Abstract
A metallopeptide-based fluorescence assay has been designed for the detection of small-molecule inhibitors of human immunodeficiency virus type 1 gp41, the viral protein involved in membrane fusion. The assay involves two peptides representing the inner N-terminal-heptad-repeat (HR1) coiled coil and the outer C-terminal-heptad-repeat (HR2) helical domains of the gp41 six-helix bundle which forms prior to fusion. The two peptides span a hydrophobic pocket previously defined in the literature. The HR1 peptide is modified with a metal-ligated dye complex, which maintains structural integrity and permits association with a fluorophore-labeled HR2 peptide to be followed by fluorescence quenching. Compounds able to disrupt six-helix bundle formation can act as fusion inhibitors, and we show that they can be detected in the assay from an increase in the fluorescence that is correlated with the potency of the compound. Assay optimization and validation have resulted in a simple quantitative competitive inhibition assay for fusion inhibitors that bind in the hydrophobic pocket. The assay has an assay quality factor (Z′) of 0.88 and can rank order inhibitors at 10 μM concentration with Kis in the range of 0.2 μM to 30 μM, an ideal range for drug discovery. Screening of a small peptidomimetic library has yielded three new low-molecular-weight gp41 inhibitors. In vitro syncytium inhibition assays confirmed that the compounds inhibited cell-cell fusion in the low micromolar range. These lead compounds provide a new molecular scaffold for the development of fusion inhibitors.
The coiled-coil region of the extracellular matrix protein gp41 of the human immunodeficiency virus (HIV) is an important target for drugs against viral fusion (4, 10, 11, 31, 49). gp41 is trimeric with a central coiled-coil region composed of three identical N-terminal heptad repeats (HR1) from the three molecules of the trimer. While the structure of the prefusion state is unknown, the fusion-competent state is a six-helix bundle structure, with three C-terminal heptad repeats (HR2) forming three antiparallel strands down the grooves of the coiled coil (3). This structure forms as a result of a conformational change in gp41, triggered by gp120 and coreceptor binding to host cell receptors (11, 44). Prevention of six-helix bundle formation has been recognized as an important mechanism for viral fusion inhibition and is the basis of fluorescence enzyme-linked immunosorbent assays (ELISAs) for fusion inhibitors (28, 33). Numerous studies have demonstrated that the coiled-coil domain is accessible to drugs during the transition to the fusion-competent state, making it an important target for entry inhibitors that prevent fusion (7, 30, 36, 40, 47, 51). A variety of fusion inhibitors have been shown to bind to the gp41 coiled coil, including T-20 (enfuvirtide) (29, 51) and other gp41 C peptides (8), D peptides (12), and small-molecule inhibitors (9, 14, 17, 26, 27, 35, 39). Synthetic C peptides over 30 residues in length have demonstrated efficacies as fusion inhibitors at low-nM concentrations, but few small molecules have so far been found, all with efficacy at μM concentrations. Given the limitations of peptides as drugs, the identification of more small-molecule lead compounds could play a significant role in antifusion drug development.
Several studies on fusion inhibition have involved a method used to stabilize the transiently exposed coiled coil in solution. Excision of HR1 does not produce a stable coiled-coil domain, due to nonspecific hydrophobic interactions between the peptides. Strategies have included covalently linking the peptides of the coiled coil (37), a 5-helix protein lacking one of the three outer helices of the six-helix bundle (17, 48), coiled-coil extension with a soluble trimeric GCN4 coiled coil (12, 13, 50), and a gp41 coiled-coil domain fused to maltose binding protein (45), the basis of a fluorescence polarization assay for peptide binding.
We have designed a stable HR1 receptor through N-terminal ferrous ion ligation. Formation of the trimeric coiled coil is facilitated by chelation of a Tris-2,2′-bipyridine-5-carboxy-peptide complex with Fe(II) and concominant hydrophobic collapse of the heptad repeat peptides (18-20, 32). The dark red ferrous complex acts as a fluorescence quencher of an appropriately labeled HR2 peptide when it binds to the coiled coil. Studies of the affinities of various HR2 peptides have demonstrated their selectivity for the correct sequence (21). Competitive inhibition of the HR1-HR2 interaction by a small molecule, with a concomitant increase in fluorescence over that at the baseline, is the basis for the screening assay described here. The assay has been optimized, validated, and used for the discovery of novel small-molecule gp41 inhibitors from a small peptidomimetic library. The assay was able to identify inhibitors in the library, detect false-positive results, and determine the quantitative binding affinities of true hits. The newly discovered inhibitors were confirmed to be active at micromolar concentrations in a gp41-mediated cell-cell fusion assay.
MATERIALS AND METHODS
General chemicals.
Unless otherwise stated, chemicals were purchased from Sigma-Aldrich (St. Louis, MO). ADS-J1 (9) was a gift from S. Jiang; the small peptidomimetic library screened was developed at UCSF to study the MDM2-p53 interaction (38). The molecular masses of compounds 11{3,5} and 11{6,11} were confirmed by liquid chromatography-mass spectrometry to be 491.1 Da (calculated, 490.53 Da) and 485.08 Da (calculated, 484.48 Da), respectively.
Peptide receptor and fluorescent peptide probe.
Peptide env2.0 (2,2′-bipyridine-5′-carboxylate-GQAVEAQQHLLQLTVWGIKQLQARILAVEKK-amide), which contains the deep hydrophobic pocket on gp41 and a linker for metal chelation at the N terminus, was prepared by solid-state synthesis (Biosynthesis, Inc.). The underlined residues occur in the wild-type gp41 sequence. 2,2′-Bipyridine-5′-carboxylate was prepared according to the methods described in the literature (22) and was attached to the N terminus of the peptide on the resin. The Fe(II) complex [Fe(II)(env2.0)3]2+ was prepared by addition of a one-third stoichiometry of freshly prepared ferrous ammonium sulfate to peptide in 25 mM Tris-acetate buffer at pH 7.0. The probe peptide C18-Aib (acetate-MTWBEWDREIBNYTSLIC-amide, where B is α-aminoisobutyric acid) and scrambled peptide C16-Scr (acetate-DYETMIKWEEIWKKRC-amide) were ordered from Biosynthesis Inc. C18-Aib was labeled at the cysteine with Lucifer yellow iodoacetamide (LY; Invitrogen), according to the manufacturer's directions, to form C18-Aib-LY.
Fluorescence intensity measurements.
Fluorescence intensities were measured in a reaction buffer containing 25 mM Tris, 25 mM sodium acetate (pH 7.0), and 0.01% Tween 20. Up to 32% dimethyl sulfoxide (DMSO) was included in the experiments to test for the effect of DMSO on the assay. Fluorescence measurements were made with 30 μl of solution in black 384-well plates (Greiner [Bio-one, Monroe, NC] or Costar [Corning, Cambridge, MA]) by using a Spectramax Gemini XPS plate reader or an Analyst HT plate reader (Molecular Devices, Sunnyvale, CA). The LY probe was detected by using excitation and emission frequencies of 425 nm and 540 nm, respectively. All fluorescence data are reported as the mean of repeat measurements, with error bars showing the standard deviations.
Assay development and optimization.
The binding constant between the receptor and the peptide probe was determined by serial dilution of 72 μM ferropeptide receptor in wells containing 1 μM C18-Aib-LY in 0 to 32% DMSO. The receptor concentration is reported in terms of the concentration of the component peptide, which is equivalent to the concentration of the binding sites. For competitive inhibition, assay plates were typically prepared with 10 μl of the inhibitor in buffer containing 12% DMSO and 0.03% Tween 20 and 10 μl each of 21.6 μM [Fe(II)(env2.0)3]2+ and 3 μM C18-Aib-LY. Fluorescence intensities were also measured in parallel for wells containing 10 μl inhibitor and 20 μl buffer to identify self-fluorescent library compounds and for a second set containing 10 μl inhibitor, 10 μl 3 μM C18-Aib-LY, and 10 μl buffer in order to identify false-positive results (see below). In all cases, wells contained a final DMSO concentration of 4% (except where indicated) and a Tween 20 concentration of 0.01%.
Assay performance was analyzed by measurements of the signal-to-noise ratio (S/N) and a screening window coefficient (Z) and Z′ factors (52), as follows:
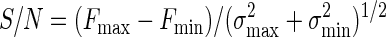 |
(1) |
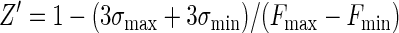 |
(2) |
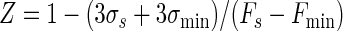 |
(3) |
where Fmax, Fmin, and FS are the mean fluorescence intensities for the free, bound, and library sample signals, respectively; and σmax, σmin, and σS are the respective standard deviations for those signals. In addition, we define a fourth parameter, Z1:
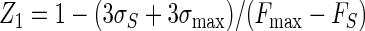 |
(4) |
which is a second screening window coefficient. The conditions Z equal to −1 and Z1 equal to −1 correspond to the minimum and the maximum values of FS for which the signal size is correlated to the inhibition constant; i.e., the signal is at least 3σmin above Fmin (Z = −1) or 3σmax is below Fmax (Z1 = −1).
Calculation of binding constants.
The dissociation constant of the receptor-probe peptide interaction (Kd) was calculated from a fit to the equation
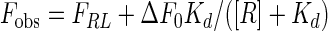 |
(5) |
where Fobs is the observed fluorescence,
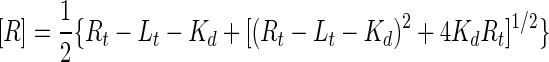 |
(6) |
and Lt is the total probe peptide concentration; Rt is the total concentration of receptor binding sites, equal to 3[Fe(II)(env2.0)3]2+; and ΔF0 is equal to Fmax − FRL, which is the difference between the fluorescence of the free probe and the fluorescence of the receptor-probe complex (FRL). The data were fit by using the Kaleidagraph program.
The inhibition constant (Ki) for competitive inhibition can be obtained in the general case from the numerical solution of five simultaneous equations (Mathcad; Mathsoft):
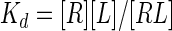 |
(7a) |
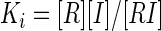 |
(7b) |
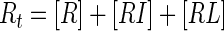 |
(7c) |
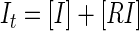 |
(7d) |
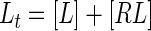 |
(7e) |
where [L], [R], and [I] are the concentrations of free probe peptide, free receptor peptide, and free inhibitor, respectively, [RL] and [RI] are the concentrations of bound probe peptide and inhibitor, and It is the total concentration of inhibitor. The solution is obtained by using the correlation between FS and the ligand concentration:
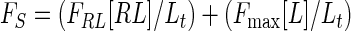 |
(7f) |
The values for FRL and Fmax were extracted from the titration curves in Fig. 1 (equation 5). Dose-response curves for competitive inhibition were also approximated by equation 5 by using
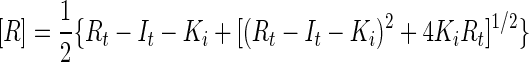 |
(8) |
Equation 8 is valid when the probe concentration is low relative to the concentrations of the receptor binding sites.
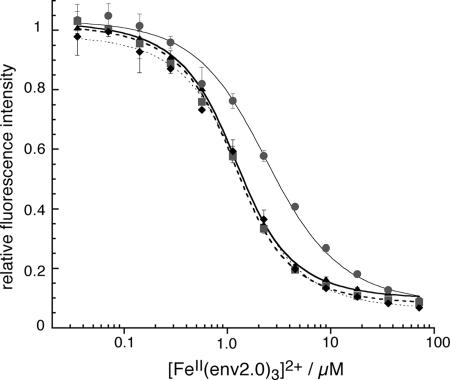
FIG. 1.
Titration of Fe(II) metallopeptide into a constant concentration (1 μM) of C18-Aib-LY. The experiment was repeated in triplicate and with different DMSO concentrations: 0% DMSO (▴), 4% DMSO (▪), 16% DMSO (⧫), and 32% DMSO (•). Experiments were conducted in 384-well plates on a Spectramax Gemini XPS plate reader by using 30 μl solution per well. The data were fit to equations 5 and 6.
Cell-cell fusion assay.
Measurement of HIV type 1 (HIV-1) fusion inhibition was performed by Changhua Ji at Roche (Palo Alto, CA) by using a screening protocol involving inducible gp160 expression in HeLa effector cells and CCR5-expressing Tat-long terminal repeat-luciferase reporter cells (24).
RESULTS
[Fe(II)(env2.0)3]2+ fluorescence intensity assay system design.
The fluorescence intensity screen was set up by using a fluorescently labeled probe peptide from HR2 and a metal ion-chelated coiled-coil peptide from HR1. The interaction of the two peptides resulted in fluorescence quenching of the probe by the metal chelate. The two peptides synthesized for the assay were env2.0, a 31-residue N-terminal bipyridylated HR1 peptide containing critical residues of the deep hydrophobic pocket on gp41 (residues 565 to 577), and C18-Aib, an 18-residue cysteine-terminated HR2 peptide containing residues Trp628, Trp631, Asp632, and Ile635 that play a key role in the specific interaction in the hydrophobic pocket (5, 6, 45). The Fe(II)-chelated receptor [Fe(II)(env2.0)3]2+ was formed by the addition of a one-third equivalent of ferrous ion to env2.0. The helical propensity of C18-Aib was increased by replacement of two residues nonessential to the binding interaction with α-aminoisobutyric acid (51). This peptide had improved solubility compared to those of similar native peptides of gp41 HR2. It is 18% helical by circular dichroism measurement. C18-Aib was labeled with LY at the C-terminal cysteine for direct dissociation constant measurements and was used without labeling as a positive control in competitive inhibition experiments. In addition, a 16-residue peptide, C16-Scr, made by scrambling the HR2 region from positions 628 to 643, was prepared as a negative control (21).
The interaction of the two peptides resulted in fluorescence quenching of the probe by the metal chelate due to overlap of the fluorescence emission with the iron(II)-bipyridyl charge transfer band. Titration of an increasing concentration of [Fe(II)(env2.0)3]2+ into a constant concentration of LY-labeled HR2 peptide yielded a Kd of 0.9 ± 0.2 μM for C18-Aib-LY (Fig. 1). The data were fit by using equations 5 and 6. The effect of DMSO on Kd was determined by carrying out the experiment with 0, 4, 16, and 32% DMSO. The binding was unaffected by DMSO concentrations up to 16%, after which some impairment occurred, with a Kd of 2.5 μM obtained with 32% DMSO.
Inhibitor detection by competitive inhibition.
Small molecules that bind to the hydrophobic pocket can, in principle, be detected by measurement of an increase in the fluorescence in a ternary system containing the probe and receptor peptides mixed with the compound of interest. Use of the assay for the detection of novel inhibitors of HIV fusion required assay evaluation and validation in order to confirm both the sensitivity and the validity of the assay with the [Fe(II)(env2.0)3]2+ model receptor system representing the intact coiled-coil domain in HIV-1. For these purposes, assay statistics and the dynamic range were evaluated and known gp41 hydrophobic pocket binders were used to test the assay response.
Assay bounds, statistics, and dynamic range.
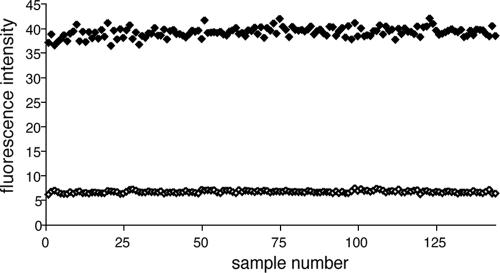
Assay behavior was evaluated by measurement of the standard statistical parameters Z and Z′ (52). Z′, which should exceed 0.5 for an assay suitable for high-throughput screening, depends on the standard deviations of both the bound and the free probe signals and on their separation. A receptor concentration on the order of or greater than the Kd is desirable for maximum sensitivity, according to the binding equation (equation 5); 7.2 μM receptor, which corresponds to a lower bound of 20% Fmax, was selected. The probe concentration was minimized within the limits of S/N; the use of lower probe concentrations simplifies data analysis (equation 8). The probe was used at a concentration of 1 μM to calibrate the assay. Figure 2 shows 144 repeat measurements of the upper and lower bounds, which correspond to the probe and the probe-receptor mixture, respectively. The Z′ factor in Fig. 2, obtained by using 4% DMSO, is 0.88. The S/N is 28.8. The assay components and the fluorescence readings were very stable over a 24-h period at 4°C.
FIG. 2.
Determination of Z′ factor for the assay. The signals obtained with 1 μM probe (⧫) and a mixture of 1 μM probe and 7.2 μM receptor (⋄) are shown. DMSO (4%) and Tween 20 (0.01%) were included in the buffer. Experiments were conducted at room temperature on a Spectramax Gemini XPS plate reader in Greiner Bio-one 384-well plates, with 30 μl used per well.
The standard deviation obtained for plate reader measurements in Fig. 2 was consistently 2% of the signal mean amplitude, and the background noise in the absence of sample was 0.2 fluorescence units. These assay statistics could be applied to the calculation of the dynamic range (2) by using the empirical relationship σS ≈ 0.02FS + 0.2. Substitution into equations 2 to 4 gave, respectively,
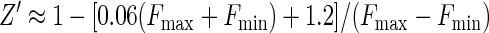 |
(9) |
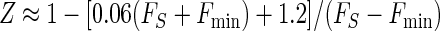 |
(10) |
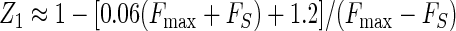 |
(11) |
Equation 9 gave a Z′ factor of 0.88 for the conditions used for Fig. 2, which is equal to the experimental value obtained. The smallest and largest FS levels which define the limits of the quantitative range, where Z is equal to −1 or Z1 is equal to −1, were calculated from equations 10 and 11 and were correlated to Ki by using equations 7a to 7f and by assuming an inhibitor concentration of 10 μM. This gave a predicted quantitative range of Ki from 30 μM (Z = −1) to 0.14 μM (Z1 = −1), which is a good range for the detection of initial hits from a library. The use of higher or lower inhibitor concentrations would shift the window of quantitative detection.
Assay validation.
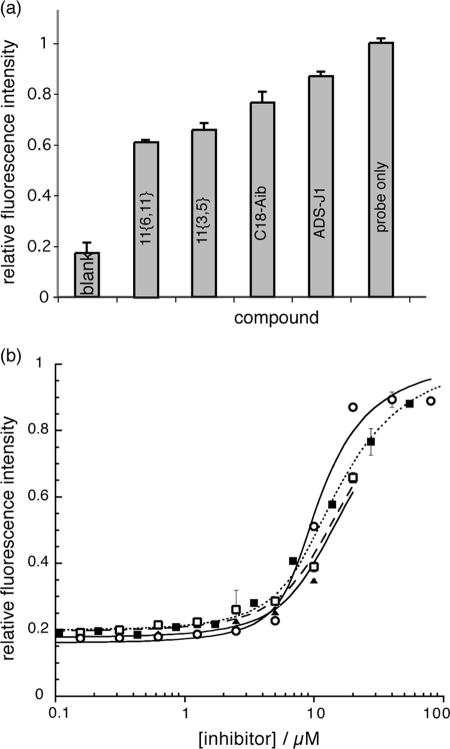
Small-molecule HIV-1 fusion inhibitors known to bind in the deep hydrophobic pocket were tested by the assay to confirm its utility for discovering compounds that can prevent viral fusion. Reports in the literature have identified several compounds which inhibit HIV-1 fusion, as measured by syncytium formation and viral infectivity assays. Of these, compound ADS-J1 (9, 46), several tea extracts (35, 39), and the cyclic D peptide D10-p1-2K (12) were tested. The results of a scan of several compounds in 384-well plates is shown in Fig. 3a. The signal recovered in the presence of a competitive inhibitor from a low of 18% in the absence of inhibitor to a level that was directly related to the Ki of the inhibitor. The Kis were estimated from the single-point scan of Fig. 3a by using equations 7a to 7f and were determined more accurately from dose-response curves (Fig. 3b). The results are listed in Table 1, together with known values from the literature. Fitting of the data to the approximate equation 8 gave Kis comparable to those obtained by equation 7, demonstrating that equation 8 provides an adequate analytical expression for competitive inhibition under these conditions, where Rt is equal to 7.2Lt.
FIG. 3.
Fluorescence intensity measurements for 30-μl solutions in the wells of 384-well plates in which each well contained 7.2 μM [Fe(II)(env2.0)3]2+, 1 μM C18-Aib-LY, and 20 μM inhibitor (a) or as a function of the concentrations of four inhibitors, compounds ADS-J1 (○), 11{6,11} (▴), 11{3,5}(□), and C18-Aib (▪) (b). Data from the dose-response curves were fit to equations 5 and 8. The experiments were repeated in triplicate. Fluorescence is given as a ratio of primary and control scans (see text) and is relative to a maximum probe fluorescence of 1.
TABLE 1.
Inhibition constants from single-point measurement and dose-response curves compared to the values in the literature for known gp41 binders and fusion inhibitors
| Compound | Molecular mass (Da) |
Ki (μM)a
|
IC50 (μM [reference]) | |
|---|---|---|---|---|
| Single-point measurement | Dose-response curve | |||
| D10-p1-2K (oxidized) | 2,125 | 20 | NDb | 44c (12) |
| D10-p1-2K (reduced) | 2,127 | None | None | |
| ADS-J1 | 1,177 | 0.44 | 0.35 ± 0.09 | 3.28 ± 0.03c (46), 2.55 ± 0.16d (46) |
| Tannin | 1,701 | 0.39 | 0.18 ± 0.08 | ∼0.4d (34) |
| C18-Aib | 2,271 | 0.60 | 0.80 ± 0.13 | ND |
| 11{6,11} | 484.5 | 1.20 | 1.34 ± 0.19 | ∼8c (this work) |
| 11{3,5} | 490.5 | 1.15 | 1.51 ± 0.16 | ∼8c (this work) |
| Gallocatechin gallate | 458.4 | None | 7.55 ± 0.37c (35), 4.32 ± 0.94c (35) | |
| Epigallocatechin gallate | 458.4 | None | Nonec (35), 19.91 ± 1.27d (35) | |
Kis are from this work. Equation 8 was used to calculate the Kis from the dose-response curve, and the data represent the means and standard deviations from three independent experiments.
ND, not available at a concentration high enough to determine the dose-response curve.
Syncytium inhibition in cell culture.
Six-helix bundle formation assay (33).
C18-Aib had a Ki equal to the Kd of C18-Aib-LY, as expected. Compound ADS-J1 and the D peptide D10-p1-2K tested positive by the assay, with Kis that agreed in order of magnitude with the 50% inhibitory concentrations (IC50s) reported in the literature. Reduced (noncyclized) D peptide tested negative, as expected, because it lacks the required tertiary structure (12). The correlation between a positive hit in the assay and in vitro antifusion activity confirmed that [Fe(II)(env2.0)3]2+ is a faithful representation of the intact HR1 domain in the virus. The assay was able to select for compounds that are active against HIV-1 fusion through binding to the gp41 hydrophobic pocket.
Several gp41 binders identified from tea extracts gave anomalous results by our assay. Tannin tested positive, with an affinity similar to that reported previously (34) (Table 1); but it was found to interact with probe peptide as well as receptor, so that consistent signal recovery was not observed. Tannin is a multiphenol compound which binds nonspecifically to proteins and peptides through the phenol groups (16). Two catechin compounds from green tea tested negative by our assay, although they had previously been identified as gp41 binders. These compounds were confirmed to be positive by an ELISA; one was confirmed to be positive by a cell fusion assay, and the other gave negative results (35). One of the ways that they could have escaped detection by our assay is if their mechanism of action does not involve binding in the deep hydrophobic pocket, since only hydrophobic pocket binders would be detected. In the ELISA for six-helix bundle formation, an alternative or overlapping site could be the target.
Identification of novel inhibitors of HIV-1 fusion.
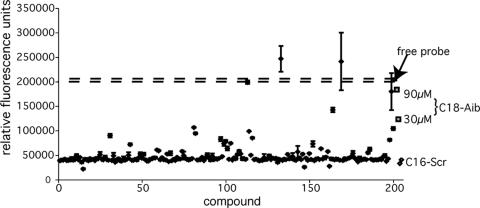
A novel small-molecule scaffold for fusion inhibitors was discovered by using the assay to screen a small peptidomimetic library, developed as helical mimetics for a p53 binding site on MDM2 (38). The library compounds consisted of two or three amide-linked aromatic rings that form a scaffold for the presentation of hydrophobic side chains at the i, i + 4, and in some cases, i + 7 positions along a putative α-helix (i is the residue number). Half of the compounds (chemset 12 [38]) contained three substituent side chains, while the remaining half of the compounds (chemset 11 [38]) were truncated after the second substituent. Two hundred compounds were available, with stock concentrations in DMSO varying from 0.5 to 3 mM. No autofluorescence at the wavelengths of measurement was observed for any of the compounds.
The results of the screen are shown in Fig. 4. The dashed lines indicate the maximum fluorescence obtained for C18-Aib-LY in the buffer solution, which contained 4% DMSO. Most compounds had no effect on the baseline fluorescence of 40,000 relative fluorescence units, indicating no binding. Surprisingly, about 8% of the compounds appeared to test positive. Further evaluation of these compounds revealed that a large percentage of them had false-positive results which could be detected by a control screen (see below). The false-positive results occurred when the compounds formed micelle-like aggregates in solution, causing spurious fluorescence enhancement of the probe peptide. Three of the compounds, compounds 11{2,4}, 11{3,5}, and 11{6,11}, were confirmed to have true-positive results; and two of them were tested further in a cell-cell fusion assay (24). The structures of the compounds are shown in Fig. 5, and the results of the biochemical and cell-cell fusion assay for two of the compounds are included in Table 1 and Fig. 3. A follow-up with compound 11{2,4} was not possible because of a short supply. Kis of 1.3 μM and 1.5 μM for compounds 11{3,5} and 11{6,11}, respectively, were determined from the fluorescence assay. Inhibition of syncytium formation occurred in the micromolar range, although the limited stock concentrations in the library precluded determination of the IC50s of these compounds due to a limit in the DMSO concentration of 1% in cell culture. Compound concentration limits also resulted in truncated dose-response curves (Fig. 3).
FIG. 4.
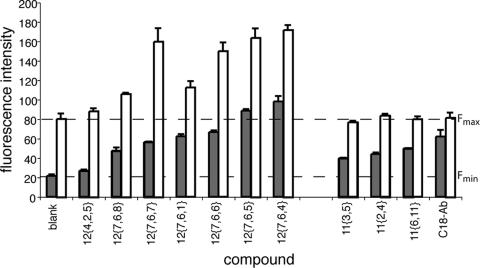
Screening of 200 peptidomimetic compounds from a library. Measurements were make in duplicate from 384-well plates in which each well contained 30 μl of 18.6 μM [Fe(II)(env2.0)3]2+, 5 μM probe, and 12 to 50 μM compound. The solutions contained 4% DMSO. Also shown are data for a positive control (C18-Aib), a negative control (C16-Scr), and 100% fluorescence (C18-Aib-LY, dashed lines). The screen was conducted on an Analyst HT plate reader. Signals above 100% correspond to those for compounds 12{7,6,4} and 12 {7,6,5} (38) and, along with the signals for many of the compounds, were false positives. True-positive signals were detected following the control experiment, whose results are demonstrated in Fig. 6.
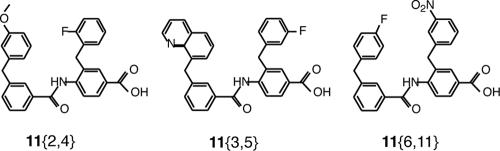
FIG. 5.
Structures of two scaffold ring compounds which had true-positive results in the biochemical assay. Compounds 11{3,5} and 11{6,11} were confirmed to be positive in viral syncytium formation assays. Compound 11{2,4} was not tested further.
Compounds 11{2,4}, 11{3,5}, and 11{6,11} are peptidomimetic compounds consisting of a 4-benzamidobenzoic acid scaffold with aromatic substituent groups at the 3 positions in orientations matched to the i and i + 4 positions of an α helix. The carboxylate moiety may form a salt bridge with Lys574 in the hydrophobic pocket (25). The compounds have built-in flexibility to vary side chain substituents and modifiers and are promising candidates for use for the development of more potent fusion inhibitors. The library contained about 100 of these structures, but most tested negative, attesting to a specific interaction with the coiled coil. This discovery demonstrates the ability of the [Fe(II)(env2.0)3]2+-C18-Aib-LY fluorescence assay to detect novel compounds with HIV-1 fusion-inhibitory properties and validates the assay as a means of selecting compounds with biological activity.
Control assay for detection of false-positive results.
The observation of spurious fluorescence enhancement of the probe peptide in the presence of certain compounds provided a facile control screen for the detection of false-positive results in the assay. Figure 6 demonstrates the detection of false-positive results in a control assay with several compounds from chemsets 11 and 12 in which the [Fe(II)(env2.0)3]2+ receptor was excluded from the wells. The three-substituent compounds from chemset 12 gave an apparent positive result in the primary assay (Fig. 6, gray bars) but clearly interacted with the probe in the control assay, altering the fluorescence yield (Fig. 6, white bars). The three compounds from chemset 11 had no effect on the fluorescence of probe peptide and were true hits.
FIG. 6.
Fluorescence intensity of 2 μM C18-Aib-LY mixed with compounds from the library in Fig. 4 in the presence (gray bars) and absence (white bars) of 6 μM [Fe(II)(env2.0)3]2+. The lower dashed line (Fmin) indicates the fluorescence intensity of the [Fe(II)(env2.0)3]2+-C18-Aib-LY mixture in the absence of compound (20 fluorescence units), and the upper dashed line (Fmax) indicates the fluorescence intensity of 2 μM C18-Aib-LY alone (100% intensity, 80 fluorescence units). Compounds from chemset 12 were aggregators and caused the probe fluorescence to rise above Fmax. The chemset 11 compounds for which the results are shown here and C18-Aib did not aggregate at the concentrations used in the assay. Compounds were diluted in buffer by a factor of 25 from stock solutions in DMSO. The experiments were repeated in duplicate.
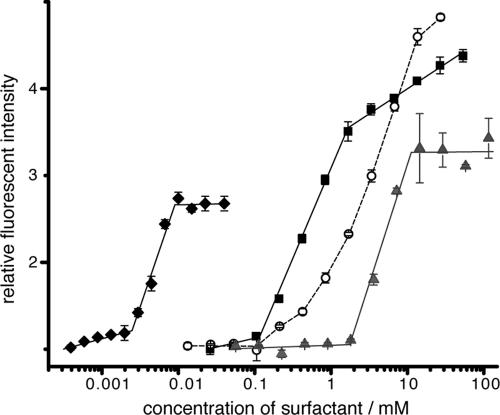
The spurious fluorescence enhancement of C18-Aib-LY in the presence of certain compounds was attributed to the formation of soluble micelle-like compound aggregates into which the peptide partitioned. Figure 7 demonstrates the large increase in fluorescence of C18-Aib-LY above the critical micelle concentration of detergents and one of the compounds from chemset 12. The dose-response curve typically featured a sudden sharp rise in fluorescence, effectively at the critical micelle concentration of the compound (43), instead of the gradual rise expected from a typical binding event. Variations in the shapes of the dose-response curves for different detergents are suggestive of differences in the C18-Aib-LY interaction with different lipids. Fluorescence enhancement resulted from the loss of peptide secondary structure and a more hydrophobic fluorophore environment in the micelles (23). The quantum yield of LY increases 3-fold when it is in DMSO and 2.5-fold when it is attached to a peptide without α-aminoisobutyric acid residues and in which all tryptophan residues have been replaced by alanine. This indicates both the effect of a hydrophobic environment on LY and the quenching effect of tryptophan within the existing secondary structure of C18-Aib-LY in aqueous buffer solution.
FIG. 7.
Titration of compound 12 {7,6,4} (38) into 2 μM probe peptide (♦) and titration of Triton X-100 (▪), Tween 20 (○), and sodium dodecyl sulfate (▴) into 1 μM probe peptide.
This observation suggests a simple method by which the fluorescence of a hydrophobic peptide such as C18-Aib-LY could be used in a control assay for the detection of aggregators in high-throughput screening (15). Aggregators present a problem in many biochemical assays because they sequester the enzymes or receptors used in the assay into micellar compound aggregates, causing false-positive results (41-43). The amphiphilic or hydrophobic character that causes this effect is difficult to predict (15). The important properties of the peptide C18-Aib-LY for the detection of micelle formation are its hydrophobicity and its existing secondary structure in aqueous buffer solution. A simple fluorescence readout and high-throughput detection make the control assay easy to implement. The effect depends on the peptide-micelle association, and the probe peptide concentration should be kept low to magnify the effect, which is saturable. The control assay also detects false-negative results caused by light scattering due to turbidity or the absorption of light by colored compounds, both of which would cause a reduction in the baseline fluorescence of the probe.
DISCUSSION
A biochemical method for the detection of gp41 inhibitors has led to the development of an efficient protocol for the screening of low-molecular-weight compounds that are able to inhibit HIV-1 fusion in cell culture. Screening of a small peptidomimetic library has resulted in the identification of a novel scaffold for low-molecular-weight HIV-1 fusion inhibitors and two compounds which tested positive in syncytium formation assays.
The biochemical assay involves two modified peptides which are simply mixed with library compounds and whose fluorescence intensity is measured. The assay protocol developed has three steps. (i) Primary screening of a compound library with receptor and probe peptides for potential HIV-1 inhibitor detection is performed first. This assay has a Z′ factor of 0.88 and an S/N of 28.8 in pH 7 buffer containing 4% DMSO and 0.01% Tween 20. The assay can tolerate DMSO concentrations up to at least 16%. The signal in this single-point screening assay is quantitatively related to the Ki of a true inhibitor. (ii) A control assay uses probe peptide to detect false-positive results and some false-negative results. A peptide with the properties of C18-Aib (hydrophobicity and residual solution structure) could be used more widely to identify compounds in libraries that may act as nonspecific or promiscuous inhibitors. (ii) Serial dilution of true-positive hits is performed by the primary assay for the more accurate determination of Ki values. The sensitive, selective, and quantitative detection of inhibitors has been demonstrated and is expected to cover a Ki range of 0.14 to 30 μM, assuming the use of 10 μM inhibitor. Inhibitors with an affinity greater than 0.14 μM can be detected quantitatively by lowering the inhibitor and receptor concentrations, while inhibitors with an affinity less than 30 μM can be detected by increasing the inhibitor concentration. Ultimately, the range of measurable Ki values is limited by the Kd in the assay to values of ∼0.1Kd to 100Kd. The compounds detected by the assay are expected to bind in the deep hydrophobic pocket of gp41, a known region of high conservation in HIV-1.
The peptidomimetic compounds discovered have molecular masses less than 500 Da and are promising hits for further development. They contain multiple sites amenable to modification, including variations of the functional groups attached to the scaffold or extension of the scaffold to create a third site for functional group attachment, i.e., at positions i − 4, i − 3, i + 7, or i + 8 along the putative α helix. Such additions would increase the area of hydrophobic contact with the coiled coil and could increase compound affinity. We are constructing and testing a diverse array of compounds based on the structural scaffold.
The fluorescence intensity assay combines speed and quality, in that it is readily adaptable to high-throughput screening, includes a control screen for false-positive results, and permits quantitative detection of true hits. The signal size for true hits in a single-point screen is directly correlated to compound potency and can be used to select compound concentrations for dose-response measurements and determination of Ki values. The method was used to screen a small peptidomimetic library and to discover three valuable inhibitory compounds. The entire screen, from high-throughput screening to quantitative analysis, can be automated and simply requires the mixing of the reagents with the compounds to be tested and direct measurement, with a possible 20-min wait for consistent results. We estimate a maximum cost for reagents of $8,000 for high-throughput screening and control screening of 100,000 compounds in 384-well plates, a cost that is likely to be reduced if the reagents are ordered in bulk and that can be reduced even further by the use of 1,536-well plates. The reproducibility of the assay between plates is within 10%.
The key component of the assay is a designed metallopeptide receptor representing the gp41 HR1 coiled-coil domain. This approach is directly applicable to all class 1 viruses, which use the coiled-coil structure in their fusion machinery. In this way, existing compound libraries can be mined for promising antiviral drug candidates.
Acknowledgments
This work was supported by NIH grant AI060361 to M.G. Additional support has been provided by Touro University. We acknowledge support from the Bay Area Screening Center (UCSF-QB3) for the use of the Analyst HT plate reader.
We are grateful to I. D. Kuntz, R. K. Guy, and F. Lu (UCSF) for making their p53-MDM2 library available for testing, for help in setting up the library screen, and for invaluable discussions. We thank S. Jiang for supplying a sample of ADS-J1 and D. Wu for preparing 2,2′-bipyridine carboxylate. We are very grateful to Chenghua Ji at Roche for testing our compounds using his newly developed syncytium formation assay.
Footnotes
Published ahead of print on 23 April 2007.
REFERENCES
- 1.Reference deleted.
- 2.Buxser, S., and S. Vroegop. 2005. Calculating the probability of detection for inhibitors in enzymatic or binding reactions in high-throughput screening. Anal. Biochem. 3401-13. [DOI] [PubMed] [Google Scholar]
- 3.Caffrey, M., M. Cai, J. Kaufman, S. J. Stahl, P. T. Wingfield, D. G. Covell, A. M. Gronenborn, and G. M. Clore. 1998. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 174572-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cammack, N. 2001. The potential for HIV fusion inhibition. Curr. Opin. Infect. Dis. 1413-16. [DOI] [PubMed] [Google Scholar]
- 5.Chan, D. C., C. T. Chutkowski, and P. S. Kim. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 9515613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89263-273. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y.-H., Y. Xiao, and M. P. Dierich. 2000. HIV-1 gp41: role in HIV entry and prevention. Immunobiology 201308-316. [DOI] [PubMed] [Google Scholar]
- 8.Chinnadurai, R., J. Munch, M. Dittmar, and F. Kirchhoff. 2005. Inhibition of HIV-1 group M and O isolates by fusion inhibitors. AIDS 191919-1922. [DOI] [PubMed] [Google Scholar]
- 9.Debnath, A. K., L. Radigan, and S. Jiang. 1999. Structure-based identification of small molecule antiviral compounds targeted to the gp41 core structure of the human immunodeficiency virus type 1. J. Med. Chem. 423203-3209. [DOI] [PubMed] [Google Scholar]
- 10.Doms, R. W., and J. P. Moore. 2000. HIV-1 membrane fusion: targets of opportunity. J. Cell Biol. 151F9-F13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70777-810. [DOI] [PubMed] [Google Scholar]
- 12.Eckert, D. M., V. N. Malashkevich, L. H. Hong, P. A. Carr, and P. S. Kim. 1999. Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell 99103-115. [DOI] [PubMed] [Google Scholar]
- 13.Eckert, D. M., V. N. Malashkevich, and P. S. Kim. 1998. Crystal structure of GCN4-pIQI, a trimeric coiled coil with buried polar residues. J. Mol. Biol. 284859-865. [DOI] [PubMed] [Google Scholar]
- 14.Ernst, J. T., O. Kutzki, A. K. Debnath, S. Jiang, H. Lu, and A. D. Hamilton. 2002. Design of a protein surface antagonist based on alpha-helix mimicry: inhibition of gp41 assembly and viral fusion. Angew. Chem. Int. Ed. Engl. 41278-281. [DOI] [PubMed] [Google Scholar]
- 15.Feng, B. Y., A. Shelat, T. N. Doman, R. K. Guy, and B. K. Shoichet. 2005. High-throughput assays for promiscuous inhibitors. Nat. Chem. Biol. 1146-148. [DOI] [PubMed] [Google Scholar]
- 16.Frazier, R. A., A. Papadopoulou, I. Mueller-Harvey, D. Kissoon, and R. J. Green. 2003. Probing protein-tannin interactions by isothermal titration microcalorimetry. J. Agric. Food Chem. 515189-5195. [DOI] [PubMed] [Google Scholar]
- 17.Frey, G., S. Rits-Volloch, X. Q. Zhang, R. T. Schooley, B. Chen, and S. C. Harrison. 2006. Small molecules that bind the inner core of gp41 and inhibit HIV envelope-mediated fusion. Proc. Natl. Acad. Sci. USA 10313938-13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghadiri, M. R., C. Soares, and C. Choi. 1992. A convergent approach to protein design. Metal ion-assisted spontaneous self-assembly of a polypeptide into a triple-helix bundle protein. J. Am. Chem. Soc. 114825-831. [Google Scholar]
- 19.Gochin, M., R. K. Guy, and M. A. Case. 2003. A metallopeptide assembly of the HIV-1 gp41 coiled coil is an ideal receptor in fluorescence detection of ligand binding. Angew. Chem. Int. Ed. Engl. 425325-5328. [DOI] [PubMed] [Google Scholar]
- 20.Gochin, M., V. Khorosheva, and M. A. Case. 2002. Structural characterization of a paramagnetic metal-ion-assembled three-stranded alpha-helical coiled coil. J. Am. Chem. Soc. 12411018-11028. [DOI] [PubMed] [Google Scholar]
- 21.Gochin, M., R. Savage, S. Hinckley, and L. Cai. 2006. A fluorescence assay for rapid detection of ligand binding affinity to HIV-1 gp41. Biol Chem. 387477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, T. L. J., and D. G. Brewer. 1981. Investigation of the 1H NMR isotropic shifts from some methyl-substituted bipyridine complexes with Ni(II) and Co(II). Can. J. Chem. 591689-1700. [Google Scholar]
- 23.Hunter, R. J. 1991. Foundations of colloid science. Oxford University Press, New York, NY.
- 24.Ji, C., J. Zhang, N. Cammack, and S. Sankuratri. 2006. Development of a novel dual CCR5-dependent and CXCR4-dependent cell-cell fusion assay system with inducible gp160 expression. J. Biomol. Screen. 1165-74. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, S., and A. K. Debnath. 2000. A salt bridge between an N-terminal coiled coil of gp41 and an antiviral agent targeted to the gp41 core is important for anti-HIV-1 activity. Biochem. Biophys. Res. Commun. 270153-157. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, S., H. Lu, S. Liu, Q. Zhao, Y. He, and A. K. Debnath. 2004. N-substituted pyrrole derivatives as novel human immunodeficiency virus type 1 entry inhibitors. Antimicrob. Agents Chemother. 484349-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang, S., Q. Zhao, and A. K. Debnath. 2002. Peptide and non-peptide fusion inhibitors. Curr. Pharm. Design 8563-580. [DOI] [PubMed] [Google Scholar]
- 28.Jin, B. S., W. K. Lee, K. Ahn, M. K. Lee, and Y. G. Yu. 2005. High-throughput screening method of inhibitors that block the interaction between 2 helical regions of HIV-1 gp41. J. Biomol. Screen. 1013-19. [DOI] [PubMed] [Google Scholar]
- 29.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 41302-1307. [DOI] [PubMed] [Google Scholar]
- 30.Kilgore, N. R., K. Salzwedel, M. Reddick, G. P. Allaway, and C. T. Wild. 2003. Direct evidence that C-peptide inhibitors of human immunodeficiency virus type 1 entry bind to the gp41 N-helical domain in receptor-activated viral envelope. J. Virol. 777669-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kliger, Y., and Y. Shai. 2000. Inhibition of HIV-1 entry before gp41 folds into its fusion-active conformation. J. Mol. Biol. 295163-168. [DOI] [PubMed] [Google Scholar]
- 32.Lieberman, M., and T. Sasaki. 1991. Iron(II) organizes a synthetic peptide into three-helix bundles. J. Am. Chem. Soc. 1131470-1471. [Google Scholar]
- 33.Liu, S., L. Boyer-Chatenet, H. Lu, and S. Jiang. 2003. Rapid and automated fluorescence-linked immunosorbent assay for high-throughput screening of HIV-1 fusion inhibitors targeting gp41. J. Biomol. Screen. 8685-693. [DOI] [PubMed] [Google Scholar]
- 34.Liu, S., S. Jiang, Z. Wu, L. Lv, J. Zhang, Z. Zhu, and S. Wu. 2002. Identification of inhibitors of the HIV-1 gp41 six-helix bundle formation from extracts of Chinese medicinal herbs Prunella vulgaris and Rhizoma cibotte. Life Sci. 711779-1791. [DOI] [PubMed] [Google Scholar]
- 35.Liu, S., H. Lu, Q. Zhao, Y. He, J. Niu, A. Debnath, S. G. Wu, and S. Jiang. 2005. Theaflavin derivatives in black tea and catechin derivatives in green tea inhibit HIV-1 entry by targeting gp41. Biochim. Biophys. Acta 1723270-281. [DOI] [PubMed] [Google Scholar]
- 36.Liu, S., S. Wu, and S. Jiang. 2007. HIV entry inhibitors targeting gp41: from polypeptides to small-molecule compounds. Curr. Pharm. Design 13143-162. [DOI] [PubMed] [Google Scholar]
- 37.Louis, J. M., I. Nesheiwat, L. Chang, G. M. Clore, and C. A. Bewley. 2003. Covalent trimers of the internal N-terminal trimeric coiled-coil of gp41 and antibodies directed against them are potent inhibitors of HIV envelope-mediated cell fusion. J. Biol. Chem. 27820278-20285. [DOI] [PubMed] [Google Scholar]
- 38.Lu, F., S. Chi, D. Kim, K. Han, I. Kuntz, and R. Guy. 2006. Proteomimetic libraries: design, synthesis, and evaluation of p53-MDM2 interaction inhibitors. J. Comb. Chem. 8315-325. [DOI] [PubMed] [Google Scholar]
- 39.Lu, L., S. W. Liu, S. B. Jiang, and S. G. Wu. 2004. Tannin inhibits HIV-1 entry by targeting gp41. Acta Pharmacol. Sin. 25213-218. [PubMed] [Google Scholar]
- 40.Markosyan, R. M., M. Xiuwen, M. Lu, F. S. Cohen, and G. B. Melikyan. 2002. The mechanism of Inhibition of HIV-1 env-mediated cell-cell fusion by recombinant cores of gp41 ectodomain. Virology 302174-184. [DOI] [PubMed] [Google Scholar]
- 41.McGovern, S., E. Caselli, N. Grigorieff, and B. Shoichet. 2002. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem. 451712-1722. [DOI] [PubMed] [Google Scholar]
- 42.McGovern, S., B. Helfand, B. Feng, and B. Shoichet. 2003. A specific mechanism of nonspecific inhibition. J. Med. Chem. 464265-4272. [DOI] [PubMed] [Google Scholar]
- 43.McGovern, S. L., and B. Shoichet. 2003. Kinase inhibitors: not just for kinases anymore. J. Med. Chem. 461478-1483. [DOI] [PubMed] [Google Scholar]
- 44.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mo, H., A. K. Konstantinidis, K. Stewart, T. Dekhtyar, T. Ng, K. Swift, E. Matayoshi, W. Kati, W. Kohlbrenner, and A. Molla. 2004. Conserved residues in the coiled-coil pocket of human immunodeficiency virus type 1 gp41 are essential for viral replication and interhelical interaction. Virology 329319-327. [DOI] [PubMed] [Google Scholar]
- 46.Naicker, K. P., S. Jiang, H. Lu, J. Ni, L. Boyer-Chatenet, L.-X. Wang, and A. K. Debnath. 2004. Synthesis and anti-HIV-1 activity of 4-[4-(4,6-bisphenylamino-[1,3,5]triazin-2-ylamino)-5-methoxy-2-methylphenylazo]-5-hydroxynaphthalene-2,7-disulfonic acid and its derivatives. Bioorg. Med. Chem. 121215-1220. [DOI] [PubMed] [Google Scholar]
- 47.Root, M., and H. Steger. 2004. HIV-1 gp41 as a target for viral entry inhibition. Curr. Pharm. Design 101805-1825. [DOI] [PubMed] [Google Scholar]
- 48.Root, M. J., M. S. Kay, and P. S. Kim. 2001. Protein design of an HIV-1 entry inhibitor. Science 291884-888. [DOI] [PubMed] [Google Scholar]
- 49.Shu, W., J. Liu, H. Ji, L. Radigen, S. Jiang, and M. Lu. 2000. Helical interactions in the HIV-1 gp41 core reveal structural basis for the inhibitory activity of gp41 peptides. Biochemistry 391634-1642. [DOI] [PubMed] [Google Scholar]
- 50.Sia, S. K., P. A. Carr, A. G. Cochran, V. N. Malashkevich, and P. S. Kim. 2002. Short constrained peptides that inhibit HIV-1 entry. Proc. Natl. Acad. Sci. USA 9914664-14669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 919770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, J., T. Chung, and K. Oldenburg. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 467-73. [DOI] [PubMed] [Google Scholar]