRp complex activity.
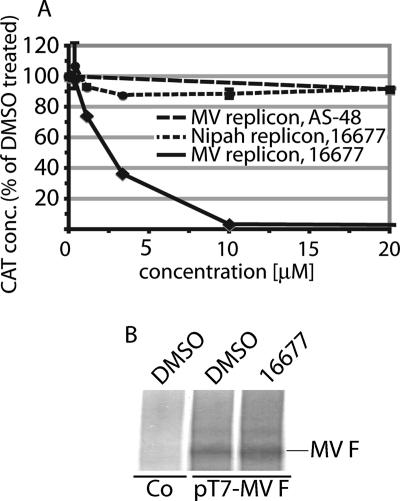
Our initial HTS protocol does not favor the identification of inhibitors of particle assembly or release. This suggests inhibition of the RdRp machinery as a likely mechanism of antiviral activity of 16677. To evaluate the functionality of the polymerase complex, a plasmid-based subinfection MV minireplicon reporter assay under the control of the T7 promoter was employed (42). In this assay, the amount of CAT reporter produced is proportional to the activity of the MV polymerase complex. Compound 16677 demonstrated a strong dose-dependent inhibition of CAT expression when its effect on minireplicon activity was examined (Fig. 9A). This was in contrast to the essentially unchanged CAT levels found in control samples that were treated with the entry inhibitor AS-48. The inhibitory activity of 16677 was specific for the MV minireplicon, since the compound had no effect on a comparable minireplicon that was derived from the related Nipah virus (20). Nipah virus belongs to the genus Henipavirus within the paramyxovirus family (5), which is related to the genus Morbillivirus and shares approximately 33% overall protein identify with MV.
FIG. 9.
16677 specifically inhibits MV RdRp complex activity. (A) A CAT reporter-based minireplicon assay demonstrates inhibition of MV polymerase complex activity by 16677. BSR T7/5 cells cotransfected with the minireplicon plasmid and plasmids encoding MV nucleoprotein (N), phosphoprotein (P) and polymerase (L), all under the control of the T7 promoter, were incubated in the presence of 16677 or the fusion inhibitor AS-48, followed by lysis and assessment of CAT concentrations (conc.). Values were determined in quadruplicate and are expressed as percentages of DMSO-treated controls. A comparable minireplicon system derived from Nipah virus, a related member of the paramyxovirus family, was not inhibited by 16677. (B) T7 polymerase is fully active in the presence of 16677. Shown is in vitro MV F transcription/translation in the presence of 50 μM 16677 or an equal volume of DMSO. Controls (Co) included DMSO and a variant of plasmid pT7-MV F that harbors the F-encoding gene in reverse orientation to the T7 promoter.
The specificity of the minireplicon assay was further confirmed when an effect of 16677 on the cellular transcription/translation machinery or the T7 polymerase function was assessed in a cell-free in vitro transcription/translation assay of MV F. Equal amounts of F protein were detected in treated and untreated samples (Fig. 9B), indicating that cellular protein biosynthesis was unimpaired by the compound. Taken together, these findings argue against interference of 16677 with the cellular protein biosynthesis machinery or T7 polymerase function as an alternative explanation for the reduction of reporter expression in the MV minireplicon assay. Our data thus highlight 16677 as a first-in-class compound of nonnucleoside inhibitors of MV RdRp complex activity.
DISCUSSION
With the goal of developing novel therapeutics against MV, we have established a robust protocol for the automated identification of MV inhibitors. Our hit identification strategy combines an automated screen with several confirmatory assays that manually assess cytotoxicity, antiviral activity, and compound structure. A screen of 34,000 compounds from a library of the Emory Chemical Biology Discovery Center has yielded four confirmed hits, providing proof of concept for the validity of this assay for the discovery of MV inhibitors.
All four hit compounds harbor a sulfonamide group, although chemically they belong to different structural classes. Future testing will reveal whether this has a specific molecular cause or merely reflects a bias of the compound library assessed in this screening exercise. Comparative structural analysis of all four hit compounds may also provide valuable information for the future generation of structure-activity relationships for hit-to-lead development.
The identification of promising small-molecule inhibitors can be hampered by promiscuous compounds that frequently emerge as putative hits (29, 30). Rather than docking to defined target areas, promiscuous compounds are thought to act nonspecifically through adsorption or absorption of target structures to larger compound aggregates. Nonspecific and noncompetitive binding ultimately leads to flat structure-activity relationships, typically in the low micromolar range, that usually render chemical efforts to improve biological activity futile (29, 30). Our observation that the addition of a large amount of BSA and an increase in the number of infectious particles does not affect the EC50 determined for our most potent hit compound, 16677, recommends this compound as a well-behaved inhibitor of MV. This is further confirmed by its high target specificity. Yields of hPIV2, a distantly related paramyxovirus, are unaffected by 16677, and CDV, a closely related member of the same genus as MV, is mildly inhibited. Selective inhibition of MV also corroborates the results of our cytotoxicity assays, since it argues against general interference with cellular functions, which would likely be nonspecific, as the underlying mechanism of antiviral activity of the compound.
Prior to any future hit-to-lead development efforts, compound 16677 shows activity in the low nanomolar range against a panel of MV field isolates, thus underscoring the therapeutic potential of the compound. Importantly, the panel of viruses examined includes the MV-Ibd isolate of genotype B3, which we have previously found to be naturally resistant to the AS-48 MV entry inhibitor class (14). The lack of cross-resistance of 16677 with the entry inhibitors suggests a different mechanism of antiviral activity for 16677 and opens potential avenues to counteract spontaneous viral resistance that may develop in the field. While we have not observed an increase in antiviral activity when 16677 and AS-48 were combined in vitro (data not shown), combined administration of inhibitors with different target structures, in analogy to the clinical experience with highly active antiretroviral therapy (3, 4, 31), has the potential to reduce the frequency with which resistant virions emerge in vivo and/or lower the overall fitness of viral variants with multiple resistances.
Consistent with the absence of cross-resistance with the MV fusion inhibitors and in contrast to the findings obtained for AS-48, compound 16677 does not prevent cell-to-cell fusion mediated by plasmid-encoded H and F MV glycoproteins and shows no inhibitory activity in an MV entry assay. Time-of-addition assays demonstrated that 16677 potently inhibits MV even when added late in the replicative cycle. These observations suggest interference with RdRp complex function as the mechanism of 16677 activity, since inhibitors acting downstream of RdRp function are not expected to interfere with eGFP synthesis in rMV-eGFP-infected cells. A minigenome reporter assay that monitors the activity of the viral RNA polymerase complex confirmed this hypothesis, since it demonstrated dose-dependent inhibition of MV minigenome expression. The fact that an analogous minigenome assay established for Nipah virus, a related member of the paramyxovirus family, was not sensitive to 16677 underscores the target specificity of the inhibitor. The EC50s of 16677 were on average 25 times higher in the minigenome assay than against the different MV isolates. It is tempting to speculate that this could reflect the approximately 24-fold difference in length between the reporter gene and the viral genome. Longer template sequences, requiring more polymerization cycles, may increase the likelihood of productive inhibitor binding when compound concentrations are low and thus result in the lower EC50s determined for 16677 against live virus.
Our functional characterization of HTS hit 16677 has revealed that the compound represents a first in class of novel, highly potent nonnucleoside inhibitors of MV RdRp complex activity. Provided that these compounds exhibit favorable safety and pK profiles, members of this inhibitor class may be suitable for the treatment of persistent MV infections. Viruses isolated from subacute sclerosing panencephalitis patients typically show defects in their envelope proteins (19, 22), which may render entry inhibitors ineffective. While several nucleoside inhibitors of MV have been reported previously, on average these show at least 10- to 100-fold lower potency than 16677, with EC50s typically in the micromolar range, and none has progressed to human trials (2). Nonnucleoside inhibitors of viral polymerases have, however, proven highly beneficial in antiretroviral therapy (11, 28, 34, 48).
At present, it is not known whether 16677 binds directly to the L polymerase, docks to one of the polymerase cofactors N or P, or interferes with the interaction of these proteins with each other, with a cellular cofactor specific for MV, or with the RNA template. Generation of resistant viral variants through stepwise adaptation, followed by mapping of characteristic sites of resistance, will guide an in-depth molecular characterization of the compound's mechanism of activity. Combined with a ligand-based structure-activity relationship, this information should constitute a viable platform for educated further lead optimization.
Conceptually, our MV-based findings may be adaptable to other clinically relevant members of the paramyxovirus family, such as the recently emerged, highly pathogenic henipaviruses (16, 49). In this scenario, they prepare the path for a better mechanistic understanding of these viruses and the development of novel therapeutic strategies against pathogens for which no vaccines are currently available.
Supplementary Material
Acknowledgments
We thank P. A. Rota for generously providing the MV and Nipah virus replicon systems, Y. Yanagi for Vero-dogSLAM cells, N. Chandrakumar for help with compound synthesis, D. Li for technical assistance with HTS, the Emory Chemical Biology Discovery Center for the use of its screening system, and A. L. Hammond for critical reading of the manuscript.
This work was supported by a research grant from the American Lung Association and Public Health Service grants AI056179 and AI071002 from NIH/NIAID (to R.K.P.).
Footnotes
Published ahead of print on 30 April 2007.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Barltrop, J. A., T. C. Owen, A. H. Cory, and J. G. Cory. 1991. 5-(3-carboxymethoxyphenyl)-2-(4,5-dimethylthiazolyl)-3-(4-sulfophenyl)tetrazolium, inner salt (MTS) and related analogs of 3-(4,5-dimethylthiazolyl)-2,5-diphenyltetrazolium bromide (MTT) reducing to purple water-soluble formazans as cell-viability indicators. Bioorg. Med. Chem. Lett. 1611-614. [Google Scholar]
- 2.Barnard, D. L. 2004. Inhibitors of measles virus. Antivir. Chem. Chemother. 15111-119. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett, J. A., R. DeMasi, J. Quinn, C. Moxham, and F. Rousseau. 2001. Overview of the effectiveness of triple combination therapy in antiretroviral-naive HIV-1 infected adults. AIDS 151369-1377. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett, J. A., M. J. Fath, R. Demasi, A. Hermes, J. Quinn, E. Mondou, and F. Rousseau. 2006. An updated systematic overview of triple combination therapy in antiretroviral-naive HIV-infected adults. AIDS 202051-2064. [DOI] [PubMed] [Google Scholar]
- 5.Bossart, K. N., L. F. Wang, M. N. Flora, K. B. Chua, S. K. Lam, B. T. Eaton, and C. C. Broder. 2002. Membrane fusion tropism and heterotypic functional activities of the Nipah virus and Hendra virus envelope glycoproteins. J. Virol. 7611186-11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourhis, J. M., B. Canard, and S. Longhi. 2006. Structural disorder within the replicative complex of measles virus: functional implications. Virology 34494-110. [DOI] [PubMed] [Google Scholar]
- 7.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.CDC. 2005. Progress in reducing measles mortality—worldwide, 1999-2003. Morb. Mortal. Wkly. Rep. 54200-203. [PubMed] [Google Scholar]
- 9.Chakrabarti, S., K. E. Collingham, K. Holder, C. D. Fegan, H. Osman, and D. W. Milligan. 2001. Pre-emptive oral ribavirin therapy of paramyxovirus infections after haematopoietic stem cell transplantation: a pilot study. Bone Marrow Transplant. 28759-763. [DOI] [PubMed] [Google Scholar]
- 10.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure 9255-266. [DOI] [PubMed] [Google Scholar]
- 11.Chou, R., R. Fu, L. H. Huffman, and P. T. Korthuis. 2006. Initial highly-active antiretroviral therapy with a protease inhibitor versus a non-nucleoside reverse transcriptase inhibitor: discrepancies between direct and indirect meta-analyses. Lancet 3681503-1515. [DOI] [PubMed] [Google Scholar]
- 12.Crennell, S., T. Takimoto, A. Portner, and G. Taylor. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 71068-1074. [DOI] [PubMed] [Google Scholar]
- 13.Dorig, R. E., A. Marcil, A. Chopra, and C. D. Richardson. 1993. The human CD46 molecule is a receptor for measles virus (Edmonston strain). Cell 75295-305. [DOI] [PubMed] [Google Scholar]
- 14.Doyle, J., A. Prussia, L. K. White, A. Sun, D. C. Liotta, J. P. Snyder, R. W. Compans, and R. K. Plemper. 2006. Two domains that control prefusion stability and transport competence of the measles virus fusion protein. J. Virol. 801524-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duprex, W. P., S. McQuaid, L. Hangartner, M. A. Billeter, and B. K. Rima. 1999. Observation of measles virus cell-to-cell spread in astrocytoma cells by using a green fluorescent protein-expressing recombinant virus. J. Virol. 739568-9575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eaton, B. T., C. C. Broder, D. Middleton, and L. F. Wang. 2006. Hendra and Nipah viruses: different and dangerous. Nat. Rev. Microbiol. 423-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrengruber, M. U., S. Hennou, H. Bueler, H. Y. Naim, N. Deglon, and K. Lundstrom. 2001. Gene transfer into neurons from hippocampal slices: comparison of recombinant Semliki Forest Virus, adenovirus, adeno-associated virus, lentivirus, and measles virus. Mol. Cell Neurosci. 17855-871. [DOI] [PubMed] [Google Scholar]
- 18.Erlenhoefer, C., W. J. Wurzer, S. Loffler, S. Schneider-Schaulies, V. ter Meulen, and J. Schneider-Schaulies. 2001. CD150 (SLAM) is a receptor for measles virus but is not involved in viral contact-mediated proliferation inhibition. J. Virol. 754499-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffin, D. E. 2001. Measles virus, 4th ed., vol. 1. Lippincott, Philadelphia, PA.
- 20.Halpin, K., B. Bankamp, B. H. Harcourt, W. J. Bellini, and P. A. Rota. 2004. Nipah virus conforms to the rule of six in a minigenome replication assay. J. Gen. Virol. 85701-707. [DOI] [PubMed] [Google Scholar]
- 21.Hethcote, H. W. 2000. The mathematics of infectious disease. SIAM Rev. 42599-653. [Google Scholar]
- 22.Hilleman, M. R. 2001. Current overview of the pathogenesis and prophylaxis of measles with focus on practical implications. Vaccine 20651-665. [DOI] [PubMed] [Google Scholar]
- 23.Karlin, D., F. Ferron, B. Canard, and S. Longhi. 2003. Structural disorder and modular organization in Paramyxovirinae N and P. J. Gen. Virol. 843239-3252. [DOI] [PubMed] [Google Scholar]
- 24.Kizhatil, K., and L. M. Albritton. 1997. Requirements for different components of the host cell cytoskeleton distinguish ecotropic murine leukemia virus entry via endocytosis from entry via surface fusion. J. Virol. 717145-7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 26.Lamb, R. A., R. G. Paterson, and T. S. Jardetzky. 2006. Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology 34430-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Longhi, S., V. Receveur-Brechot, D. Karlin, K. Johansson, H. Darbon, D. Bhella, R. Yeo, S. Finet, and B. Canard. 2003. The C-terminal domain of the measles virus nucleoprotein is intrinsically disordered and folds upon binding to the C-terminal moiety of the phosphoprotein. J. Biol. Chem. 27818638-18648. [DOI] [PubMed] [Google Scholar]
- 28.MacArthur, R. D., R. M. Novak, G. Peng, L. Chen, Y. Xiang, K. H. Hullsiek, M. J. Kozal, M. van den Berg-Wolf, C. Henely, B. Schmetter, and M. Dehlinger. 2006. A comparison of three highly active antiretroviral treatment strategies consisting of non-nucleoside reverse transcriptase inhibitors, protease inhibitors, or both in the presence of nucleoside reverse transcriptase inhibitors as initial therapy (CPCRA 058 FIRST Study): a long-term randomised trial. Lancet 3682125-2135. [DOI] [PubMed] [Google Scholar]
- 29.McGovern, S. L., E. Caselli, N. Grigorieff, and B. K. Shoichet. 2002. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem. 451712-1722. [DOI] [PubMed] [Google Scholar]
- 30.McGovern, S. L., and B. K. Shoichet. 2003. Kinase inhibitors: not just for kinases anymore. J. Med. Chem. 461478-1483. [DOI] [PubMed] [Google Scholar]
- 31.Murphy, E. L., A. C. Collier, L. A. Kalish, S. F. Assmann, M. F. Para, T. P. Flanigan, P. N. Kumar, L. Mintz, F. R. Wallach, and G. J. Nemo. 2001. Highly active antiretroviral therapy decreases mortality and morbidity in patients with advanced HIV disease. Ann. Intern. Med. 13517-26. [DOI] [PubMed] [Google Scholar]
- 32.Naniche, D., G. Varior-Krishnan, F. Cervoni, T. F. Wild, B. Rossi, C. Rabourdin-Combe, and D. Gerlier. 1993. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 676025-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palokangas, H., K. Metsikko, and K. Vaananen. 1994. Active vacuolar H+ ATPase is required for both endocytic and exocytic processes during viral infection of BHK-21 cells. J. Biol. Chem. 26917577-17585. [PubMed] [Google Scholar]
- 34.Pauwels, R., K. Andries, J. Desmyter, D. Schols, M. J. Kukla, H. J. Breslin, A. Raeymaeckers, J. Van Gelder, R. Woestenborghs, J. Heykants, et al. 1990. Potent and selective inhibition of HIV-1 replication in vitro by a novel series of TIBO derivatives. Nature 343470-474. [DOI] [PubMed] [Google Scholar]
- 35.Plemper, R. K., J. Doyle, A. Sun, A. Prussia, L. T. Cheng, P. A. Rota, D. C. Liotta, J. P. Snyder, and R. W. Compans. 2005. Design of a small-molecule entry inhibitor with activity against primary measles virus strains. Antimicrob. Agents Chemother. 493755-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plemper, R. K., K. J. Erlandson, A. S. Lakdawala, A. Sun, A. Prussia, J. Boonsombat, E. Aki-Sener, I. Yalcin, I. Yildiz, O. Temiz-Arpaci, B. Tekiner, D. C. Liotta, J. P. Snyder, and R. W. Compans. 2004. A target site for template-based design of measles virus entry inhibitors. Proc. Natl. Acad. Sci. USA 1015628-5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plemper, R. K., A. L. Hammond, D. Gerlier, A. K. Fielding, and R. Cattaneo. 2002. Strength of envelope protein interaction modulates cytopathicity of measles virus. J. Virol. 765051-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plemper, R. K., A. S. Lakdawala, K. M. Gernert, J. P. Snyder, and R. W. Compans. 2003. Structural features of paramyxovirus F protein required for fusion initiation. Biochemistry 426645-6655. [DOI] [PubMed] [Google Scholar]
- 39.Schlosser, M., J. N. Volle, F. Leroux, and K. Schenk. 2002. Switchable reactivity: the site-selective functionalization of trifluoromethyl-substituted pyrazoles. Eur. J. Org. Chem. 20022913-2920. [Google Scholar]
- 40.Seki, F., N. Ono, R. Yamaguchi, and Y. Yanagi. 2003. Efficient isolation of wild strains of canine distemper virus in Vero cells expressing canine SLAM (CD150) and their adaptability to marmoset B95a cells. J. Virol. 779943-9950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shigeta, S., S. Mori, M. Baba, M. Ito, K. Honzumi, K. Nakamura, H. Oshitani, Y. Numazaki, A. Matsuda, T. Obara, et al. 1992. Antiviral activities of ribavirin, 5-ethynyl-1-β-d-ribofuranosylimidazole-4-carboxamide, and 6′-(R)-6′-C-methylneplanocin A against several ortho- and paramyxoviruses. Antimicrob. Agents Chemother. 36435-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sidhu, M. S., J. Chan, K. Kaelin, P. Spielhofer, F. Radecke, H. Schneider, M. Masurekar, P. C. Dowling, M. A. Billeter, and S. A. Udem. 1995. Rescue of synthetic measles virus minireplicons: measles genomic termini direct efficient expression and propagation of a reporter gene. Virology 208800-807. [DOI] [PubMed] [Google Scholar]
- 43.Spearman, C. 1908. The method of right and wrong cases (constant stimuli) without Gauss's formula. Br. J. Psychol. 2227-242. [Google Scholar]
- 44.Sun, A., A. Prussia, W. Zhan, E. E. Murray, J. Doyle, L. T. Cheng, J. J. Yoon, E. V. Radchenko, V. A. Palyulin, R. W. Compans, D. C. Liotta, R. K. Plemper, and J. P. Snyder. 2006. Nonpeptide inhibitors of measles virus entry. J. Med. Chem. 495080-5092. [DOI] [PubMed] [Google Scholar]
- 45.Talaga, P. 2004. Compound decomposition: a new drug discovery tool? Drug Discov. Today 951-53. [DOI] [PubMed] [Google Scholar]
- 46.Tatsuo, H., N. Ono, K. Tanaka, and Y. Yanagi. 2000. SLAM (CDw150) is a cellular receptor for measles virus. Nature 406893-897. [DOI] [PubMed] [Google Scholar]
- 47.van den Hof, S., M. A. Conyn-van Spaendonck, and J. E. van Steenbergen. 2002. Measles epidemic in the Netherlands, 1999-2000. J. Infect. Dis. 1861483-1486. [DOI] [PubMed] [Google Scholar]
- 48.van Leth, F., P. Phanuphak, K. Ruxrungtham, E. Baraldi, S. Miller, B. Gazzard, P. Cahn, U. G. Lalloo, I. P. van der Westhuizen, D. R. Malan, M. A. Johnson, B. R. Santos, F. Mulcahy, R. Wood, G. C. Levi, G. Reboredo, K. Squires, I. Cassetti, D. Petit, F. Raffi, C. Katlama, R. L. Murphy, A. Horban, J. P. Dam, E. Hassink, R. van Leeuwen, P. Robinson, F. W. Wit, and J. M. Lange. 2004. Comparison of first-line antiretroviral therapy with regimens including nevirapine, efavirenz, or both drugs, plus stavudine and lamivudine: a randomised open-label trial, the 2NN Study. Lancet 3631253-1263. [DOI] [PubMed] [Google Scholar]
- 49.Wang, L., B. H. Harcourt, M. Yu, A. Tamin, P. A. Rota, W. J. Bellini, and B. T. Eaton. 2001. Molecular biology of Hendra and Nipah viruses. Microbes Infect. 3279-287. [DOI] [PubMed] [Google Scholar]
- 50.Wolfson, L. J., P. M. Stebel, M. Gacic-Dobo, E. J. Hoekstra, J. W. McFarland, and B. S. Hersh. 2007. Has the 2005 measles mortality reduction goal been achieved? A natural history modelling study. Lancet 369191-200. [DOI] [PubMed] [Google Scholar]
- 51.Yin, H. S., R. G. Paterson, X. Wen, R. A. Lamb, and T. S. Jardetzky. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. USA 1029288-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin, H. S., X. Wen, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 43938-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan, P., T. B. Thompson, B. A. Wurzburg, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2005. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13803-815. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, J. H., T. D. Y. Chung, and K. R. Oldenburg. 1999. A simple statistical parameter for used in evaluation and validation of high throughput screening assays. J. Biomol. Screen 467-73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.