Abstract
Seven blaIMP-1-harboring Acinetobacter sp. isolates and one Pseudomonas putida clinical isolate were recovered from hospitalized patients. All isolates possessed a class 1 integron, named In86, carrying the same cassette array [blaIMP1, aac(6′)-31, and aadA1], which was plasmid located in five of the isolates. This report describes the ability of nonfermentative nosocomial pathogens to acquire and disseminate antimicrobial resistance determinants.
Metallo-β-lactamases (MβLs) and aminoglycoside-modifying enzymes (AgMEs) represent a new challenge to antimicrobial therapy of nosocomial infections, since they confer phenotypic resistance to nearly all clinically available β-lactams and aminoglycosides, respectively (1). Several MβL and aminoglycoside resistance genes in nosocomial isolates recovered from Latin American countries have been described previously (7, 8, 9); however, there is limited information regarding the dissemination of these genes in this region. In the present study, we describe a new blaIMP-1-carrying integron that contains a new aminoglycoside resistance gene and its dissemination among genetically unrelated clinical isolates recovered from a Brazilian hospital.
As part of the SENTRY Antimicrobial Surveillance Program (12), gram-negative bacilli recovered from Latin American hospitals between March 2001 and April 2003 were tested for antimicrobial susceptibility by reference methods according to standard guidelines (3, 4). Strains showing combined resistance to ceftazidime (MIC, ≥16 μg/ml), imipenem (MIC, ≥16 μg/ml), and meropenem (MIC, ≥16 μg/ml) were routinely screened for MβL genes by standards PCRs (2, 9, 11). Seven Acinetobacter sp. isolates and one Pseudomonas putida clinical isolate recovered from a 600-bed tertiary university hospital located in São Paulo, Brazil, were found to harbor blaIMP-1 and were further evaluated in the present study (Table 1).
TABLE 1.
Clinical and antimicrobial profile and molecular information from the clinical isolates harboring blaIMP-1-carrying In86 evaluated in the present study
| Parameter | Value
|
|||||||
|---|---|---|---|---|---|---|---|---|
| A. baumannii 694 | A. baumannii 696 | A. baumannii 9043 | A. baumannii 695 | A. baumannii 501 | Acinetobacter sp. strain 5227 | Acinetobacter sp. strain 5248 | P. putida 12346 | |
| Sample collection date (mo/day/yr) | 05/28/2002 | 06/04/2002 | 06/22/2001 | 06/02/2002 | 05/06/2001 | 08/14/2002 | 09/28/2002 | 07/23/2001 |
| Source | SSTIb | SSTIb | BSIc | SSTIb | BSIc | LRTId | LRTId | BSIc |
| MIC (μg/ml)a | ||||||||
| β-Lactam | ||||||||
| Aztreonam | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 |
| Ampicillin | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 |
| Amoxicillin-clavulanate | >16 | >16 | >16 | >16 | 16 | >16 | >16 | >16 |
| Aztreonam | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 |
| Ceftazidime | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 |
| Piperacillin | >128 | >128 | >128 | 128 | 32 | >128 | >128 | 64 |
| Piperacillin-tazobactam | 32 | 64 | >64 | 64 | 64 | >64 | 64 | 64 |
| Ticarcillin | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| Ticarcillin-clavulanate | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| Cefepime | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 |
| Imipenem | >8 | >8 | >8 | >8 | >8 | >8 | >8 | >8 |
| Meropenem | >8 | >8 | >8 | >8 | >8 | >8 | >8 | >8 |
| Aminoglycoside | ||||||||
| Amikacin | >32 | >32 | >32 | 32 | 32 | >32 | 32 | >32 |
| Gentamicin | >8 | >8 | >8 | >8 | >8 | >8 | >8 | >8 |
| Netilmycin | 32 | 32 | >32 | 8 | >32 | >32 | 16 | >32 |
| Tobramycin | 16 | 16 | 16 | >16 | >16 | 16 | 8 | >16 |
| Quinolone | ||||||||
| Levofloxacin | 0.25 | 0.25 | >4 | >4 | 0.25 | 2 | 4 | >4 |
| Gatifloxacin | 0.25 | 0.06 | >4 | 4 | 0.12 | 1 | 4 | >4 |
| Ciprofloxacin | 1 | 0.5 | >2 | >4 | 0.12 | 2 | >4 | >2 |
| Polymyxin B | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 |
| blaIMP-1 location | Plasmidial | Plasmidial | Plasmidial | Plasmidial | Plasmidial | Chromosomal | Plasmidial | Chromosomal |
| Ribotyping | 258.105.2 | 258.105.2 | 105.815.4 | 105.815.4 | 252.43.4 | 258.148.6 | 258.148.2 | NAe |
| PFGE | PSA48A | PSA48A | PSA48B | PSA48B1 | PSA48C | PSA48D | PSA48E | NAe |
Interpretive criteria for the antimicrobial tested were those published by the Clinical and Laboratory Standard Institute (CLSI) (formerly National Committee for Clinical Laboratory Standards) (4).
SSTI, skin and soft tissue infection.
BSI, bloodstream infection.
LRTI, lower respiratory tract infection.
NA, not applicable.
In general, the evaluated isolates showed a decreased susceptibility or resistance phenotype to mostly all the antimicrobial agents tested, showing susceptibility only to polymyxin B, and some isolates also showed susceptibility to quinolones (Table 1). A genotypic comparison of the blaIMP-1-harboring Acinetobacter sp. isolates was performed using the RiboPrinter microbial characterization system and pulsed field-gel electrophoresis (PFGE), as previous described (12). Acinetobacter sp. isolates 694 and 696 showed identical PFGE patterns, while isolates 9043 and 695 differed from each other in less than five bands (similar PFGE patterns), and they were considered to belong to the same ancestor. The remaining Acinetobacter spp. showed distinct PFGE patterns and were considered to be unrelated (16). In summary, five different clones were observed among the seven Acinetobacter sp. isolates (Table 1).
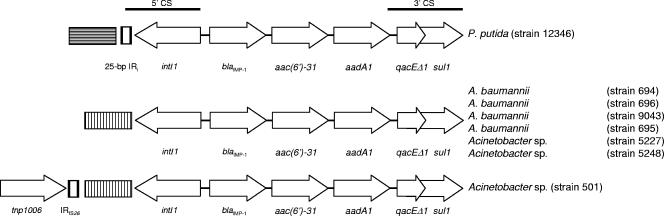
The blaIMP-1-containing integrons were amplified by PCR and sequenced using primers targeting the 5′ conserved sequences (CS) and 3′ CS of class 1 integrons (2, 9). All isolates possessed the same class 1 integron cassette arrangement, designated In86. This integron harbored blaIMP-1 at the first position downstream of the 5′ CS, followed by an open reading frame of 519 bp identified as a new AgME gene cassette. This gene, designated aac(6′)-31, was followed by another AgME gene cassette, namely, aadA1 (Fig. 1). aac(6′)-31 potentially encoded a protein of 173 amino acids (19.1 kDa), which exhibited the highest identity (82.1%) to AAC(6′)-Ib′ (GenBank accession number CAE48336) (Fig. 2), encoded by the blaIMP-16-carrying Pseudomonas aeruginosa integron isolated from Brasília, Brazil (9), 630 miles from São Paulo, suggesting that these two genes could be derived from a common ancestor.
FIG. 1.
Schematic representation of the blaIMP-1-containing class 1 integron found in P. putida and Acinetobacter sp. Inserted genes are indicated by boxes, and the arrows indicate their transcriptional orientations. Boxes with horizontal and vertical bars represent the inserted DNA sequences found upstream of the In86 25-bp IRi in the P. putida strain and in the In86 5′ CS in the Acinetobacter sp. strains, respectively.
FIG. 2.
Comparison of the deduced amino acid sequences of the most similar proteins compared to AAC(6′)-31. Differences in the amino acid sequences are noted by the insertion of a single letter representing the amino acid change within that particular sequence. GenBank accession numbers for each sequence are as follows: AAA25685 for AAC(6′)-Ib′ (6) and CAE48336 for AAC(6′)-Ib" (9).
To evaluate the flanking DNA sequences upstream of In86, these regions were sequenced using a random primer PCR approach as previously described (15). The 5′ CS of In86 in the P. putida isolate contained the integrase gene, which was bound by a Tn402-like 25-bp IRi sequence. The DNA sequence upstream of the 25-bp IRi did not show any homology with previously deposited sequences in the GenBank database (Fig. 1 and 3). However, all the Acinetobacter sp. isolates showed the same DNA insertion just downstream of the In86 integrase gene, which was inserted between the i2 and i3 19-bp repeats that are usually present in the internal region of the IRi (13) (Fig. 1 and 3). This sequence also did not possess homology with previously reported DNA sequences. Moreover, Acinetobacter sp. isolate 501 showed a second DNA insertion just upstream of the previous one, which was revealed to be a terminal inverted repeat, IS1006.1, located 138 bp upstream of the IRi. IS1006.1 was closely related to the terminal repeat of IS26, IS1006, IS1007, IS1009, and IS1010 followed by the exact last 39 nucleotides of the tnp1006 gene (Fig. 3). This structure showed the highest identity to a similar region in the Acinetobacter lwoffii plasmid pKLH202 (GenBank accession number AJ486857) (5). These findings suggest that the integron found in the P. putida strain was likely to be the progenitor blaIMP-1-carrying integron circulating in this nosocomial environment.
FIG. 3.
Nucleotides and amino acid sequences of the In86 5′ CS found in the studied strains. Asterisks and horizontal arrows indicate the stop codon and the transcription direction of the partial open reading frames, respectively. The corresponding predicted protein translation is reported below the DNA sequence. The IS26 15-bp inverted repeat is boxed, and the IRi and the i1 to i4 inverted repeats typical of the 5′ CS of Tn402-like elements are also indicated. The hypothetical 8-bp direct repeat of IS26 is in boldface type.
Repeated electroporation and conjugation experiments of putative plasmid DNA extracts from the blaIMP-1-containing strains, performed as previously described (9), failed. Despite several preparations, the presence of plasmid DNA was not observed in P. putida and in Acinetobacter sp. isolate 5227, suggesting a chromosomal location. The remaining isolates showed several plasmids (data not shown), which were recognized by a blaIMP-1-specific probe in a Southern blot experiment (14), suggesting that In86 was plasmid located in those isolates (data not shown). The blaIMP-1 probe recognized plasmids showing similar sizes, apart from Acinetobacter sp. isolate 501, which seemed to possess a slightly smaller blaIMP-1-carrying DNA plasmid. These hybridization profiles were in agreement with the sequencing results, which showed an identical structure downstream of the integrase gene in those Acinetobacter sp. isolates except for Acinetobacter sp. isolate 501, suggesting a distinct genetic locus in the latter strain.
To determine the functionality of aac(6′)-31, the gene was amplified by PCR and ligated into a pPCRScriptCam SK(+) vector to construct the recombinant plasmid pIMPAR-31 as previously described (9). This plasmid was subsequently transferred into Escherichia coli DH5α cells, and the recombinant strain was tested for susceptibility against several aminoglycosides (Table 2). aac(6′)-31 expressed in E. coli DH5α cells conferred decreased susceptibility to all aminoglycosides evaluated, including gentamicin, tobramycin, kanamycin, amikacin, neomycin, netilmicin, sisomicin, and isepamicin. MICs were 8- to ≥32-fold higher than those for E. coli DH5α (Table 2). This phenotype has not been described previously but was observed only when different AgME genes were expressed in combinations. This suggests that the expression of aac(6′)-31 may be sufficient to confer resistance to all clinically available aminoglycosides, and this gene may gradually replace other aac(6′) family genes commonly found among nonfermentative pathogens. A similar situation was previously observed among bacterial strains recovered from Turkey (10).
TABLE 2.
Aminoglycoside susceptibility profiles of E. coli DH5α-harboring recombinant plasmid pIMPAR-31 and the recipient strain E. coli DH5α
| E. coli plasmid or strain | MIC (μg/ml)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Gentamicin | Amikacin | Kanamycin | Neomycin | Netilmicin | Sisomicin | Isepamicin | Tobramycin | |
| pIMPAR-31 | 4 | 8 | 16 | 8 | 4 | 8 | 4 | 4 |
| DH5α | 0.25 | 0.5 | 0.5 | ≤0.25 | 0.5 | ≤0.25 | 0.12 | 0.25 |
The presence of five genetically unrelated Acinetobacter sp. strains and one P. putida strain, sharing a common blaIMP-1-carrying integron during the study period, shows the ability of nonfermentative nosocomial strains to acquire and subsequently spread antimicrobial resistance determinants. This may be the reason for the continuing recovery of blaIMP-1-harboring P. aeruginosa and Acinetobacter sp. strains in this hospital.
Nucleotide sequence accession numbers.
The nucleotide sequences of the blaIMP-1-containing integron described in this paper have been submitted to the EMBL/GenBank/DNA Data Bank of Japan sequence databases and assigned the accession numbers AM283489 (P. putida), AM283490 (Acinetobacter baumannii strain 501), and AJ640197 (Acinetobacter sp. strains 695, 696, 5227, 9043, and 5248).
Acknowledgments
We thank Rosa Maria Silva and Renata Cristina Picão for their excellent technical contribution to the manuscript.
H. S. Sader and R. N. Jones have received research/education grants in the last 3 years from AB BIODISK, Abbott, AlamX, Arpida, AstraZeneca, Avexa, Basilea, Bayer, Becton Dickinson, Beninger-Ingelheim, bioMerieux, Bristol-Myers Squibb, Cadence, Cerexa, Chiron, Cognigen, Cubist, Daiichi, Elan, Elanco, Enanta, GlaxoSmithKline, Intrabiotics, Johnson & Johnson, LG Chemicals, Merck, Micrologix, Novartis, Optimer, Ordway, Oscient, Osmotics, Peninsula, Pfizer, Replidyne, Schering-Plough, Sequoia, Serenex, Shionogi, Theravance, TREK Diagnostics, Vicuron, and Wyeth.
Footnotes
Published ahead of print on 30 April 2007.
REFERENCES
- 1.Bush, K., and G. H. Miller. 1998. Bacterial enzymatic resistance: β-lactamases and aminoglycoside-modifying enzymes. Curr. Opin. Microbiol. 1509-515. [DOI] [PubMed] [Google Scholar]
- 2.Castanheira, M., M. A. Toleman, R. N. Jones, F. J. Schmidt, and T. R. Walsh. 2004. Molecular characterization of a β-lactamase gene, blaGIM-1, encoding a new subclass of metallo-β-lactamase. Antimicrob. Agents Chemother. 484654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Document M7-A7. CLSI, Wayne, PA.
- 4.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing, 16th informational supplement M100-S16. CLSI, Wayne, PA.
- 5.Kholodii, G., S. Mindlin, Z. Gorlenko, M. Petrova, J. Hobman, and V. Nikiforov. 2004. Translocation of transposition-deficient (TndPKLH2-like) transposons in the natural environment: mechanistic insights from the study of adjacent DNA sequences. Microbiology 150979-992. [DOI] [PubMed] [Google Scholar]
- 6.Lambert, T., M. C. Ploy, and P. Courvalin. 1994. A spontaneous point mutation in the aac(6′)-Ib′ gene results in altered substrate specificity of aminoglycoside 6′-N-acetyltransferase of a Pseudomonas fluorescens strain. FEMS Microbiol. Lett. 115297-304. [DOI] [PubMed] [Google Scholar]
- 7.Lincopan, N., J. A. McCulloch, C. Reinert, V. C. Cassettari, A. C. Gales, and E. M. Mamizuka. 2005. First isolation of metallo-β-lactamase-producing multiresistant Klebsiella pneumoniae from a patient in Brazil. J. Clin. Microbiol. 43516-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendes, R. E., M. Castanheira, P. Garcia, M. Guzman, M. A. Toleman, T. R. Walsh, and R. N. Jones. 2004. First isolation of blaVIM-2 in Latin America: report from the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 481433-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendes, R. E., M. A. Toleman, J. Ribeiro, H. S. Sader, R. N. Jones, and T. R. Walsh. 2004. Integron carrying a novel metallo-β-lactamase gene, blaIMP-16, and a fused form of aminoglycoside-resistant gene aac(6′)-30/aac(6′)-Ib′: report from the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 484693-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Over, U., D. Gur, S. Unal, and G. H. Miller. 2001. The changing nature of aminoglycoside resistance mechanisms and prevalence of newly recognized resistance mechanisms in Turkey. Clin. Microbiol. Infect. 7470-478. [DOI] [PubMed] [Google Scholar]
- 11.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribeiro, J., R. E. Mendes, R. Domingos, E. Franca, S. Silbert, R. N. Jones, and H. S. Sader. 2006. Microbiological and epidemiological characterization of imipenem-resistant Pseudomonas aeruginosa strains from a Brazilian tertiary hospital: report from the SENTRY Antimicrobial Surveillance Program. J. Chemother. 18461-467. [DOI] [PubMed] [Google Scholar]
- 13.Riccio, M. L., L. Pallecchi, J. D. Docquier, S. Cresti, M. R. Catania, L. Pagani, C. Lagatolla, G. Cornaglia, R. Fontana, and G. M. Rossolini. 2005. Clonal relatedness and conserved integron structures in epidemiologically unrelated Pseudomonas aeruginosa strains producing the VIM-1 metallo-β-lactamase from different Italian hospitals. Antimicrob. Agents Chemother. 49104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 15.Sorensen, A. B., M. Duch, P. Jorgensen, and F. S. Pedersen. 1993. Amplification and sequence analysis of DNA flanking integrated proviruses by a simple two-step polymerase chain reaction method. J. Virol. 677118-7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]