Abstract
Novel N4-hydroxy- and 5-methyl-modified β-l-deoxycytidine analogues were synthesized and evaluated as anti-hepatitis B virus (HBV) agents. Their in vitro efficiencies were investigated in HepG2.2.15 cells stably transfected with HBV. β-l-2′,3′-Didehydro-2′,3′-dideoxy-N4-hydroxycytidine (β-l-Hyd4C) was most effective in reducing secreted HBV DNA (50% effective concentration [EC50], 0.03 μM), followed by β-l-2′,3′-dideoxy-3′-thia-N4-hydroxycytidine (EC50, 0.51 μM), β-l-2′,3′-dideoxy-N4-hydroxycytidine (EC50, 0.55 μM), and β-l-5-methyl-2′-deoxycytidine (EC50, 0.9 μM). The inhibition of the presumed target, the HBV DNA polymerase, by the triphosphates of some of the β-l-cytidine derivatives was also assessed. In accordance with the cell culture data, β-l-Hyd4C triphosphate was the most active inhibitor, with a 50% inhibitory concentration of 0.21 μM. The cytotoxicities of some of the 4-NHOH-modified β-l-nucleosides were dramatically lower than those of the corresponding cytidine analogues with the unmodified 4-NH2 group. The 50% cytotoxic concentrations for β-l-Hyd4C in HepG2 and HL-60 cells were 2,500 μM and 3,500 μM, respectively. In summary, our results demonstrate that at least β-l-Hyd4C can be recommended as a highly efficient and extremely selective inhibitor of HBV replication for further investigations.
Complete and sustained suppression of viral replication remains the most important goal in the treatment of patients with chronic hepatitis B virus (HBV) infection. Nucleoside and nucleotide analogues have greatly improved the disease outcome for these patients and have also prevented hepatic decompensation or the development of hepatocellular carcinoma. While β-l-2′,3′-dideoxy-3′-thiacytidine (3TC; lamivudine) has been approved for the treatment of HBV infections, a series of further β-l-nucleosides are under clinical investigation as inhibitors of hepadnavirus infections. These include β-l-2′,3′-dideoxy-3′-thia-5-fluorocytidine (FTC), β-l-2′,3′-didehydro-2′,3′-dideoxy-5-fluorocytidine (β-l-Fd4C), β-l-2′-fluoro-5-methyl-arabinosyluracil (β-l-FMAU), and β-l-thymidine (β-l-dT) (40).
The most important advantage associated with some, but not all, of these β-l-nucleosides seems to be a much higher selectivity for the viral target than for the cellular targets, resulting in reduced cytotoxicity. This quality of β-l-nucleosides stimulated the search for new β-l-nucleosides but also the synthesis of enantiomeric isomers of effective β-d-nucleosides (7).
Some of the unmodified pyrimidine and purine β-l-nucleosides were synthesized for the first time more than 35 years ago (12, 28, 34) but were considered to be metabolized very poorly in mice (14). A systematic biochemical investigation of the β-l-enantiomers was initiated later and demonstrated the anti-human immunodeficiency virus (HIV) activity of β-l-2′,3′-dideoxycytidine (β-l-ddC) in a cellular system (18). In addition, it was shown that β-l-dT might be phosphorylated by herpes simplex virus type 1 (HSV-1) thymidine kinase and further, by cellular enzymes, to β-l-dTTP (35).
As a consequence, the effects of β-l-dTTP on various cellular DNA polymerases, TdT, the Klenow fragment of Escherichia coli DNA polymerase I, and viral DNA polymerases such as HSV-1 DNA polymerase and HIV reverse transcriptase were investigated (6, 32, 38). We showed that β-l-TTP does not influence the activity of any of the cellular DNA polymerases (α, β, γ, δ, or ɛ) or of HIV reverse transcriptase, but we demonstrated for the first time a strong inhibition of human and duck HBV DNA polymerases by β-l-dTTP (concentrations resulting in 50% inhibition of HBV DNA polymerase activity [IC50], 0.46 μM and 1.0 μM, respectively) (37). Investigations on the cellular level and in vivo experiments have confirmed that β-l-dT is a promising and highly selective inhibitor of HBV replication (3). This is also true for β-l-deoxycytidine (β-l-CdR) and β-l-deoxyadenosine (β-l-AdR) (3).
On the basis of these findings, we synthesized a series of β-l-N4-hydroxydeoxycytidine analogues and various β-l-5-methyl-deoxycytidine derivatives, evaluated their potential as inhibitors of HBV replication in cell culture, and tested the triphosphates of some of the β-l-nucleosides directly on HBV DNA polymerase.
Among the newly synthesized compounds, β-l-2′,3′-didehydro-2′,3′-dideoxy-N4-hydroxycytidine (β-l-Hyd4C) emerged as a strong and highly selective inhibitor of HBV replication.
MATERIALS AND METHODS
Compounds.
The N4-hydroxy- and 5-methyl-modified β-l-deoxycytidine analogues were synthesized by methods described previously (E. Matthes, M. von Janta-Lipinski, H. Will, H. Sirma, and A. Funk, 21 October 2005, European patent application PCT/EP2005/011555; E. Matthes, M. von Janta-Lipinski, H. Will, H. Sirma, and L. Lin, 13 September 2004, worldwide patent application WO2005/026186 A1). 1H and 13C nuclear magnetic resonance (NMR) spectra were recorded on a Bruker DRX 600 instrument with tetramethylsilane as an internal standard. ASCA GmbH, Berlin, Germany, performed combustion analyses. High-performance liquid chromatography (HPLC) analyses were performed on a Shimadzu 10AVP system using a reverse-phase analytical column (Zorbax, 300SB-C18; inner diameter, 5 μm; 4.5 by 250 mm), eluting with a linear gradient of 100% A (water) to 100% B (acetonitrile) over 15 min at a flow rate of 1 ml/min.
Data are given for the most effective compounds of each modification: β-l-Hyd4C, representative of the β-l-N4-hydroxydeoxycytidine analogues, and β-l-5-methyl-2′-deoxycytidine (β-l-MetCdR), representative of the β-l-5-methyl-deoxycytidine derivatives.
1-(2,3-Dideoxy-β-l-glycero-pent-2-enofuranosyl)-4-hydroxyamino-pyrimidin-2(1H)-one (β-l-Hyd4C) was obtained as a white amorphous material. 1H NMR (dimethyl sulfoxide [DMSO]-d6) δ 10.96, 10.01 (2H, 2s, NH4, OH-4), 7.64 (1H, s, H-6), 6.58 (1H, d, H-1′), 6.37 (1H, dd, H-3′), 6.23 (1H, dd, H-2′), 5.53 (1H, d, H-5), 5.21 (1H, t, OH-5′), 4.20 to 4.12 (1H, m, H-4′), 3.75 to 3.52 (2H, m, H-5′, H-5‴). 13C NMR (DMSO-d6) δ 35.22 (C-2′), 67.13 (C-5′), 69.27(C-3′), 82.64 (C-1′), 86.47 (C-4′), 112.28 (C-5), 135.62 (C-6), 151.74 (C-2), 165.26 (C-4). UV (H2O, pH 7), λmax 234 nm (ɛ 8,600), 270 nm (ɛ 2,160). HPLC retention time = 8.93 min (purity, 99%). Analysis (C9H11N3O4) was as follows. Calculated: C, 48.00; H, 4.92; N, 18.66. Found: C, 48.21; H, 4.85; N, 18.49.
1-(2-Deoxy-β-l-ribofuranosyl)-5-methylcytosine (β-l-MetCdR) was characterized as a hydrochloride (white crystals from methanol with a few drops of HCl) with a melting point of 151 to 153°C (decomposition). UV (H2O, pH 7) λmax 271 nm (ɛ 10,560), λmin 247 nm (ɛ 1,930); UV (H2O, pH 1) λmax 281 nm (ɛ 12,300), λmin 249 nm (ɛ 5,170). HPLC retention time = 8.44 min (purity, 97%). Analysis (C10H15N3O4, HCl) was as follows. Calculated: C, 43.25; H, 5.81; N, 15.13. Found: C, 43.37; H, 5.68; N, 15.02. 1H NMR (DMSO-d6) δ 7.61 (1H, s, H-6), 7.26 (1H, br s, NH2), 6.77 (1H, broad singlet, NH2), 6.17 to 6.15 (1H, d, H-1′), 5.21 (1H, s, OH-3′), 5.07 (1H, s, OH-5′), 4.20 (1H, d, H-3′), 3.74 (1H, d, H-3′), 3.57 to 3.42 (2H, m, H-5′, H-5‴), 2.07 to 2.04 (1H, m, H-2′), 1.97 to 1.92 (1H, m, H-2‴), 1.85 (3H, s, CH3). 13C NMR (DMSO-d6) δ 13.3 (CH3), 40.2 (C-2′), 61.4 (C-5′), 70.4 (C-3′), 84.6 (C-1′), 87.1 (C-4′), 101.27 (C-5), 138.30 (C-6), 155.20 (C-2), 165.31 (C-4).
β-l-dT was synthesized as described elsewhere (37). 3TC and β-l-ddC were synthesized according to published procedures (2, 17). FTC was kindly provided by R. F. Schinazi, Emory University, Atlanta, GA.
Nucleoside 5′-triphosphates of β-l-deoxycytidine nucleosides were synthesized via the monophosphates (39) as described previously for the corresponding β-d-counterparts (10) and were separated, purified, and characterized by matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry as described elsewhere (37).
Unlabeled deoxynucleoside triphosphates were purchased from Roche Diagnostics, Mannheim,Germany, and [3H]dCTP (52 Ci/mmol) was purchased from Amersham Biosciences, GmbH, Freiburg, Germany.
Estimations of anti-HBV activities of novel β-l-nucleoside analogues in HepG2.2.15 cells.
The antiviral activities of the new compounds were estimated in HepG2.2.15 cells, a human hepatoblastoma cell line with a replication-competent HBV genome that produces infectious progeny viruses (31).
The cells were cultured under standardized conditions in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum as described previously (15). Initially, about 105 cells were plated in 1 ml medium in each well of a 12-well plate. Antiviral drugs were added at various concentrations with daily medium changes. Cell culture supernatants and cells were harvested at treatment day 3, 6, or 9 as indicated, and cell culture supernatants were subjected to PCR analysis of HBV DNA (29).
For this purpose, the cell culture medium was cleared of cell debris, digested with proteinase K, and used for PCR without further manipulation, employing subgenomic forward primer F1 (5′-CTC CAG TTC AGG AAC AGT AAA CCC-3′) and the corresponding reverse primer R1 (5′-TTG TGA GCT CAG AAA GGC CTT GTA AGT TGG CG-3′). The following conditions were used: 10 cycles of 94°C for 40 s, 60°C for 90 s, and 70°C for 2 min; 30 cycles of 94°C for 40 s, 60°C for 90 s, and 70°C for 3.5 min; and 1 cycle of 70°C for 10 min. Serial dilutions of a cloned HBV genome served for calibration of the PCR assay.
Intracellular HBV DNA was investigated after Southern blotting (29). The intracellular viral DNA was purified from the cells after lysis in a buffer containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 0.45% Tween, and 0.45% NP-40. Proteins were digested with proteinase K, and viral DNA was extracted with phenol-chloroform. Ethanol-precipitated DNA was dissolved in water and loaded on an agarose gel for Southern blotting.
PCR products were separated on 1% agarose gels, stained with ethidium bromide, and quantified by a Fluor-S MultiImager (Bio-Rad). For calibration of the PCR, serial dilutions of plasmid pTHBV with known genome equivalents (GE) were included in each PCR run. This standard showed a detection limit of <5 × 103 GE and linearity of the PCR between 5 × 103 and 5 × 105 GE. From dose-response curves, the nucleoside analogue concentrations required for 50% reduction of HBV DNA levels in the medium (50% effective concentrations [EC50]) were estimated.
To investigate the duration of the antiviral effects of certain nucleoside analogues, we performed washout experiments. Cells were treated with the indicated drugs at a concentration corresponding to 5 times the EC50 for 6 days. Subsequently, the drugs were removed, and cells were further incubated for 3 and 6 days. Cells and culture supernatants were harvested, and the amount of HBV DNA was determined.
Estimation of cytotoxicity in HepG2 and HL-60 cells.
Human myeloid leukemia (HL-60) and HepG2 cells were seeded at a density of 50,000 per well and incubated in RPMI medium or Dulbecco's modified Eagle medium, respectively, for 3 days with varying concentrations of the compounds under humidified 5% CO2-air at 37°C. Cell growth was monitored as described previously (33) by a colorimetric reaction based on the capacity of cells to reduce 3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) to formazan. Cytotoxicity levels were expressed as concentrations that inhibited cell growth by 50% (CC50).
Endogenous HBV DNA polymerase assay.
To determine endogenous HBV DNA polymerase activity, about 125 ml of serum from patients with untreated HBV infections with a titer of >107 HBV GE/ml (kindly provided by F. van Bömmel, Charité, Berlin, Germany) was centrifuged at 3,000 rpm. Virus particles in the cleared serum were sedimented using a Beckman SW28 rotor (25,000 rpm for 60 min). The virus pellet was suspended in 7 ml of TKM buffer (50 mM Tris-HCl, pH 7.5, 50 mM KCl, 5 mM MgCl2), layered over a step gradient of 10 ml each of 0.3 M, 0.6 M, and 0.9 M sucrose in TKM buffer, and centrifuged at 25,000 rpm for 20 h.
The purified virus pellet was suspended in 250 μl TKM buffer, sonified, divided into aliquots, and frozen at −80°C (4).
The HBV DNA polymerase assay mixture contained in 30 μl about 2 × 108 to 4 × 108 virus particles (quantified by a VERSANT HBV DNA 3.0 assay with a System 340 bDNA analyzer; Bayer, Wuppertal, Germany), and the assay was performed in 42 mM Tris-HCl (pH 7.5), 34 mM MgCl2, 340 mM KCl, 22 mM β-mercaptoethanol, 0.4% Igepal, 70 μM (each) dTTP, dATP, and dGTP, 1 μCi [3H]dCTP (corresponding to 0.64 μM dCTP), and varying concentrations of the triphosphates of some of the novel β-l-deoxycytidine derivatives and of two reference nucleosides (20). After a 1-h incubation at 37°C, during which time the incorporation of [3H]dCMP into HBV DNA had a linear course (27), 20-μl samples were processed for tritium measurement as described previously (22).
Using the concentration-dependent inhibition curves of HBV DNA synthesis, the IC50 of the β-l-nucleoside triphosphate inhibitors were determined.
RESULTS
Comparative effects of novel β-l-deoxycytidine analogues on HBV replication in vitro.
The HBV producer cell line HepG2.2.15 was used to evaluate the concentration-dependent antiviral activities of the novel N4-hydroxy- and 5-methyl-β-l-deoxycytidine derivatives in comparison to those of well known inhibitors of HBV replication such as 3TC, FTC, β-l-ddC, and β-l-dT. The structures of the most effective compounds, β-l-Hyd4C and β-l-MetCdR, are given in Fig. 1. Antiviral effects were measured by analysis of levels of extracellular HBV DNA from released virions, which decreased in a dose-dependent manner for all compounds tested. The experimental data presented in Fig. 2 demonstrate that β-l-Hyd4C was a more effective inhibitor than 3TC. In order to study the effects of the analogues on the amount of intracellular HBV DNA, Southern blot analysis of the lysed cells was performed. As expected, β-l-Hyd4C proved to be the most effective inhibitor of viral DNA synthesis in treated cells as well (data not shown).
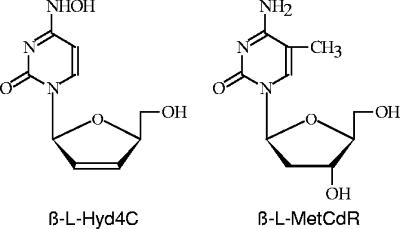
FIG. 1.
Structures of the most effective compounds of the synthesized N4-hydroxy- and 5-methyl-β-l-deoxycytidine analogues: β-l-Hyd4C and β-l-MetCdR.
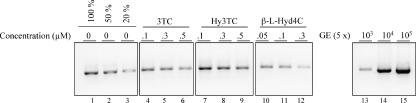
FIG. 2.
Dose-dependent inhibition of HBV DNA release from HepG2.2.15 cells 6 days after incubation with 0.1, 0.3, and 0.5 μM 3TC, 0.1, 0.3, and 0.5 μM Hy3TC, and 0.05, 0.1, and 0.3 μM β-l-Hyd4C. HBV DNA was detected in the medium by PCR. Serial dilutions of a cloned HBV genome (GE) served for calibration. For convenient quantification, dilutions of the PCR product from the untreated control corresponding to 100%, 50%, and 20% of the signal were used. “5 ×” means that 5 × 10y GE (5×) were used for PCR.
The EC50 for all compounds synthesized and tested are summarized in Table 1. The EC50 for 3TC, FTC, β-l-ddC, and β-l-dT were slightly higher than those described previously (3). β-l-Hyd4C was by far the most potent compound in this in vitro system, with an EC50 of 0.03 μM. The EC90 was estimated to be 0.6 μM.
TABLE 1.
Evaluation of novel β-l-deoxycytidine derivatives
| Compound | Anti-HBV activity in HepG2.2.15 cells (EC50)a | Inhibition of growth (CC50)a of:
|
|
|---|---|---|---|
| HepG2 cells | HL-60 cells | ||
| Known inhibitors | |||
| 3TC | 0.1 ± 0.025 | 1,200 ± 265 | 1,400 ± 310 |
| FTC | 0.1 ± 0.028 | 765 ± 145 | 845 ± 174 |
| β-l-ddC | 0.2 ± 0.046 | 85 ± 18.3 | 94 ± 19.5 |
| β-l-dT | 0.3 ± 0.082 | 2,100 ± 438 | 1,760 ± 385 |
| β-l-N4-HyCdR derivatives | |||
| β-l-Hyd4C | 0.03 ± 0.009 | 2,500 ± 572 | 3,500 ± 794 |
| Hy3TC | 0.51 ± 0.13 | 1,320 ± 254 | 1,450 ± 305 |
| β-l-HyddC | 0.55 ± 0.15 | 2,250 ± 487 | 1,600 ± 362 |
| β-l-HyCdR | 2.38 ± 0.56 | 545 ± 144 | 460 ± 98 |
| β-l-HyFCdR | 4.50 ± 1.3 | 920 ± 231 | 1,370 ± 289 |
| HyFTC | 5.0 ± 1.3 | 855 ± 196 | 770 ± 163 |
| β-l-HyMetCdR | 5.04 ± 1.7 | 490 ± 123 | 600 ± 184 |
| β-l-FHyCdR | 25 ± 7.1 | 820 ± 168 | 4,400 ± 910 |
| β-l-N3HyCdR | 50 ± 14.6 | 1,160 ± 241 | 2,000 ± 431 |
| β-l-5-MetCdR derivatives | |||
| β-l-MetCdR | 0.9 ± 0.24 | >2,000 | >2,000 |
| β-l-N3MetCdR | 13 ± 3.9 | 1,750 ± 197 | 1,900 ± 410 |
| β-l-araMetC | 50 ± 15.0 | >2,000 | >2,000 |
| β-l-FMetCdR | >50 | 1,800 ± 350 | 1,650 ± 396 |
| β-l-ddMetC | >50 | 1,550 ± 348 | 950 ± 211 |
| β-l-d4MetC | >50 | 1,400 ± 343 | 600 ± 167 |
Results, in micromolar concentrations, are means ± standard deviations for at least three independent experiments.
β-l-2′,3′-Dideoxy-3′-thia-N4-hydroxycytidine (Hy3TC), β-l-2′,3′-dideoxy-N4-hydroxycytidine (β-l-HyddC), and β-l-MetCdR gave EC50 between 0.51 μM and 0.9 μM. A second group of compounds with EC50 between 2.38 μM and 13 μM includes β-l-N4-hydroxy-2′-deoxycytidine (β-l-HyCdR), β-l-5-fluoro-N4-hydroxy-2′-deoxycytidine (β-l-HyFCdR), β-l-2′,3′-dideoxy-3′-thia-5-fluoro-N4-hydroxycytidine (HyFTC), β-l-5-methyl-N4-hydroxy-2′-deoxycytidine (β-l-HyMetCdR), and β-l-3′-azido-2′, 3′-dideoxy-5-methylcytidine (β-l-N3MetCdR). A third group of nucleoside analogues including β-l-2′,3′-dideoxy-3′-fluoro-N4-hydroxycytidine (β-l-FHyCdR), β-l-3′-azido-2′,3′-dideoxy-N4-hydroxycytidine (β-l-N3HyCdR), β-l-5-methylcytosine arabinoside (β-l-araMetC), β-l-2′,3′-dideoxy-3′-fluoro-5-methylcytidine (β-l-FMetCdR), β-l-2′,3′-dideoxy-5-methylcytidine (β-l-ddMetC), and β-l-2′,3′-didehydro-2′,3′-dideoxy-5-methylcytidine (β- l-d4MetC) were inactive (EC50, 25 μM to >50 μM).
Cellular toxicity of the β-l-nucleosides.
The compounds were tested in vitro for their cytotoxicity. Table 1 shows that they exhibited no significant antiproliferative activity against HepG2 and HL-60 cells. The most active anti-HBV compounds (β-l-Hyd4C, Hy3TC, β-l-HyddC, and β-l-MetCdR) have no effect on proliferation. Remarkably, for β-l-Hyd4C, CC50 of 2,500 μM for HepG2 cells and 3,500 μM for HL-60 cells were found.
Time dependence and recovery of the antiviral effects of the new analogues.
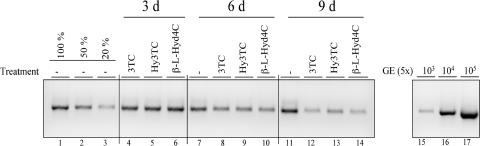
For the compounds β-l-Hyd4C and Hy3TC, we investigated the time dependency of the antiviral effect in cell culture and measured the extracellular progeny virus DNA in the culture medium of treated HepG2.2.15 cells in comparison with those for 3TC. As judged from the HBV DNA-specific PCR, the amount of HBV DNA in the supernatant of treated cells decreased over time (Fig. 3). The antiviral activities of the compounds were evident at day 6 and continued at day 9.
FIG. 3.
Time-dependent inhibition of HBV DNA release from HepG2.2.15 cells by 3TC, Hy3TC, and β-l-Hyd4C. The cells were treated with the indicated drugs for 3, 6, and 9 days. 3TC was used at 0.3 μM, Hy3TC at 0.5 μM, and β-l-Hyd4C at 0.1 μM. HBV DNA of the supernatants was analyzed by PCR. Serial dilutions of a cloned HBV genome served for calibration of the PCR assay (GE). For convenient quantification, dilutions of the PCR product from the untreated control corresponding to 100%, 50%, and 20% of the signal were used. −, no treatment.
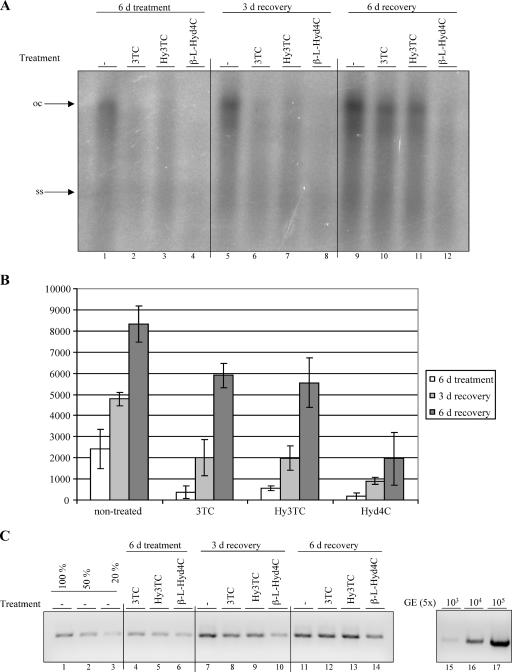
To test how long the antiviral activities of the compounds lasted upon removal of the drugs, we performed a washout assay. Cells were treated with 3TC, Hy3TC, or β-l-Hyd4C at concentrations 5 times the evaluated EC50 for 6 days. Then the drugs were removed, and cells were further cultivated for 3 and 6 days. The amounts of extracellular and intracellular HBV DNA were investigated for each time point by PCR and Southern blotting, respectively. As judged from both assays, time-dependent increases in intracellular and extracellular viral DNA levels were observed in 3TC- and Hy3TC-treated cells after removal of the drugs (Fig. 4A to C). The recovery was strongest at day 6 compared to day 3. This shows that viral replication recovers from the drug treatment as expected, and the data are consistent with published data (24).
FIG. 4.
Recovery from the antiviral effect after cessation of drug treatment. 3TC was used at 1.5 μM, Hy3TC at 2.5 μM, and β-l-Hyd4C at 0.5 μM. (A) Southern blot analysis of HBV DNA of HepG2.2.15 cells treated for 6 days and incubated for a further 3 or 6 days in drug-free medium. Arrows indicate the open circular (oc) and single-stranded (ss) forms of HBV DNA. (B) Quantitative evaluation of HBV DNA of these cells by phosphorimaging. Means and standard deviations from two independent experiments are shown. The persistence of antiviral activity of β-l-Hyd4C upon removal of the drug was statistically significant compared to that for an untreated culture (P < 0.0266). (C) PCR analysis of viral DNA in the supernatants of the same cells. Serial dilutions of a cloned HBV genome and dilutions of PCR products of untreated cells served for the calibration and quantification of the PCR signals. d, day; −, no treatment.
In contrast to those for 3TC and Hy3TC, recovery after β-l-Hyd4C treatment was slower and was incomplete even 6 days after removal. This indicates that the antiviral activity of this drug is more pronounced and is sustained over a longer time than those of 3TC and Hy3TC. The delayed resumption of HBV replication was not caused by a decreased number of cells, because the antiproliferative activity of β-l-Hyd4C is lower than that of 3TC or Hy3TC (Table 1).
Inhibition of HBV DNA polymerase.
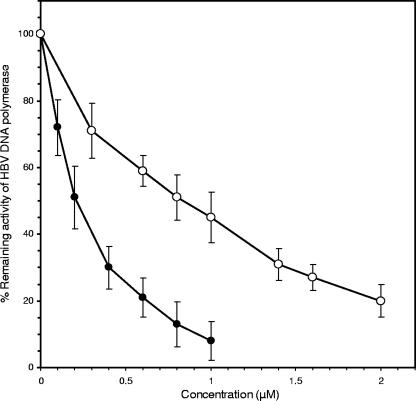
To determine the direct effects of some of the novel β-l-nucleosides at the presumed viral target, we synthesized their triphosphates and examined them as inhibitors of HBV DNA polymerase. From concentration-dependent inhibition curves, as shown for β-l-Hyd4C triphosphate and β-l-MetCdR triphosphate (Fig. 5), we estimated the IC50 given in Table 2. Table 2 demonstrates that β-l-Hyd4C triphosphate inhibited the presumed viral target more effectively than did 3TC triphosphate and β-l-dTTP (IC50, 0.21 μM versus 0.30 and 0.46 μM, respectively). Also, the triphosphates of β-l-N3MetCdR, β-l-MetCdR, and β-l-HyddC were found to be strong inhibitors of HBV DNA polymerase activity (IC50, 0.85 to 0.99 μM), whereas the triphosphates of β-l-HyMetCdR, β-l-HyCdR, β-l-d4MetC, β-l-ddMetC, and β-l-araMetC were less effective (IC50, 4.0 to 10.8 μM).
FIG. 5.
Inhibition of HBV DNA polymerase activity by β-l-Hyd4C triphosphate (•) and β-l-MetCdR triphosphate (○). Means and standard deviations from three independent experiments are given.
TABLE 2.
Inhibition of HBV DNA polymerase by triphosphates of novel β-l-deoxycytidine analogues in comparison to 3TC triphosphate and β-l-dTTP
| Compound | IC50 (μM)a for the triphosphate |
|---|---|
| Known inhibitors | |
| 3TC (lamivudine) | 0.30 ± 0.05 |
| β-l-dT | 0.46 ± 0.08b |
| β-l-N4-HyCdR derivatives | |
| β-l-Hyd4C | 0.21 ± 0.04 |
| β-l-HyddC | 0.99 ± 0.16 |
| β-l-HyMetCdR | 4.0 ± 0.63 |
| β-l-HyCdR | 6.0 ± 0.84 |
| β-l-5-MetCdR derivatives | |
| β-l-N3MetCdR | 0.85 ± 0.17 |
| β-l-MetCdR | 0.90 ± 0.16 |
| β-l-FMetCdR | 1.9 ± 0.42b |
| β-l-d4MetC | 6.2 ± 0.99 |
| β-l-ddMetC | 6.5 ± 1.1 |
| β-l-araMetC | 10.8 ± 1.9 |
Results are means ± standard deviations from at least three independent experiments.
Result from reference 37.
DISCUSSION
In view of the experience that some nucleosides with the unnatural β-l configuration were more powerful and selective antiviral agents than their corresponding β-d isomers, we synthesized and evaluated the corresponding opposite optical isomers of some of the β-d-MetCdR derivatives that we studied several years ago. These investigations have shown that the triphosphates of some of the β-d-MetCdR derivatives are extremely powerful inhibitors of HBV DNA polymerase in vitro (e.g., β-d-3′-FMetdCTP [IC50, 0.03 μM]) (20), though they proved to be less active under cell culture and in vivo conditions (19, 21).
Now we found for the corresponding β-l analogues that β-l-FMetCdR, β-l-ddMetC, β-l-d4MetC, and β-l-araMetC as well as their triphosphates were unable to inhibit either HBV replication in HepG2.2.15 cells or cell-free HBV DNA polymerase activity (EC50, ≥50 μM; IC50, 1.9 μM to 10.8 μM).
In contrast, β-l-N3MetdCTP efficiently inhibited HBV DNA polymerase activity (IC50, 0.85 μM) but was nearly inactive as the nucleoside in HepG2.2.15 cells (EC50, 13 μM). Additionally, we synthesized β-l-MetCdR, which gained interest after the discovery of β-l-CdR as an efficient anti-HBV agent (EC50, 0.24 μM) (3).
We show here that β-l-MetCdR is a potent anti-HBV agent in cell culture (EC50, 0.9 μM) and that its triphosphate is an efficient inhibitor of HBV DNA polymerase activity (IC50, 0.9 μM).
It remains to be seen whether the monophosphate β-l- MetdCMP could be deaminated to β-l-dTMP inside cells, a reaction that might produce two anti-HBV active compounds: β-l-MetdCTP and β-l-dTTP. At least for β-l-dCMP, deamination in HepG2 cells has been described (8).
A further modification we considered promising is the replacement of the 4-NH2 group by a 4-NHOH group in certain β-l-deoxycytidine analogues.
The corresponding β-d enantiomer of deoxycytidine, β-d-N4-hydroxydeoxycytidine, was synthesized and investigated 40 years ago. It was found that this compound could be phosphorylated in L5178Y leukemic cells and that the monophosphate inhibited thymidylate synthase (23).
The corresponding β-d-ribo derivative, β-d-N4-hydroxycytidine, was shown to be a strong mutagen for bacterial cells, phages, and Neurospora crassa (5, 13, 25). Later, β-d-N4-hydroxycytidine was demonstrated to be an efficient inhibitor of bovine viral diarrhea virus as well as in the hepatitis C virus replicon system and of severe acute respiratory syndrome-associated coronavirus (1, 36). These studies initiated the synthesis of further β-d- and β-l-N4-hydroxycytidine analogues with a 3′-deoxy and a 2′-fluoro modification, respectively (11, 33).
We describe here the synthesis of novel 4-NHOH-modified β-l-deoxynucleosides and their inhibitory effects on HBV replication.
Of these compounds, β-l-Hyd4C emerged as the most effective (EC50, 0.03 μM), followed by Hy3TC and β-l-HyddC (EC50, 0.51 μM and 0.55 μM, respectively). The corresponding derivatives with the unmodified 4-NH2 group, β-l-d4C, 3TC, and β-l-ddC, suppressed HBV replication more efficiently (EC50, 0.01 μM, 0.017 μM, and 0.01 μM, respectively) (16, 17), indicating a decline in antiviral activity associated with the 4-NHOH modification.
Surprisingly, the cytotoxicity of these compounds seems to be much more decreased than their anti-HBV activity. Thus, β-l-Hyd4C and β-l-HyddC displayed extremely low antiproliferative activities, as demonstrated by the CC50 of 2,500 μM for β-l-Hyd4C and 2,250 μM for β-l-HyddC in HepG2 cells. In comparison, the corresponding derivatives β-l-d4C and β-l-ddC with the unmodified 4-NH2 group have CC50 of 20 μM and 70 μM in CEM cells (16, 17). As a consequence, the selectivity index of both 4-NHOH-modified compounds increased dramatically.
Comparative analysis of the potencies of the antiviral substances showed that time-dependent inhibition of DNA replication in the cells occurs after treatment with all drugs tested. Prolongation of the treatment up to 9 days would reduce progeny virus formation in a manner even more pronounced than that observed after 6 days. The washout experiments suggest that the antiviral effect of β-l-Hyd4C seems to persist to a certain degree even after removal of the drug. The recovery of the level of HBV DNA seems to be delayed more effectively by β-l-Hyd4C than by 3TC, as shown for cells incubated 6 days in drug-free medium.
It could be argued that the 4-NHOH group of the β-l-deoxycytidine derivatives might be reduced inside the cells to the corresponding NH2 group, which was described as a primary catabolic step for the ribonucleoside β-d-N4-hydroxycytidine in Salmonella enterica serovar Typhimurium and liver cells (9, 26). Such a reaction of our β-l-N4-hydroxydeoxycytidine analogues should produce drugs with much higher cytotoxicity. The extremely low cytotoxicity observed in our study allows us to assume that this kind of modification is unlikely.
This view is supported further by the fact that Hy3TC is inactive against HIV replication (E. Matthes et al., unpublished data). If the NHOH group of Hy3TC were reduced to the NH2 group, the resulting 3TC should display the well-known highly efficient inhibition of HIV replication (EC50, 0.002 μM) (30).
Furthermore, our results obtained with the presumed target, HBV DNA polymerase, demonstrate that the 4-NHOH group itself is responsible for the strong inhibition by triphosphates of β-l-Hyd4C and β-l-HyddC (IC50, 0.21 μM and 0.99 μM).
Although the cellular metabolism of both groups of β-l-deoxycytidine derivatives presented here remains to be investigated, the results obtained with β-l-Hyd4C and some other derivatives indicate a unique and high selectivity index, which makes them candidates for further investigations as potential anti-HBV agents.
Acknowledgments
We thank N. Lohrengel for excellent technical support and F. van Bömmel, Charité, Berlin, Germany for providing the HBV serum samples.
The Max Delbrück-Centrum is part of the Helmholtz Gesellschaft and is supported by the Bundesministerium für Bildung und Forschung. The Heinrich-Pette-Institute is supported by the Freie und Hansestadt Hamburg and the Bundesministerium für Gesundheit und Soziale Sicherung. This work was supported by grants from the DFG and by the German Competence Network for Viral Hepatitis (Hep-Net), funded by the German Ministry of Education and Research (BMBF), grant TP13.1.
Footnotes
Published ahead of print on 2 April 2007.
REFERENCES
- 1.Barnard, D. L., V. D. Hubbard, J. Burton, D. F. Smee, J. D. Morrey, M. J. Otto, and R. S. Sidwell. 2004. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARS CoV) by calpain inhibitors and β-d-N4-hydroxycytidine. Antivir. Chem. Chemother. 1515-22. [DOI] [PubMed] [Google Scholar]
- 2.Beach, J. W., L. S. Jeong, A. J. Alves, D. Pohl, H. O. Kim, C. N. Chang, S. L. Doong, R. F. Schinazi, Y. C. Cheng, and C. K. Chu. 1992. Synthesis of enantiomerically pure (2′R,5′S)-(−)-1-(2-hydroxymethyloxathiolan-5-yl)cytosine as a potent antiviral agent against hepatitis B virus (HBV) and human immunodeficiency virus (HIV). J. Org. Chem. 572217-2219. [Google Scholar]
- 3.Bryant, M. L., E. Bridges, L. Placidi, A. Faraj, A.-G. Loi, C. Pierra, D. Dukhan, G. Gosselin, J.-L. Imbach, B. Hernandez, A. Juodawlkis, B. Tennent, B. Korba, P. Cote, P. Marion, E. Cretton-Scott, R. Schinazi, and J. P. Sommadossi. 2001. Antiviral l-nucleosides specific for hepatitis B virus infection. Antimicrob. Agents Chemother. 45229-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis, M. G., J. E. Wilson, N. A. VanDraanen, W. H. Miller, G. A. Freeman S. M. Daluge, F. L. Boyd, A. E. Aulabaugh, G. R. Painter, and L. R. Boone. 1996. DNA polymerase activity of hepatitis B virus particles: differential inhibition by l-enantiomers of nucleotide analogs. Antivir. Res. 30133-145. [DOI] [PubMed] [Google Scholar]
- 5.de Serres, F. J., and H. E. Brockman. 1993. Comparison of the spectra of genetic damage in N4-hydroxycytidine-induced ad-3 mutations between nucleotide excision repair-proficient and-deficient heterocaryons of Neurospora crassa. Mutat. Res. 285145-163. [DOI] [PubMed] [Google Scholar]
- 6.Focher, F., G. Maga, A. Bendiscioli, M. Capobianco, F. Colonna, A. Garbesi, and S. Spadari. 1995. Stereospecificity of human DNA polymerases α, β, γ, δ, and ɛ, HIV reverse transcriptase, HSV-1 DNA polymerase, calf thymus terminal transferase and Escherichia coli DNA polymerase I in recognizing d- and l-thymidine 5-triphosphate as substrate. Nucleic Acids Res. 232840-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gumina, G., Y. Chong, H. Choo, G.-Y. Song, and C. K. Chu. 2002. l-Nucleosides: antiviral activity and molecular mechanism. Curr. Top. Med. Chem. 21065-1086. [DOI] [PubMed] [Google Scholar]
- 8.Hernandez-Santiago, B., L. Placidi, E. Cretton-Scott, A. Faraj, E. G. Bridges, M. L. Bryant, J. Rodriguez-Orengo, J.-L. Imbach, G. Gosselin, C. Pierra, D. Dukhan, and J. P. Sommadossi. 2002. Pharmacology of β-l-thymidine and β-l-2′-deoxycytidine in HepG2 cells and primary human hepatocytes: relevance to chemotherapeutic efficacy against hepatitis B virus. Antimicrob. Agents Chemother. 461728-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hernandez-Santiago, B., T. Beltran, L. Stuyver, C. K. Chu, and R. Schinazi. 2004. Metabolism of the anti-hepatitis C virus nucleoside β-d-N4-hydroxycytidine in different liver cells. Antimicrob. Agents Chemother. 484636-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoard, D. E., and D. G. Ott. 1965. Conversion of mono-and oligodeoxyribonucleotides to 5′-triphosphates. J. Am. Chem. Soc. 871785-1788. [DOI] [PubMed] [Google Scholar]
- 11.Hollecker, L., H. Choo, Y. Chong, C. K. Chu, S. Lostia, T. R. McBrayer, L. J. Stuyver, J. C. Mason, J. Du, S. Rachakonda, J. Shi, R. F. Schinazi, and K. A. Watanabe. 2004. Synthesis of β-enantiomers of N4-hydroxy-3′-deoxypyrimidine nucleosides and their evaluation against bovine viral diarrhoea and hepatitis C virus in cell culture. Antivir. Chem. Chemother. 1443-55. [DOI] [PubMed] [Google Scholar]
- 12.Holy, A., and F. Sorm. 1969. Preparation of some β-l-ribonucleosides, their 2′,3′-cyclic phosphates. Collect. Czech. Chem. Commun. 343383-3391. [Google Scholar]
- 13.Janion, C., and E. Kajtaniak. 1979. Mutagenesis induced in amber P22 phages by base analogues. Mutat. Res. 62191-195. [DOI] [PubMed] [Google Scholar]
- 14.Jurovcik, M., and A. Holy. 1976. Metabolism of pyrimidine l-nucleosides. Nucleic Acids Res. 32143-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korba, B. E., and J. L. Gerin. 1992. Use of a standardized cell culture assay to assess activities of nucleoside analogs against hepatitis B virus replication. Antivir. Res. 1955-70. [DOI] [PubMed] [Google Scholar]
- 16.Lin, T.-S., M.-Z. Luo, M.-C. Liu, S. B. Pai, E. Gullen, G. E. Dutschman, and Y.-C. Cheng. 1994. Synthesis and biological evaluation of 2′,3′-dideoxy-l-pyrimidine nucleosides as potential antiviral agents against human immunodeficiency virus (HIV) and hepatitis B virus (HBV). J. Med. Chem. 37798-803. [DOI] [PubMed] [Google Scholar]
- 17.Lin, T.-S., M.-Z. Luo, M.-C. Liu, Y.-L. Zhu, E. Gullen, G. E. Dutschman, and Y.-C. Cheng. 1996. Design and synthesis of 2′,3′-dideoxy-2′,3′-didehydro-β-l-cytidine (β-l-d4C) and 2′,3′-dideoxy-2′,3′-didehydro-β-l-5-fluorocytidine (β-l-Fd4C), two exceptionally potent inhibitors of human hepatitis B virus (HBV) and potent inhibitors of human immunodeficiency virus (HIV) in vitro. J. Med. Chem. 391757-1759. [DOI] [PubMed] [Google Scholar]
- 18.Mansuri, M. M., V. Farina, J. E. Starrett, D. A. Benigni, V. Brankovan, and J. C. Martin. 1991. Preparation of the geometric isomers of DDC, DDA, D4C, and D4T as potential anti-HIV agents. Bioorg. Med. Chem. Lett. 165-68. [Google Scholar]
- 19.Matthes, E., P. Langen, M. von Janta-Lipinski, H. Will, H. C. Schröder, H. Merz, B. E. Weiler, and W. E. G. Müller. 1990. Potent inhibition of hepatitis B virus production in vitro by modified nucleosides. Antimicrob. Agents Chemother. 341986-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthes, E., K. Reimer, M. von Janta-Lipinski, H. Meisel, and C. Lehmann. 1991. Comparative inhibition of hepatitis B virus DNA polymerase and cellular DNA polymerases by triphosphates of sugar-modified 5-methyl-deoxycytidines and other nucleoside analogues. Antimicrob. Agents Chemother. 351254-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthes, E., M. von Janta-Lipinski, H. Will, H. C. Schröder, H. Merz, R. Steffen, and W. E. G. Müller. 1992. Inhibition of hepatitis B virus production by modified 2′,3′-dideoxythymidine and 2′,3′-dideoxy-5-methylcytidine derivatives. Biochem. Pharmacol. 431571-1577. [DOI] [PubMed] [Google Scholar]
- 22.Meisel, H., K. Reimer, M. von Janta-Lipinski, D. Bärwolff, and E. Matthes. 1990. Inhibition of hepatitis B virus DNA polymerase by 3′-fluorothymidine triphosphate and other modified nucleoside triphosphate analogs. J. Med. Virol. 30137-141. [DOI] [PubMed] [Google Scholar]
- 23.Nelson, D. J., and C. E. Carter. 1966. Inhibition of the biosynthesis of thymidylic acid by 4-N-hydroxy-2′-deoxycytidine in L5178Y leukemic cells. Mol. Pharmacol. 2248-258. [PubMed] [Google Scholar]
- 24.Pai, S. B., S.-H. Liu, Y.-L. Zhu, C. K. Chu, and Y.-C. Cheng. 1996. Inhibition of hepatitis B virus by a novel l-nucleoside, 2′-fluoro-5-methyl-β-l-arabinofuranosyl uracil. Antimicrob. Agents Chemother. 40380-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popowska, E., and C. Janion. 1974. N4-hydroxycytidine—a new mutagen of a base analogue type. Biochem. Biophys. Res. Commun. 56459-466. [DOI] [PubMed] [Google Scholar]
- 26.Popowska, E., and C. Janion. 1977. The N4-hydroxycytidine reduction system in toluenized cells of Salmonella typhimurium. Acta Biochim. Pol. 24197-205. [PubMed] [Google Scholar]
- 27.Reimer, K., E. Matthes, M. von Janta-Lipinski, and H. Meisel. 1991. Inhibition of hepatitis B virus DNA polymerase by thymidine triphosphate analogues in vitro. Antivir. Chem. Chemother 2249-253. [Google Scholar]
- 28.Robins, M. J., T. A. Khwaja, and R. K. Robins. 1970. Purine nucleosides. XXIX. The synthesis of 2′-deoxy-l-adenosine and 2′-deoxy-l-guanosine and their alpha anomers. J. Org. Chem. 35636-639. [DOI] [PubMed] [Google Scholar]
- 29.Schildgen, O., H. Sirma, A. Funk, C. Olotu, U. C. Wend, H. Hartmann, M. Helm, J. K. Rockstroh, W. R. Willems, H. Will, and W. H. Gerlich. 2006. Variant of hepatitis B virus with primary resistance to adefovir. N. Engl. J. Med. 3541807-1812. [DOI] [PubMed] [Google Scholar]
- 30.Schinazi, R. F., A. McMillan, D. Cannon, et al. 1992. Selective inhibition of human immunodeficiency viruses by racemates and enantiomers of cis-5-fluoro-1-[2-(hydroxymethyl)-1,3-oxathiolan-5-yl]-cytosine. Antimicrob. Agents Chemother. 362423-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sells, M. A., M. L. Chen, and G. Acs. 1987. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B DNA. Proc. Natl. Acad. Sci. USA 841005-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semizarov, D. G., A. A. Arzumanov, N. B. Dyatkina, A. Meyer, S. Vichier-Guerre, G. Gosselin, B. Rayners, J.-L. Imbach, and A. A. Krayevski. 1997. Stereoisomers of deoxynucleoside 5′-triphosphates as substrates for template-dependent and-independent DNA polymerases. J. Biol. Chem. 2729556-9560. [DOI] [PubMed] [Google Scholar]
- 33.Shi, J., J. Du, T. Ma, K. W. Pankiewicz, S. E. Patterson, P. M. Tharnish, T. R. McBrayer, L. J. Stuyver, M. J. Otto, C. K. Chu, R. F. Schinazi, and K. A. Watanabe. 2005. Synthesis and antiviral activity of a series of d- and l-2′-deoxy-2′-fluororibonucleosides in the subgenomic HCV replicon system. Bioorg. Med. Chem. 131641-1652. [DOI] [PubMed] [Google Scholar]
- 34.Smejkal, J., and F. Sorm. 1964. Nucleic acid components and their analogues. 53. Preparation of 1-(2-deoxy-β-l-ribofuranosyl)thymine (l-thymidine). Collect. Czech. Chem. Commun. 292809-2813. [Google Scholar]
- 35.Spadari, S., G. Maga, F. Focher, G. Ciarrocchi, R. Manservigi, F. Arcamone, M. Capobianco, A. Carcuro, F. Colonna, S. Iotti, and A. Garbesi. 1992. l-Thymidine is phosphorylated by herpes simplex type 1 thymidine kinase and inhibits viral growth. J. Med. Chem. 354214-4220. [DOI] [PubMed] [Google Scholar]
- 36.Stuyver, L. J., T. Whitaker, T. R. McBrayer, B. I. Hernandez-Santiago, S. Lostia, P. M. Tharnish, M. Ramesh, C. K. Chu, R. Jordan, J. Shi, S. Rachakonda, K. A. Watanabe, M. J. Otto, and R. F. Schinazi. 2003. Ribonucleoside analogue that blocks replication of bovine viral diarrhea and hepatitis C viruses in culture. Antimicrob. Agents Chemother. 47244-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Janta-Lipinski, M., B. Costisella, H. Ochs, U. Hübscher, P. Hafkemeyer, and E. Matthes. 1998. Newly synthesized l-enantiomers of 3′-fluoro-modified β-2′-deoxyribonucleoside triphosphates inhibit hepatitis B DNA polymerase but not the five cellular DNA polymerases α, β, γ, δ, and ɛ. J. Med. Chem. 412040-2046. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi, T., N. Iwanami, K. Shudo, and M. Saneyoshi. 1994. Chiral discrimination of enantiomeric 2′-deoxythymidine-5′-triphosphate by HIV-reverse transcriptase and eukaryotic DNA polymerases. Biochem. Biophys. Res. Commun. 2001023-1027. [DOI] [PubMed] [Google Scholar]
- 39.Yoshikawa, M., T. Kato, and T. Takenishi. 1967. A novel method for phosphorylation of nucleosides to 5′-monophosphates. Tetrahedron Lett. 505065-5068. [DOI] [PubMed] [Google Scholar]
- 40.Younger, H. M., A. J. Bathgate, and P. C. Hayes. 2004. Nucleoside analogues for the treatment of chronic hepatitis B. Aliment. Pharmacol. Ther. 201211-1230. [DOI] [PubMed] [Google Scholar]