Abstract
We characterize in vitro resistance to and demonstrate the in vivo efficacy of two antisense phosphorodiamidate morpholino oligomers (PMOs) against West Nile virus (WNV). Both PMOs were conjugated with an Arg-rich peptide. One peptide-conjugated PMO (PPMO) binds to the 5′ terminus of the viral genome (5′-end PPMO); the other targets an essential 3′ RNA element required for genome cyclization (3′ conserved sequence I [3′ CSI] PPMO). The 3′ CSI PPMO displayed a broad spectrum of antiflavivirus activity, suppressing WNV, Japanese encephalitis virus, and St. Louis encephalitis virus, as demonstrated by reductions in viral titers of 3 to 5 logs in cell cultures, likely due to the absolute conservation of the 3′ CSI PPMO-targeted sequences among these viruses. The selection and sequencing of PPMO-resistant WNV showed that the 5′-end-PPMO-resistant viruses contained two to three mismatches within the PPMO-binding site whereas the 3′ CSI PPMO-resistant viruses accumulated mutations outside the PPMO-targeted region. The mutagenesis of a WNV infectious clone demonstrated that the mismatches within the PPMO-binding site were responsible for the 5′-end PPMO resistance. In contrast, a U insertion or a G deletion located within the 3′-terminal stem-loop of the viral genome was the determinant of the 3′ CSI PPMO resistance. In a mouse model, both the 5′-end and 3′ CSI PPMOs (administered at 100 or 200 μg/day) partially protected mice from WNV disease, with minimal to no PPMO-mediated toxicity. A higher treatment dose (300 μg/day) caused toxicity. Unconjugated PMOs (3 mg/day) showed neither efficacy nor toxicity, suggesting the importance of the peptide conjugate for efficacy. The results suggest that a modification of the peptide conjugate composition to reduce its toxicity yet maintain its ability to effectively deliver PMO into cells may improve PMO-mediated therapy.
The genus Flavivirus in the family Flaviviridae is composed of more than 70 viruses (27). Many flaviviruses are arthropod borne and cause significant human diseases. Among these, the four serotypes of dengue virus (DENV), yellow fever virus (YFV), West Nile virus (WNV), Japanese encephalitis virus (JEV), and tick-borne encephalitis virus are categorized as emerging global pathogens (7). The World Health Organization has estimated the numbers of annual human cases of DENV (52), YFV (54), and JEV (53) infections to be more than 50 million, 200,000, and 50,000, respectively. Since 1999, WNV has spread rapidly throughout the Western Hemisphere, including the contiguous United States, Canada, Mexico, the Caribbean, and parts of Central and South America (24); from 1999 to 2006, the Centers for Disease Control and Prevention reported 23,786 human cases of WNV infection and 934 deaths caused by the disease in the United States alone (Centers for Disease Control and Prevention website, http://www.cdc.gov/ncidod/dvbid/westnile/index.htm). Vaccines for humans are presently available only for YFV, JEV, and tick-borne encephalitis virus infections (7); no clinically approved vaccine or antiviral therapy for humans is available for WNV and DENV infections. A number of approaches to the development of efficacious vaccines and therapies for flavivirus infections are being actively pursued (24).
Flavivirus virions are spherical and contain a single-stranded, plus-sense RNA genome of approximately 11 kb (27). The genomic RNA carries a single long open reading frame flanked by a 5′ untranslated region (5′ UTR) and a 3′ UTR. The open reading frame encodes a polyprotein which is processed by viral and cellular proteases into three structural proteins (capsid, premembrane or membrane, and envelope) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). The structural proteins are involved primarily in viral particle formation; the nonstructural proteins are required for viral RNA replication (27), virion assembly (25, 29), and the evasion of host immune responses (28, 34). Upon the translation of the genomic RNA in host cells, viral proteins are assembled into replication complexes through interactions with viral RNA and host factors in a finely modulated fashion. The replication complexes then transcribe the plus-strand genome into a complementary minus-strand RNA, which in turn serves as the template for the synthesis of more plus-strand genomic RNA (27).
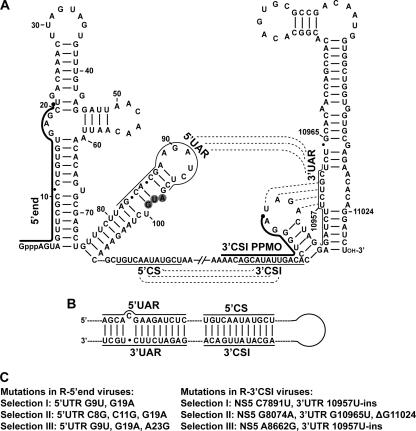
The 5′ and 3′ UTRs of the flavivirus genome contain conserved sequence (CS) elements and can form conserved stem-loop structures (5, 6, 31, 41). As depicted using a WNV sequence (Fig. 1A), three RNA interactions were previously reported: (i) the mosquito-borne flavivirus-conserved 5′ CS element (located at the N-terminal coding region of the capsid gene) forms a perfect 12-nucleotide base pairing with the 3′ conserved sequence I (3′ CSI) element (located in the 3′ UTR) (18), (ii) a 5′ upstream AUG region (5′ UAR) element may interact with a 3′ UAR sequence (located at the bottom portion of the 3′ stem-loop of the genome) (2), and (iii) a pseudoknot may be formed within the 3′-terminal two-stem-loop structure (42). Among the above-listed interactions, the base pairings mediated by the 5′ CS-3′ CSI and 5′ UAR-3′ UAR interactions were previously shown to cyclize the flavivirus genome (Fig. 1B) (2, 4, 10, 21, 23, 30, 33). Besides the RNA-RNA interactions, host and viral proteins were previously reported to bind to the 3′ UTR of the flavivirus genome (3, 9, 12, 26, 46, 47). Mutagenesis analyses using infectious cDNA clones and replicons of various flaviviruses have shown that the conserved RNA sequences and structures are critical for viral replication (4, 8, 10, 20, 21, 23, 30, 32, 33, 48, 49, 55, 57). These results suggest that RNA elements within the 5′ and 3′ UTRs are critical for viral multiplication and, therefore, represent rational targets for antiviral therapy.
FIG. 1.
Sequences and structures of the PPMO-targeted WNV RNA regions. (A) Terminal stem-loop structures and potential RNA-RNA interactions within the WNV genome. Secondary structures formed by the 5′- and 3′-terminal sequences of the WNV genome are depicted. Two PPMOs, the 5′-end PPMO and the 3′ CSI PPMO, are indicated as thick lines with filled circles (representing an Arg-rich peptide [11]) at the 5′ ends. The AUG initiation codon of the open reading frame is shaded in gray. Three RNA interactions are denoted by dashed lines, the 5′ UAR-3′ UAR and 5′ CS-3′ CSI interactions and a pseudoknot base pairing (located at the 3′-terminal stem-loops). The sequences involved in the 5′ UAR-3′ UAR and 5′ CS-3′ CSI interactions are indicated by thin lines. Nucleotide positions are numbered based on the full-length sequence of the WNV genome. Gppp indicates a gap structure of viral genome. (B) Potential genome cyclization of WNV through the 5′ UAR-3′ UAR and 5′ CS-3′ CSI base pairings. (C) Adaptive mutations derived from viruses resistant to the 5′-end PPMO (left panel) or the 3′ CSI PPMO (right panel).
In support of the above-mentioned hypothesis, we previously found that a panel of phosphorodiamidate morpholino oligomers (PMOs) whose sequences are complementary to RNA elements located in the 5′ and 3′ termini of the WNV genome (including the 5′ UAR, 5′ CSI, 3′ CSI, and 3′ UAR and the pseudoknot sequences depicted in Fig. 1A) had various antiviral activities in cell culture (11). PMOs are uncharged, water-soluble, nuclease-resistant antisense agents that are usually synthesized to a length of about 20 subunits and contain purine and pyrimidine bases attached to a backbone composed of morpholine rings joined by phosphorodiamidate intersubunit linkages (45). All of our previous PMOs contained a 5′-terminal Arg-rich peptide conjugate. The 5′ peptide conjugate greatly facilitates the delivery of the molecules into cultured cells (11). Among the tested peptide-conjugated PMOs (PPMOs), two exhibited the greatest potency: one targeting the 5′-terminal 20 nucleotides of the viral genome (5′-end PPMO) and another targeting the conserved 3′ CSI element (3′ CSI PPMO); both PPMOs reduced viral titers by several logs at a 5 μM concentration without cytotoxicity (11). In agreement with our results, Kinney and coworkers showed that PPMOs targeting the 5′-end and 3′ CSI regions of DENV RNA also potently inhibit the replication of all four serotypes of DENV in cell culture (22). These results suggest that PPMOs are potential antiviral agents for flavivirus disease therapy. However, the in vivo efficacy of these inhibitors has not been determined.
Besides flaviviruses, PPMOs have also previously been reported to inhibit other plus-sense RNA viruses (mouse hepatitis virus [36], severe acute respiratory syndrome coronavirus [35], equine arteritis virus [50], and coxsackievirus B3 [56]) and minus-sense RNA viruses (hematopoietic necrosis virus [1], Ebola virus [15], and influenza virus [17]) in cell culture. Furthermore, PPMOs have previously exhibited potency against Ebola virus (15) and coxsackievirus B3 (56) in mouse models. Interestingly, PMOs without any peptide conjugation were recently shown to be efficacious against Ebola virus in mouse, guinea pig, and monkey models (51).
The goals of this study were to analyze potential PPMO-resistant flavivirus in cell culture and to examine the in vivo efficacy of both PMOs and PPMOs against flavivirus. Using WNV as a model, we selected viruses in cell culture that were resistant to the 5′-end or 3′ CSI PPMO. The 5′-end-PPMO-resistant viruses accumulated mutations within the PPMO-binding regions; these mismatch mutations were responsible for the 5′-end PPMO resistance. Surprisingly, the 3′ CSI PPMO escape viruses contained mutations outside the PPMO-binding site; one nucleotide insertion or deletion at the 3′-terminal stem-loop of the genomic RNA was the determinant of the 3′ CSI PPMO resistance phenotype. In an evaluation of in vivo efficacy, we found that unconjugated PMOs (3 mg/day) were nontoxic in mice but did not exhibit any efficacy in WNV-infected mice. In contrast, PPMOs (200 μg/day) showed minimal toxicity in uninfected mice and partially protected infected mice from WNV disease, even when the treatment started at day 5 postinfection.
MATERIALS AND METHODS
Viruses and cells.
WNV was produced from a full-length cDNA clone of a strain isolated in 2000 in New York, strain 3356 (GenBank accession no. AF404756), by the electroporation of baby hamster kidney (BHK) cells with in vitro-transcribed RNA as previously described (44). Viral stock titers were determined by plaque assays on Vero cells (37). For cell culture assays, virus was diluted in BA-1 diluent (M199 [Mediatech Inc.] supplemented with 1 μg of amphotericin B [Invitrogen]/ml, 0.05 M Tris, 1% bovine serum albumin, 0.35 g of sodium bicarbonate/liter, 100 U of penicillin/ml, and 100 μg of streptomycin/ml). Besides WNV, other flaviviruses, including St. Louis encephalitis virus (SLEV; Kern217.3.1.1), DENV type 2 (DENV-2; New Guinea), YFV (17D), and JEV (Nakayama), were used in antiviral assays.
BHK cells and African green monkey kidney cells (Vero cells; American Type Culture Collection) were grown in 5% CO2 at 37°C with MEM (minimal essential medium [Invitrogen], 2 mM l-glutamine, 1.5 g of sodium bicarbonate/liter, 100 U of penicillin/ml, and 100 μg of streptomycin/ml) supplemented with 10% fetal bovine serum (FBS). Confluent cell monolayers and virus-inoculated cultures were maintained in MEM supplemented with 2% FBS.
PMOs and PPMOs.
Three PMOs of 20 to 22 bases in length were synthesized: the 5′-end PMO targeting the first 20 nucleotides of the WNV genome (5′-GCTCACACAGGCGAACTACTC-3′; an extra C [underlined] was added to the 3′ end to potentially interact with the 7-methylguanylate of the 5′ cap and to further stabilize its hybridization with the viral genome), the 3′ CSI PMO targeting the 3′ CSI region (5′-TCCCAGGTGTCAATATGCTGTT-3′), and the scramble PMO containing a random sequence as a nonviral negative control (5′-AGTCTCGACTTGCTACCTCA-3′). For each PMO sequence, two versions were prepared and analyzed: one PMO was 5′-terminally conjugated with an Arg-rich peptide (5′-end, 3′ CSI, and scramble PPMOs); another PMO did not contain any conjugate (5′-end, 3′ CSI, and scramble PMOs). The components of the peptide conjugate and the chemistry of the PMO backbone were described previously (11).
Generation of PPMO-resistant viruses.
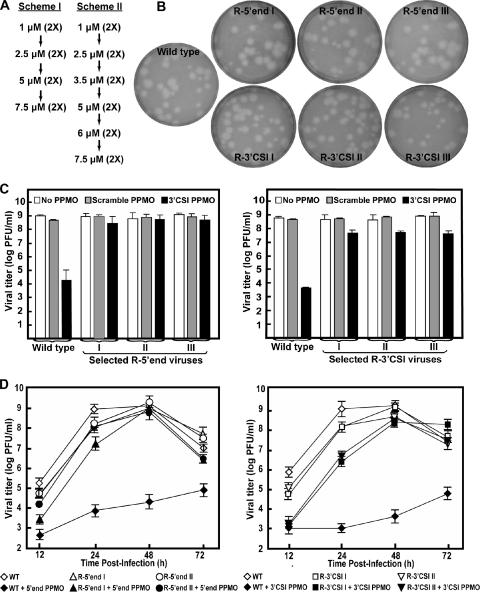
WNV was passaged in Vero cells with increasing concentrations of the 3′ CSI or 5′-end PPMO. Three selections for each PPMO were independently performed. For the first passage, three wells of a Vero cell monolayer in a six-well plate were inoculated at a multiplicity of infection (MOI) of 0.1 with wild-type virus that was diluted in 100 μl of 1 μM 3′ CSI or 5′-end PPMO and incubated for 1 h at 37°C. After the incubation, the cell monolayers were washed three times with BA-1 diluent, and 3 ml of MEM plus 1 μM 3′ CSI or 5′-end PPMO were added to each well. The supernatants were harvested at 42 h postinfection, and each well represented an independent selection (selections I, II, and III). Each selection was blindly passaged by adding 100 μl of the supernatant to new Vero cell monolayers. Viruses were passaged twice for each PPMO concentration. Initially, we performed the selection by following scheme I, with PPMO concentrations increasing from 1 to 2.5, 5.0, and finally 7.5 μM (Fig. 2A). We chose 7.5 μM as the highest concentration for the selection of resistant viruses because the PPMOs showed some cytotoxicity at 10 μM (11). Scheme I did not efficiently yield resistant viruses. Therefore, we modified the procedure to allow for smaller increments of increase in PPMO concentrations, from 1 to 2.5, 3.5, 5.0, 6.0, and 7.5 μM (Fig. 2A, scheme II). During scheme II selection, supernatants were harvested when a cytopathic effect was evident, which in all cases occurred before 96 h postinfection. Supernatants from the second passage at 7.5 μM PPMO were subjected to resistance and sequencing analyses.
FIG. 2.
Selection and characterization of PPMO-resistant WNV. (A) Two schemes for the selection of PPMO-resistant viruses. (B) Plaque morphologies of wild-type and PPMO-resistant viruses. Plaque assays for the resistant and wild-type viruses were performed on Vero cells in the absence of PPMOs. The results of three independent selections for 5′-end-PPMO- and 3′ CSI PPMO-resistant viruses are presented. (C) Resistance phenotypes of the PPMO escape isolates. The left and right panels represent results for 5′-end-PPMO-resistant and 3′ CSI PPMO-resistant viruses, respectively. The results are expressed as the means and standard deviations derived from three independent experiments. See the experimental details in Materials and Methods. (D) Growth kinetics of the PPMO-resistant and wild-type (WT) viruses in the presence and absence of PPMO. Vero cells were infected (MOI of 0.1) for 1 h, washed with PBS, and incubated for the indicated times in medium with or without PPMO at 7.5 μM. The averages of results from three experiments are presented.
Sequencing of drug-resistant viruses.
To identify resistance mutations, we extracted viral RNA from culture supernatants by using RNeasy kits (QIAGEN). Viral RNA was then amplified by reverse transcription-PCR using one-step reverse transcription-PCR kits (Invitrogen). The PCR products were directly subjected to DNA sequencing. Sequences of the 5′- and 3′-terminal nucleotides of the resistant viruses were determined by 5′ and 3′ rapid amplification of cDNA ends (RACE), respectively. The 5′ RACE was performed using the FirstChoice RLM-RACE kit (Ambion). The 3′ RACE was carried out as previously described (49).
Preparation of recombinant WNV containing resistance mutations.
An overlapping-PCR technique was employed to prepare DNA fragments containing specific mutations. The PCR products were then cloned into a full-length cDNA clone of the epidemic WNV isolate 3356, pFLWNV (39). Four types of infectious cDNA mutant plasmids were constructed. (i) The G9U G19A mutant plasmid was generated by cloning the mutant PCR DNA (in which the mutations were engineered through overlap PCR) into the pFLWNV vector at the BamHI site (immediately preceding the T7 RNA promoter) and the ClaI site (nucleotide position 1087; GenBank accession no. AF404756). (ii) The C7891U and G8074A mutant plasmids were prepared by cloning the mutant PCR products into the pFLWNV vector at the two AvrII sites (positions 6316 and 8400). (iii) For constructing mutants containing a U nucleotide insertion (position 10957), a G10965U change, or a G deletion (position 11024), mutant PCR products were cloned into the pFLWNV vector at the SpeI site (position 8022) and the XbaI site (immediately following the 3′ end of the genome). (iv) To prepare the double mutants carrying the C7891U change and the U insertion (U-ins) and the G8074A and G10965U mutations, the AvrII-AvrII fragment was swapped between the C7891U mutant plasmid (derived from type ii plasmids) and the U-ins plasmid (derived from type iii plasmids) and between the G8074A and G10965U plasmids, respectively. All mutant cDNAs were verified by DNA sequencing.
RNA synthesis and transfection were performed as reported previously (43). Briefly, cDNA plasmids were linearized with XbaI and transcribed into genome-length RNA by using a T7 polymerase-based mMessage mMachine kit (Ambion). BHK cells (107 cells in 800 μl of cold phosphate-buffered saline [PBS]) were electroporated with 10 μg of recombinant RNA. Transfected cells were allowed to recover for 10 min at room temperature, resuspended in 24 ml of MEM supplemented with 10% FBS, and cultured in a T-150 flask in 5% CO2 at 37°C. Culture supernatant was collected when an apparent cytopathic effect was observed (day 5 posttransfection), aliquoted, and stored at −80°C.
Antiviral assays.
Vero cells (106 per well of a 12-well plate) were infected at an MOI of 0.1 with the viruses indicated below diluted in BA-1 for 1 h at 37°C, with gentle rocking every 15 min to ensure even levels of infection. Immediately following incubation, 1.5 ml of MEM containing 2% FBS was added to the infected cells with no or a 7.5 μM concentration of the PPMO indicated below. Viral supernatants were collected at 42 h postinfection, and viral titers were determined using plaque assays. The plaque assays for WNV, SLEV, DENV-2, and YFV were performed as described previously (39). The plaque assay for JEV was identical to that for YFV (39).
Mice.
Five-week-old female C3H/HeN (C3H) mice were purchased from Taconic, acclimatized for at least 1 week, and given food and water ad libitum. Mice were 6 to 7 weeks of age at the initiation of all studies. Mice inoculated with WNV were housed in a biosafety level 3 facility. All studies were approved by the Institutional Animal Care and Use Committee of the Wadsworth Center and followed criteria established by the National Institutes of Health.
Toxicity analysis of PMOs and PPMOs in mice.
Toxicity studies were performed with uninfected mice to evaluate PMOs and PPMOs. All compounds were reconstituted in a vehicle consisting of endotoxin-free PBS (tissue culture grade; Invitrogen) with 1% FBS. For the PMOs, eight mice were inoculated intraperitoneally (i.p.) daily with 3 mg of the scramble PMO in 0.1 ml for seven treatments and four mice were inoculated i.p. with 0.1 ml of the vehicle alone. For the PPMOs, eight mice per group were inoculated i.p. daily with 200 or 300 μg of the scramble PPMO in 0.1 ml for up to nine treatments, and four mice were inoculated i.p. with 0.1 ml of the vehicle alone. Toxicity in mice was evaluated by weight loss, changes in organ weight, and abnormal clinical observations (including ruffled fur, hunching, ataxia, gait abnormalities, weakness, diarrhea, changes in activity levels or responses, and increased respiratory rates). All mice were observed and weighed daily throughout the course of the study. Two days after the last treatment, the mice were sacrificed and examined for gross pathology. The following organs were removed in their entirety and weighed to the nearest 0.01 g: the brain, heart, lung, liver, and spleen and both kidneys. Treatment was stopped for the group receiving 300 μg of the scramble PPMO after five treatments due to an average loss of more than 5% of body weight, and these mice were monitored and weighed for an additional 3 days.
Efficacy analysis of PMOs and PPMOs in mice.
On day 0, mice were inoculated subcutaneously (s.c.) with diluent (mock) or 103 PFU of WNV in the left rear footpad as previously described (38). The diluent for virus and the vehicle for PMOs and PPMOs consisted of PBS with 1% FBS as described above. For the PMOs, mice were inoculated i.p. daily with 3 mg of the PMO in 0.1 ml on days 0 to 6 (seven treatments); the initial PMO treatment began at 2 to 3 h post-WNV inoculation. Four groups of eight WNV-inoculated mice each were treated with the vehicle alone or the 5′-end, 3′ CSI, or scramble PMO and one group of four mock-inoculated mice was treated with the vehicle alone. For the PPMOs, two different dosing regimens were tested. In the first regimen, mice were injected i.p. daily with 100 or 200 μg of the PPMO in 0.1 ml on days 0 to 8 (nine treatments). Seven groups of eight WNV-inoculated mice each were treated with the vehicle alone or the 5′-end PPMO, the 3′ CSI PPMO, or the scramble PPMO at one of the two doses, and one group of four mock-inoculated mice was treated with the vehicle alone. In the second regimen, mice were injected i.p. daily with 200 μg of PPMO in 0.1 ml on days 5 to 15 (11 treatments) postinfection. Three groups of eight WNV-inoculated mice each were treated with the vehicle alone, the 5′-end PPMO, or the scramble PPMO, and one group of four mock-inoculated mice was treated with the vehicle alone. All mice were observed for clinical disease at least once per day for the entire study, and all mice were weighed daily for at least 14 days postinoculation and at least three times per week for the duration of the study.
Clinical scores were assigned on a scale of 0 to 6 as follows: 0, normal health; 1, ruffled fur; 2, hunching, mild to moderate weakness, or ataxia; 3, severe weakness or neurologic signs; 4, paralysis; 5, moribund state; 6, death or euthanization. Mice that exhibited severe disease (a score greater than 2) were euthanized. Morbidity was defined as the presence of clinical signs of disease for at least 2 days and/or the loss of more than 9% of body weight. Surviving mice were sacrificed, bled by cardiac puncture, and necropsied at 35 to 37 days postinoculation. Sera were tested for WNV-specific antibodies by using an enzyme-linked immunosorbent assay adapted for mouse serum from a previously described protocol (14). Briefly, sera were tested at 1:100 against WNV antigen and negative control antigen. The average optical density of the WNV antigen divided by the optical density of the negative control antigen (the positive/negative [P/N] ratio) was determined for each sample. A P/N ratio of greater than or equal to 2.0 was defined as a seropositive result, and a P/N ratio of less than 2.0 was defined as a seronegative result. All WNV-inoculated mice were seropositive, with P/N ratios ranging from 6.4 to 22.0, and all mock-inoculated mice were seronegative, with P/N ratios ranging from 0.7 to 1.3.
Statistics.
Survival curves were analyzed with a log rank test by using GraphPad Prism (GraphPad Software, Inc., San Diego, CA). A chi-square analysis was used to evaluate the morbidity and mortality percentages, and Student's t test was used to compare organ weights, days of disease onset, and survival times (Microsoft Office Excel; Microsoft Corporation).
RESULTS
A single PPMO targeting the conserved 3′ CSI RNA element potently inhibits WNV, JEV, and SLEV.
One attractive feature of the 3′ CSI PPMO is the potential to inhibit a broad spectrum of flaviviruses, since the 3′ CSI PPMO-targeted sequence is highly conserved, yet has some variation, among mosquito-borne flaviviruses (18). Our previously described 3′ CSI PPMO (11) was designed to target the 3′ CSI element of WNV without trying to match the PPMO to bind to the 3′ CSI regions of other flaviviruses. The alignment of sequences from various flaviviruses allowed us to redesign a 3′ CSI PPMO which was perfectly complementary to the 3′ CSI-containing elements from WNV, JEV, and SLEV; however, the new 3′ CSI PPMO still had mismatches with the corresponding sequences from DENV and YFV (Table 1). Antiviral assays with cell cultures were performed using the new 3′ CSI PPMO at 7.5 μM, a concentration nontoxic to cells (11, 22). The inhibitor reduced WNV, JEV, and SLEV titers by 98,300-, 20,000-, and 1,200-fold, respectively. In contrast, DENV-2 and YFV titer reductions were 10- and 3-fold, respectively, probably due to the mismatches between the PPMO and the targeted RNA sequences (Table 1). As a control, the scramble PPMO at the same concentration did not substantially inhibit any tested viruses (Table 1). Taken together, the results demonstrated that the redesigned 3′ CSI PPMO may potently inhibit WNV, JEV, and SLEV. This PPMO was therefore used throughout the following experiments.
TABLE 1.
Activities of the 3′ CSI PPMO against mosquito-borne flavivirusesa
| Virus | Targeted sequenceb | Viral titer (PFU/ml) after treatment with:
|
3′ CSI PPMO-mediated reduction (n-fold)c | Viral titer (PFU/ml) after treatment with scramble PPMO | Scramble PPMO-mediated reduction (n-fold)c | |
|---|---|---|---|---|---|---|
| No PPMO | 3′ CSI PPMO | |||||
| WNV | AACAGCATATTGACACCTGGGA | 5.8 × 108 | 5.9 × 103 | 98,300 | 4.1 × 108 | 1.4 |
| JEV | AACAGCATATTGACACCTGGGA | 2.8 × 107 | 1.4 × 103 | 20,000 | 2.0 × 107 | 1.4 |
| SLEV | AACAGCATATTGACACCTGGGA | 5.5 × 106 | 4.5 × 103 | 1,200 | 4.2 × 106 | 1.3 |
| DENV-2 | AACAGCATATTGAC-GCTGGGA | 6.1 × 106 | 6.0 × 105 | 10 | 1.9 × 107 | 0.3 |
| YFV | GGGACCATATTGACGCCAGGGA | 1.7 × 107 | 6.1 × 106 | 3 | 4.6 × 106 | 3.7 |
Vero cells were infected with the indicated virus (MOI of 0.1), treated with a redesigned 3′ CSI PPMO (7.5 μM), and assayed for viral titers at 40 to 42 h postinfection.
Sequences from various mosquito-borne flaviviruses targeted by the 3′ CSI PPMO. Nucleotide changes relative to the WNV sequence are underlined, and a nucleotide deletion is indicated by a hyphen. Sequences were derived from GenBank entries with accession numbers AF404756, NC_001437, CQ897117, U87411, and U17066 for WNV, JEV, SLEV, DENV-2, and YFV viruses, respectively.
The reduction (n-fold) in viral titer was calculated as follows: viral titer without PPMO treatment/viral titer with PPMO treatment.
Selection and characterization of PPMO-resistant WNV.
We selected drug-resistant viruses by culturing WNV in the presence of increasing concentrations of the 5′-end or 3′ CSI PPMO (Fig. 2A). Since our previous results indicated the 50% effective concentration of the 3′ CSI PPMO to be 0.2 μM (11), we started the selection of resistant virus at 1 μM 3′ CSI PPMO (five times the 50% effective concentration). In three independent selection trials per PPMO, scheme I did not reveal any 5′-end-PPMO-resistant virus and only one 3′ CSI PPMO-resistant virus was selected. Scheme II yielded resistant WNV in all six independent selections, three viruses resistant to the 5′-end PPMO (R-5′ end I, II, and III) and three viruses resistant to the 3′ CSI PPMO (R-3′ CSI I, II, and III).
Plaque morphologies of the PPMO-resistant and wild-type viruses were compared (Fig. 2B). The R-5′ end, R-3′ CSI, and wild-type viruses exhibited similar plaque sizes when analyzed in agar containing no PPMO. The resistance phenotypes of the R-5′ end and R-3′ CSI viruses were confirmed by incubation with the corresponding PPMO (Fig. 2C). In the presence of 7.5 μM 5′-end PPMO, the titer of the wild-type virus was reduced by nearly 5 logs and those of the three R-5′ end isolates were reduced by less than 1 log (Fig. 2C, left panel). When treated with 7.5 μM 3′ CSI PPMO, the wild-type virus exhibited a decrease in titer of about 5 logs; in contrast, the titers of the three R-3′ CSI viruses were reduced by approximately 1 log (Fig. 2C, right panel). As negative controls, neither the wild-type nor the resistant viruses were substantially affected when the viruses were incubated with 7.5 μM scramble PPMO.
To further characterize the replication and resistance phenotypes of the selected viruses, we compared the growth kinetics of the wild-type and resistant viruses in the presence or absence of the appropriate PPMOs (Fig. 2D). We chose viruses from selections I and II from both R-5′ end and R-3′ CSI groups for these experiments because sequence results showed that resistant viruses from selections I and II were more distinct from each other and that virus from selection III was similar to that from selection I (see details below) (Fig. 1C). In the absence of PPMOs, the difference in viral titer between the R-5′ end and wild-type viruses was less than 1 log from 12 to 72 h postinfection (Fig. 2D, left panel). The difference in titer between the R-3′ CSI and wild-type viruses was more dramatic at 12 to 48 h postinfection but decreased from 48 to 72 h postinfection (Fig. 2D, right panel). In the presence of a PPMO, all resistant viruses showed higher titers than the wild-type virus; at 48 h postinfection, the difference in titers between the resistant and wild-type viruses reached a maximum of 4 to 5 logs. Note that the resistant viruses grew more slowly in the presence of the PPMO than in the absence of the PPMO between 12 and 24 h postinfection but that they reached similar titers at 48 or 72 h postinfection (Fig. 2D). Overall, the results demonstrate that PPMO-resistant viruses could be selected in cell culture and that compared to the wild-type virus, the resistant viruses did not have significant defects in viral amplification.
Identification of mutations in PPMO-resistant viruses.
We sequenced the complete genomes of all six resistant viruses to identify potential escape mutations. For the R-5′ end viruses (Fig. 1C, left panel), two to three mismatch mutations were found within the PPMO-binding region; a G19A substitution was shared by all three selected viruses. R-5′ end viruses from selections I and III shared G9U and G19A mutations, and R-5′ end virus from selection III had an additional A23G substitution (adjacent to the PPMO-binding site). R-5′ end II had three changes, C8G, C11G, and G19A. No other mutation was observed throughout the genomes of the R-5′ end viruses.
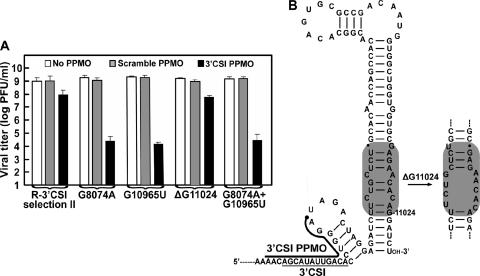
For R-3′ CSI isolates, adaptive changes in all three escape viruses were found to be located outside the PPMO-binding region (Fig. 1C, right panel). R-3′ CSI viruses from selections I and III shared a U-ins at position 10956 or 10957, resulting in a three-U tract (U10956, U10957, and U-ins), and the U-ins was therefore arbitrarily named 10957U-ins. In addition, R-3′ CSI viruses from selections I and III had one extra nonsynonymous change in the NS5 gene. R-3′ CSI virus from selection II also had a nonsynonymous substitution, G8074A, in the NS5 gene and two other mutations, G10965U and ΔG11024 (a G deletion at nucleotide position 11024), downstream of the PPMO-targeted region (Fig. 1A).
Mismatch mutations within the PPMO-targeted region are responsible for resistance to the 5′-end PPMO.
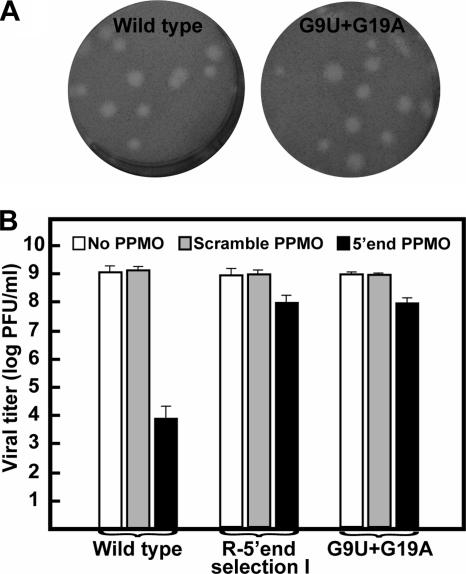
The mutations listed in Fig. 1C represent consensus sequences of the resistant viruses. The consensus mutations may be responsible for the PPMO resistance; alternatively, quasispecies within the resistant viral population may act together to confer drug resistance. To distinguish the two possibilities, we engineered a G9U G19A construct, derived from R-5′ end selection I (Fig. 1C, left panel), into an infectious cDNA clone of WNV. The resulting recombinant G9U G19A virus exhibited plaque morphology similar to that of the wild type (Fig. 3A). Resistance analysis showed that, in the presence of 7.5 μM 5′-end PPMO, the titer of recombinant G9U G19A virus was reduced by about 1 log whereas the titer of wild-type virus was reduced by more than 5 logs (Fig. 3B). Similar titers of recombinant G9U G19A and wild-type viruses in the absence of PPMOs or in the presence of the scramble PPMO were obtained (Fig. 3B). The drug sensitivity of recombinant G9U G19A virus was almost identical to that of the original R-5′ end virus from selection I. The results demonstrate that the mismatch changes within the PPMO-targeted region were responsible for the 5′-end PPMO resistance.
FIG. 3.
Analysis of mutations recovered from 5′-end-PPMO-resistant viruses. (A) Plaque morphologies of wild-type and recombinant G9U G19A viruses. (B) Resistance analyses of wild-type virus, R-5′ end virus from selection I, and recombinant G9U G19A virus. See Materials and Methods for experimental details.
A single mutation downstream of the PPMO-binding site confers resistance to the 3′ CSI PPMO.
We prepared two panels of recombinant viruses to determine the mutation(s) that was critical for the 3′ CSI PPMO resistance. The first panel of virus was designed to analyze mutations recovered from selections I and III; both selections yielded the 10957U-ins (Fig. 1C, right panel). Using R-3′ CSI virus from selection I as an example, we prepared three WNV variants that contained the mutation(s) C7891U, 10957U-ins, or C7891U and 10957U-ins. In the absence of PPMOs, the three mutant viruses showed plaque morphologies similar to that of the wild-type virus (data not shown) and grew to titers equivalent to that of the wild-type virus (Fig. 4A). Remarkably, resistance analysis showed that recombinant 10957U-ins virus had a resistance level similar to that of the original R-3′ CSI virus from selection I; the addition of C7891U to the 10957U-ins, in the case of the C7891U 10957U-ins virus, did not further enhance resistance, and the C7891U virus did not show any resistance. As a negative control, treatment with the scramble PPMO did not affect any of the tested viruses (Fig. 4A). The results strongly suggest that the 10957U-ins is a determinant of the 3′ CSI PPMO resistance in viruses recovered from selections I and III. Thermodynamic folding using MFold indicated that the 10957U-ins did not dramatically change the 5′ UAR-3′ UAR interaction (Fig. 4B); however, the 10957U-ins resulted in an internal loop within the helix region of the 3′-terminal stem-loop structure (Fig. 4C).
FIG. 4.
Verification of 10957U-ins as a determinant of 3′ CSI PPMO resistance. (A) Resistance phenotypes of wild-type virus, R-3′ CSI virus from selection I, and three recombinant viruses (C7891U, 10957U-ins, and C7891U 10957U-ins viruses). The experimental procedures were as described in Materials and Methods. The data are means ± standard deviations (n ≥ 3). (B) Effects of 10957U-ins on the thermodynamically predicted 5′ UAR-3′ UAR interaction. (C) 10957U-ins-mediated disturbance of an internal stem region of the 3′ stem-loop structure of the WNV genome. The 3′ CSI PPMO and the 3′ CSI are indicated by thick and thin lines, respectively. In panels B and C, regions containing the 10957U-ins-mediated structural changes are highlighted in gray.
A second panel of recombinant virus was prepared to examine mutations recovered from R-3′ CSI virus from selection II. The recombinant viruses contained either a single mutation (G8074A, G10965U, or ΔG11024) or double mutations (G8074A and G10965U) (Fig. 5A). All mutant viruses showed plaque morphologies similar to that of the wild-type virus (data not shown). Antiviral assay results showed that G8074A, G10965U, and G8074A G10965U viruses were not PPMO resistant: the 3′ CSI PPMO (7.5 μM) suppressed viral titers by 4 to 5 logs, whereas the scramble PPMO did not reduce viral titers. In contrast, the ΔG11024 virus exhibited significant resistance: the 3′ CSI PPMO inhibited viral titers by 1 to 2 logs, whereas the scramble PPMO did not substantially suppress viral titers (Fig. 5A). Given that titers of R-3′ CSI virus from selection II could be reduced by 1 to 2 logs when the virus was treated with 7.5 μM 3′ CSI PPMO (Fig. 1C, right panel), we conclude that ΔG11024 is the major determinant of the 3′ CSI PPMO resistance. RNA folding using MFold indicated that ΔG11024 did not affect the 5′ UAR-3′ UAR interaction (data not shown) but could lead to an internal loop within the helix region of the 3′-terminal stem-loop structure (Fig. 5B). Note that identical RNA loops associated with the 10957U-ins and ΔG11024 mutations were thermodynamically predicted (compare Fig. 4C with 5B). Overall, the results demonstrate that a single mutation, 10957U-ins or ΔG11024 downstream of the PPMO-binding site, is the determinant of the 3′ CSI PPMO resistance.
FIG. 5.
Effects of ΔG11024 on WNV resistance to the 3′ CSI PPMO. (A) Resistance profiles of R-3′ CSI virus from selection II and four recombinant viruses (G8074A, G10965U, ΔG11024, and G8074A G10965U viruses). See Materials and Methods for details. (B) Effects of ΔG11024 on the MFold-predicted 3′ stem-loop structure of the WNV genome. Structural changes caused by ΔG11024 are highlighted in gray.
Toxicity of PMOs and PPMOs in mice.
Although antisense PPMOs potently inhibit WNV (reference 11 and data presented above) and DENV (22) in cell culture, their efficacy against flaviviruses in vivo has not been reported. Our previous results showed that only the PPMOs, but not the PMOs, suppress WNV in cell culture (11); however, recent studies with Ebola virus demonstrated that both virus-specific PPMOs (15) and PMOs (51) are efficacious in animal models. The latter results prompted us to examine the in vivo efficacy of both PPMOs and PMOs in our WNV mouse model. We initially evaluated the toxicity of PMOs and PPMOs in uninfected mice and determined subtoxic doses for subsequent efficacy evaluation in vivo.
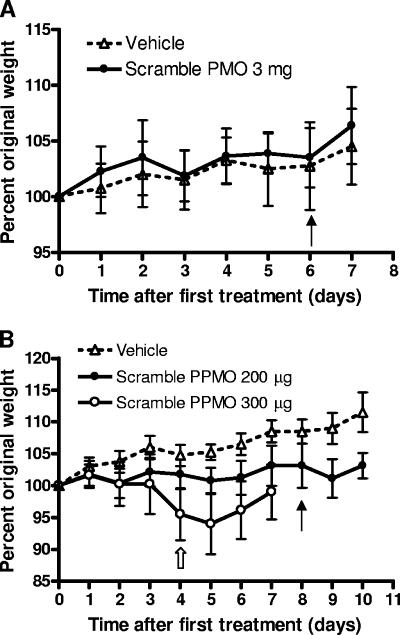
The scramble PMO and PPMO were used for toxicity analyses with C3H mice. For the PMO, there was no evidence of toxicity: no weight loss (Fig. 6A) or change in organ weight (Table 2) was observed when the PMO was administered at 3 mg/day (equivalent to approximately 150 mg/kg of body weight/day) for 7 days. In addition, no abnormal behavior of the PMO-treated mice was observed. In contrast, dose-dependent toxicity in mice treated with the PPMO was observed (Fig. 6B). There was weight loss in mice treated with the scramble PPMO at 300 μg/day (equivalent to about 15 mg/kg/day), resulting in the cessation of treatment after day 4; the body weights started to rebound 2 days after the last treatment (Fig. 6B). The group treated with the scramble PPMO at 200 μg/day (about 10 mg/kg/day) exhibited mild adverse effects, with decreased weight gain compared to that of the controls (Fig. 6B). No significant difference in organ weights in the treated group was observed, but the treated group showed a trend toward higher spleen weights than the controls (Table 2). Abnormal gaits and hunching were observed in mice for up to 2 h after treatment with both 300- and 200-μg doses of the scramble PPMO but not in the control mice treated with the vehicle alone. These signs indicate that the PPMO may temporarily irritate animals after treatment. No gross abnormalities were observed during necropsies after the toxicity studies; however, abnormalities in the livers of 4 of 21 surviving mice treated with the PPMO on days 0 to 8 were observed at the end of one of the efficacy studies (day 35). The livers were small and had rounded edges. Upon histopathology analysis, the livers were found to be normal except for capsular thickening, which was confirmed as fibrosis by trichrome staining (data not shown). In summary, these results demonstrate that mice can tolerate a much higher dose of the PMO than of the PPMO. The PMO is nontoxic to mice at a dose of 3 mg/day, whereas the PPMO at 200 μg/day begins to show mild toxicity in mice. The results also indicate that the Arg-rich peptide within the PPMO molecule is the major component that causes toxicity.
FIG. 6.
Body weights of mice treated with PMOs or PPMOs. Six-week-old C3H female mice were treated, weighed, and observed daily. Averages of the percentages of original weights are shown, and error bars represent standard deviations of ±1. (A) Mice were inoculated i.p. daily with the vehicle alone (n = 4) or 3 mg of the scramble PMO (n = 8) for seven treatments. The solid arrow indicates the last day of treatment. (B) Mice were inoculated i.p. daily with the vehicle alone (n = 4) for nine treatments, 200 μg of the scramble PPMO (n = 8) for nine treatments, or 300 μg of the scramble PPMO (n = 8) for five treatments. The solid arrow indicates the last day of treatment for groups receiving the vehicle alone and 200 μg of the scramble PPMO. The hollow arrow indicates the last day of treatment for the group receiving 300 μg of the scramble PPMO, which was discontinued after only five treatments due to an average loss among the animals of more than 5% of body weight.
TABLE 2.
Weights of organs after PMO and PPMO treatment did not differ from those from vehicle-treated controlsa
| Treatment | No. of mice | Dose | No. of days treated | Avg wt (g) (SD) of:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Brains | Hearts | Lungs | Livers | Spleens | Kidneys | ||||
| Vehicle | 4 | NA | 7 | 0.40 (0.02) | 0.09 (0.01) | 0.11 (0.04) | 1.02 (0.04) | 0.06 (0.01) | 0.26 (0.01) |
| Scramble PMO | 8 | 3 mg | 7 | 0.41 (0.02) | 0.09 (0.01) | 0.12 (0.02) | 1.05 (0.10) | 0.06 (0.01) | 0.24 (0.03) |
| Vehicle | 4 | NA | 9 | 0.38 (0.02) | 0.11 (0.02) | 0.13 (0.02) | 1.11 (0.09) | 0.07 (0.02) | 0.24 (0.03) |
| Scramble PPMO | 8 | 200 μg | 9 | 0.38 (0.02) | 0.09 (0.03) | 0.12 (0.02) | 1.16 (0.12) | 0.13 (0.06) | 0.27 (0.03) |
Six-week-old C3H female mice were treated i.p. daily for 7 or 9 days. Two days after the end of treatment, all mice were sacrificed, and the entire brain, heart, lung, liver, and spleen and both kidneys of each mouse were weighed. NA, not applicable. No statistical differences between the drug- and vehicle-treated groups were observed.
In vivo efficacy of PMOs and PPMOs.
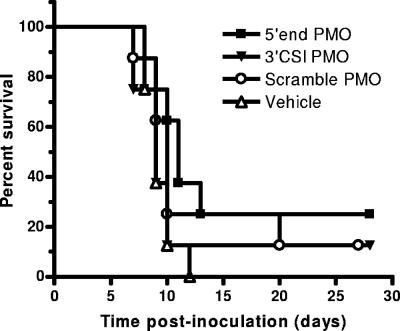
Based on the above-cited toxicity results, we examined the efficacy of the PMOs and PPMOs in protecting mice from WNV disease. Three PMOs and corresponding PPMOs were tested, namely, the 5′-end PMO and PPMO, the 3′ CSI PMO and PPMO, and the scramble PMO and PPMO. In the first experiment, we began treatment of the mice on the same day as viral inoculation. The PMOs (3 mg/day for 7 days) did not show any efficacy compared to the controls. There was no difference in survival (Fig. 7) or morbidity (data not shown) rates.
FIG. 7.
Unconjugated PMOs do not protect mice from WNV disease. Six-week-old C3H female mice were inoculated s.c. in the left rear footpad with 103 PFU of WNV on day 0. Groups of eight mice were treated with 3 mg of the 5′-end PMO, 3 mg of the 3′ CSI PMO, 3 mg of the scramble PMO, or the vehicle alone. Treatments were administered i.p. daily on days 0 to 6.
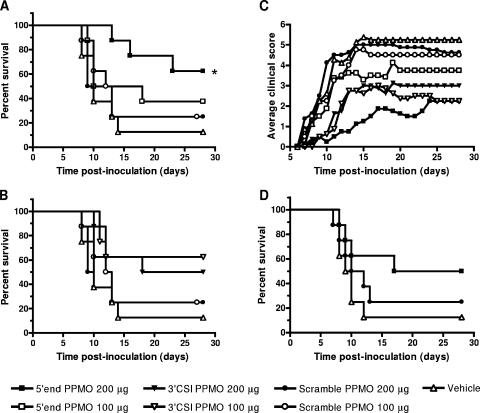
Mice that were treated with PPMOs on days 0 to 8 at 100 or 200 μg/day were partially protected from disease (Fig. 8A, B, and C). For the group treated with 200 μg of the 5′-end PPMO, the survival rate over time was statistically significant compared to that of the control group treated with 200 μg of the scramble PPMO (Fig. 8A) (log rank test; P = 0.04). The group treated with 200 μg of the 5′-end PPMO also exhibited a reduced morbidity rate, increased survival times, and a delay in disease onset that were statistically significant (Table 3). In addition, the efficacy of treatment with the 5′-end PPMO was dose dependent since the 100-μg dose was less efficacious, but the group treated with 100 μg of the 5′-end PPMO still had a slightly lower mortality rate than the control groups and a significant delay in disease onset (Table 3). In the 3′ CSI PPMO-treated groups, dosed at 100 or 200 μg/day, the mice exhibited slightly decreased mortality rates and significant delays in disease onset compared to controls, and the higher dose also corresponded to longer survival times (Table 3). The groups treated with 200 μg of the 5′-end PPMO/day, 200 μg of the 3′ CSI PPMO/day, or 100 μg of the 3′ CSI PPMO/day exhibited not only delayed disease onset but also decreased clinical scores during early disease compared to the other groups (Fig. 8C). These data demonstrate that both the 5′-end and 3′ CSI PPMOs provide partial protection from WNV disease; however, the best candidate is the 5′-end PPMO at 200 μg/day.
FIG. 8.
PPMOs partially protect mice from WNV disease. Six-week-old C3H female mice were inoculated s.c. in the left rear footpad with 103 PFU of WNV on day 0. Groups of eight mice were treated i.p. with 100 or 200 μg of PPMOs or the vehicle alone. (A and B) Survival rates among mice treated daily on days 0 to 8 with the 5′-end PPMO and the 3′ CSI PPMO, respectively. The survival curves for groups treated with the vehicle alone and the scramble PPMO are the same in panels A and B and are repeated for ease of comparison. The asterisk indicates that the survival curve for the group treated with the 5′-end PPMO at 200 μg was significantly different from that for the group treated with the scramble PPMO at 200 μg (log rank test; P = 0.04). (C) Average clinical scores for groups treated daily with the indicated PPMOs on days 0 to 8. Clinical scores were assigned a scale of 0 (normal) to 6 (dead or euthanized). The scale of clinical scores is further defined in Materials and Methods. (D) Survival rates of mice treated daily from days 5 to 15 after viral inoculation.
TABLE 3.
Partial protection against WNV disease in mice treated with conjugated PPMOsa
| Inoculum | Antiviral treatment
|
Disease outcome
|
|||||
|---|---|---|---|---|---|---|---|
| Treatment | Dose (μg) | Treatment days | Morbidity (no. of sick mice/total no. in group) | Mortality (no. of mice that died/total no. in group) | Avg day of disease onset (SD)b | Avg survival time (SD)c | |
| Diluent | Vehicle | NA | 0 to 8 | 0/4 | 0/4 | NA | NA |
| WNV | 5′-end PPMO | 200 | 0 to 8 | 5/8* | 3/8 | 9.8 (1.48)*** | 17.3 (5.13)*** |
| 5′-end PPMO | 100 | 0 to 8 | 8/8 | 5/8 | 9.1 (1.55)** | 11.4 (3.71) | |
| 3′ CSI PPMO | 200 | 0 to 8 | 8/8 | 4/8 | 10.8 (2.92)** | 13.0 (3.46)* | |
| 3′ CSI PPMO | 100 | 0 to 8 | 8/8 | 3/8 | 11.3 (2.49)*** | 11.3 (0.58) | |
| Scramble PPMO | 200 | 0 to 8 | 8/8 | 6/8 | 7.6 (0.92) | 9.7 (0.46) | |
| Scramble PPMO | 100 | 0 to 8 | 8/8 | 6/8 | 7.6 (0.52) | 11.0 (2.00) | |
| Vehicle | NA | 0 to 8 | 8/8 | 7/8 | 8.1 (1.55) | 10.43 (2.3) | |
| Diluent | Vehicle | NA | 5 to 15 | 0/4 | 0/4 | NA | NA |
| WNV | 5′-end PPMO | 200 | 5 to 15 | 8/8 | 4/8 | 7.6 (0.74) | 11.0 (4.08) |
| Scramble PPMO | 200 | 5 to 15 | 8/8 | 6/8 | 7.4 (0.52) | 9.8 (2.32) | |
| Vehicle | NA | 5 to 15 | 8/8 | 7/8 | 7.5 (0.53) | 9.3 (1.50) | |
Six-week-old C3H female mice were inoculated s.c. in the left rear footpad with 103 PFU of WNV or diluent alone on day 0. The mice were then inoculated i.p. daily on days 0 to 8 or days 5 to 15 with the indicated treatments. Mice were monitored for weight loss and clinical signs. Surviving mice were bled on days 35 to 37 postinoculation and tested for WNV-specific antibody by enzyme-linked immunosorbent assay; all WNV-inoculated mice were seropositive, and mock-inoculated mice were seronegative. NA, not applicable. Levels of statistical significance in comparisons of results for the indicated group to those for the group receiving the scramble conjugate at the same dose are noted as follows: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005.
Average day of disease onset postinoculation for mice that became sick only.
Average survival time in days for mice that died.
In a second in vivo experiment, the best candidate was tested for efficacy in protecting mice after virus has replicated and spread. Mice were treated with 200 μg of the 5′-end PPMO/day on days 5 to 15 postinfection. Clinical signs of disease in mice were first observed on days 7 to 9; therefore, mice were treated prior to disease onset but after the peak level of viremia and neuroinvasion (K. A. Bernard, unpublished data). The group treated with the 5′-end PPMO exhibited a lower mortality rate and longer survival times than the control groups, but the differences were not statistically significant (Fig. 8D and Table 3). These results suggest that treatment after viral replication and neuroinvasion is possible and that, with improvements, the PPMO may be useful as a therapeutic agent after disease onset.
The potential emergence of PPMO-resistant viruses in vivo was examined with two mice with severe WNV disease that were euthanized late in infection (day 18). One mouse had been treated with 100 μg of the 5′-end PPMO on days 0 to 8, and the other had been treated with 200 μg of the 3′ CSI PPMO on days 0 to 8. RNA was isolated from the brain of each mouse, and consensus sequencing of the 5′ and 3′ UTRs by RACE did not reveal any mutations. In addition, viruses were isolated from the brains of both mice by passaging 20% brain homogenate on Vero cells. The virus stocks were tested for resistance to the corresponding PPMO at 7.5 μM in cell culture; no resistance phenotype was observed (data not shown). These results indicate that the morbidity and mortality of the PPMO-treated mice was not due to the emergence of resistant viruses.
DISCUSSION
The development of an efficacious therapy for infections with WNV and other flaviviruses is a public health priority. We and others have previously found that PPMOs targeting various RNA elements within the flavivirus genome are potent inhibitors of WNV and DENV (19, 22) in cell culture. The present study has extended these findings by demonstrating that the PPMOs could partially protect mice from WNV disease. In addition, we showed that PPMO-resistant viruses could be selected in cell culture and that the escape isolates had accumulated nucleotide changes within or outside of the PPMO-binding site. To the best of our knowledge, this is the first study to demonstrate the in vivo efficacy of and in vitro resistance to the PPMO-mediated inhibition of flavivirus infections.
The in vivo efficacy of PMOs against WNV disease required conjugation to an Arg-rich peptide: although unconjugated PMOs were nontoxic, the treatment of infected mice with the PMOs did not exhibit any efficacy, despite the high dose level of 150 mg/kg. In contrast, PPMOs caused toxicity at a dose of 15 mg/kg/day, but 10 mg/kg/day had minimal toxicity and produced significant efficacy, as evidenced by increased survival rates and a reduction in disease-related symptoms in WNV-infected mice. The results indicate that the conjugated peptide may play two counteractive roles: it improves the potency, most likely through the enhancement of PMO delivery into cells (11), but it can also cause toxicity. Therefore, a reformulation of the peptide to reduce its toxicity while maintaining its PMO delivery characteristics will be a future direction to improve therapeutic PMO technology.
The PPMOs most efficacious against flaviviruses in cell culture are the 5′-end and 3′ CSI PPMOs (11, 22). Although another PPMO, hybridizing to the top of the 3′-terminal stem-loop of the DENV-2 genome, was recently reported to have potent antiviral activity (19), the corresponding WNV PPMO was not as efficacious as the 5′-end and 3′ CSI PPMOs (11). For the present study, we redesigned the 3′ CSI PPMO sequence so that it showed perfect complementarity to the 3′ CSI regions of WNV, JEV, and SLEV; this redesigned PPMO showed an improved spectrum of activity against WNV, JEV, and SLEV but not against DENV-2 or YFV. In likewise fashion, a single PPMO, targeting an identical 3′ CSI element of DENV, was shown to potently suppress all four serotypes of DENV but not WNV (22). These results suggest that, due to sequence variations in the 3′ CSI regions, it is not possible to design a single PPMO that can potently inhibit all viruses within the Flavivirus genus. However, viruses with identical 3′ CSI regions (such as WNV, JEV, and SLEV) can be efficaciously suppressed by a single PPMO.
The mechanism of action of the 5′-end PPMO was previously suggested to be the suppression of the formation of the 43S preinitiation complex and ribosome scanning during viral translation (11). A recent study showed that the 5′-end PPMO can also inhibit WNV RNA cap methylation (13), which is essential for the efficient translation of viral RNA (40, 58). As the 5′-terminal region of the DENV-2 genome was previously shown to function as a promoter for minus-strand RNA synthesis (16), the 5′-end PPMO may also suppress RNA replication by blocking the 5′ promoter sequence or structure. Besides DENV and WNV (19, 22), corresponding 5′-end PPMOs are potent inhibitors of YFV and SLEV in cell culture (P.-Y. Shi, unpublished results). The results strongly suggest that targeting the 5′-terminal nucleotides of a viral genome may be a general strategy for the development of PPMO-mediated flavivirus disease therapy.
The successful selection of PPMO escape viruses in cell culture allowed us to study the drug resistance profiles. The 5′-end-PPMO-resistant isolates contained mutations within the PPMO-targeted region. This finding agreed with a previous observation of three contiguous mismatch changes within the PPMO-binding site in drug-resistant severe acute respiratory syndrome coronavirus (35). Using an infectious cDNA clone of WNV, we demonstrated that the mismatch mutations within the 5′-end-PPMO-targeted region were responsible for the PPMO resistance (Fig. 3). In contrast, the 3′ CSI PPMO escape WNV accumulated changes outside the PPMO-biding site. NS5 mutations were recovered from all three 3′ CSI PPMO-selected viruses (Fig. 1C, right panel); analyses of recombinant WNV suggest that none of the NS5 mutations contribute to the resistance phenotype (Fig. 4A and 5A). Mutagenesis results demonstrate that 10957U-ins (Fig. 4A) or ΔG11024 (Fig. 5A), downstream of the 3′ CSI sequence, is the determinant of drug resistance. We previously showed that mutations within the 3′ CSI, which disrupt the base pairing between the 3′ CSI and the 5′ CS, suppress WNV replicon RNA synthesis (30). Similarly, the 3′ CSI PPMO was previously found to specifically inhibit the RNA synthesis of the WNV replicon (11). A question that arises from this study is how the 10957U-ins or ΔG11024 mutation could rescue the RNA synthesis suppressed by the 3′ CSI PPMO interference with 5′ CS-3′ CSI genome cyclization.
Alvarez and coworkers recently reported that, besides the 5′ CS-3′ CSI interaction, a second long-distance RNA-RNA interaction, known as the 5′ UAR-3′ UAR interaction, also contributes to the DENV-2 genome cyclization and that the interactions of both the 5′ CS and 3′ CSI and the 5′ UAR and 3′ UAR are essential for efficient DENV-2 replication (2). In the present study, the 10957U-ins recovered from the 3′ CSI PPMO-resistant viruses resided within the 3′ UAR. One possible explanation for the resistance may be that the 10957U-ins dramatically enhanced the 5′ UAR-3′ UAR interaction, thus compensating for the PPMO-mediated interference with 5′ CS-3′ CSI-mediated genome cyclization. However, MFold analysis suggests that the 10957U-ins does not enhance the 5′ UAR-3′ UAR interaction (Fig. 4B). Therefore, that explanation seems unlikely. An alternative explanation for this unexpected result is that the 10957U-ins leads to a local conformational change within the 3′ stem-loop RNA structure; this conformational change is compensatory for the PPMO-mediated blockage of the 5′ CS-3′ CSI interaction. In support of this possibility, thermodynamic folding of the 3′-terminal stem-loop of WNV RNA suggests that the 10957U-ins leads to an internal loop (Fig. 4C). Strikingly, the ΔG11024 mutation was predicted to create an identical loop at the 3′ stem-loop of the WNV genome (Fig. 5B). Experiments are in progress to examine the role of this internal loop in rescuing viral RNA synthesis suppressed by the 3′ CSI PPMO. As the molecular details of how the 5′ CS-3′ CSI and 5′ UAR-3′ UAR interactions regulate the flavivirus life cycle are presently unknown, the study of the 3′ CSI PPMO resistance mutations may provide an opportunity for insight into these events.
Acknowledgments
We thank Chrystal Chadwick and Kim Kent for technical assistance. We are grateful to the Chemistry Group at AVI BioPharma for the synthesis of the PMO and PPMO used in this study. We also thank Wadsworth Center's Molecular Genetics Core for DNA sequencing, the Cell Culture Facility for the maintenance of BHK and Vero cells, and the Pathology Core for histology support.
The work was supported in part by grants 1U01AI061193 and 1R43AI065156 and contract N01-AI-25490 from the National Institutes of Health. The biosafety level 3 animal facility at the Wadsworth Center, which is funded in part by the Northeast Biodefense Center's animal core (NIH/NIAID U54-AI057158), was used for this work.
Footnotes
Published ahead of print on 7 May 2007.
REFERENCES
- 1.Alonso, M., D. A. Stein, E. Thomann, H. M. Moulton, J. C. Leong, P. Iversen, and D. V. Mourich. 2005. Inhibition of infectious haematopoietic necrosis virus in cell cultures with peptide-conjugated morpholino oligomers. J. Fish Dis. 28399-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez, D. E., M. F. Lodeiro, S. J. Luduena, L. I. Pietrasanta, and A. V. Gamarnik. 2005. Long-range RNA-RNA interactions circularize the dengue virus genome. J. Virol. 796631-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwell, J. L., and M. A. Brinton. 1997. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol. 716433-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bredenbeek, P. J., E. A. Kooi, B. Lindenbach, N. Huijkman, C. M. Rice, and W. J. Spaan. 2003. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 841261-1268. [DOI] [PubMed] [Google Scholar]
- 5.Brinton, M. A., and J. H. Dispoto. 1988. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology 162290-299. [DOI] [PubMed] [Google Scholar]
- 6.Brinton, M. A., A. V. Fernandez, and J. H. Dispoto. 1986. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology 153113-121. [DOI] [PubMed] [Google Scholar]
- 7.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1125. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott William & Wilkins, Philadelphia, PA.
- 8.Cahour, A., A. Pletnev, M. Vazielle-Falcoz, L. Rosen, and C. J. Lai. 1995. Growth-restricted dengue virus mutants containing deletions in the 5′ noncoding region of the RNA genome. Virology 20768-76. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C. J., M. D. Kuo, L. J. Chien, S. L. Hsu, Y. M. Wang, and J. H. Lin. 1997. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J. Virol. 713466-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corver, J., E. Lenches, K. Smith, R. Robison, T. Sando, E. Strauss, and J. Strauss. 2003. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J. Virol. 772265-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deas, T. S., I. Binduga-Gajewska, M. Tilgner, P. Ren, D. A. Stein, H. M. Moulton, P. L. Iversen, E. B. Kauffman, L. D. Kramer, and P.-Y. Shi. 2005. Inhibition of flavivirus infections by antisense oligomers specifically suppressing viral translation and RNA replication. J. Virol. 794599-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Nova-Ocampo, M., N. Villegas-Sepulveda, and R. M. del Angel. 2002. Translation elongation factor-1alpha, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology 295337-347. [DOI] [PubMed] [Google Scholar]
- 13.Dong, H., D. Ray, S. Ren, B. Zhang, F. Puig-Basagoiti, Y. Takagi, C. Ho, H. Li, and P. Shi. 2007. Distinct RNA elements confer specificity to flavivirus RNA cap methylation events. J. Virol. 814412-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebel, G., A. Dupuis, D. Nicholas, D. Young, J. Maffei, and L. Kramer. 2002. Detection by enzyme-linked immunosorbent assay of antibodies to West Nile virus in birds. Emerg. Infect. Dis. 8979-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enterlein, S., K. L. Warfield, D. L. Swenson, D. A. Stein, J. L. Smith, C. S. Gamble, A. D. Kroeker, P. L. Iversen, S. Bavari, and E. Muhlberger. 2006. VP35 knockdown inhibits Ebola virus amplification and protects against lethal infection in mice. Antimicrob. Agents Chemother. 50984-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filomatori, C., M. Lodeiro, D. Alvarez, M. Samsa, L. Pietrasanta, and A. Gamarnik. 2006. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 202238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge, Q., M. Pastey, D. Kobasa, P. Puthavathana, C. Lupfer, R. Bestwick, P. Iversen, J. Chen, and D. Stein. 2006. Inhibition of multiple subtypes of influenza A virus in cell cultures with morpholino oligomers. Antimicrob. Agents Chemother. 503724-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn, C. S., Y. S. Hahn, C. M. Rice, E. Lee, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 19833-41. [DOI] [PubMed] [Google Scholar]
- 19.Holden, K., D. Stein, T. Pierson, A. Ahmed, K. Clyde, P. Iversen, and E. Harris. 2006. Inhibition of dengue virus translation and RNA synthesis by a morpholino oligomer targeted to the top of the terminal 3′ stem-loop structure. Virology 344439-452. [DOI] [PubMed] [Google Scholar]
- 20.Khromykh, A., N. Kondratieva, J. Sgro, A. Palmenberg, and E. Westaway. 2003. Significance in replication of the terminal nucleotides of the flavivirus genome. J. Virol. 7710623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 756719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinney, R., C. Huang, B. Rose, A. Kroeker, T. Dreher, P. Iversen, and D. Stein. 2005. Inhibition of dengue virus serotypes 1 to 4 in Vero cell cultures with morpholino oligomers. J. Virol. 795116-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kofler, R. M., V. M. Hoenninger, C. Thurner, and C. W. Mandl. 2006. Functional analysis of the tick-borne encephalitis virus cyclization elements indicates major differences between mosquito-borne and tick-borne flaviviruses. J. Virol. 804099-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer, L., J. Li, and P. Shi. 2007. West Nile virus. Lancet Neurol. 6171-181. [DOI] [PubMed] [Google Scholar]
- 25.Kummerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 764773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, W., Y. Li, N. Kedersha, P. Anderson, M. Emara, K. Swiderek, G. Moreno, and M. Brinton. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 7611989-12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott William & Wilkins, Philadelphia, PA.
- 28.Liu, W., X. Wang, V. Mokhonov, P. Shi, R. Randall, and A. Khromykh. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 791934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, W. J., H. B. Chen, and A. A. Khromykh. 2003. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J. Virol. 777804-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo, L., M. Tilgner, K. Bernard, and P.-Y. Shi. 2003. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus using a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 7710004-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandl, C. W., H. Holzmann, C. Kunz, and F. X. Heinz. 1993. Complete genomic sequence of Powassan virus: evaluation of genetic elements in tick-borne versus mosquito-borne flaviviruses. Virology 194173-184. [DOI] [PubMed] [Google Scholar]
- 32.Mandl, C. W., H. Holzmann, T. Meixner, S. Rauscher, P. F. Stadler, S. L. Allison, and F. X. Heinz. 1998. Spontaneous and engineered deletions in the 3′ noncoding region of tick-borne encephalitis virus: construction of highly attenuated mutants of a flavivirus. J. Virol. 722132-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Men, R., M. Bray, D. Clark, R. M. Chanock, and C. J. Lai. 1996. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 703930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 10014333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neuman, B., D. Stein, A. Kroeker, M. Churchill, A. Kim, P. Kuhn, P. Dawson, H. Moulton, R. Bestwick, P. Iversen, and M. Buchmeier. 2005. Inhibition, escape, and attenuated growth of severe acute respiratory syndrome coronavirus treated with antisense morpholino oligomers. J. Virol. 799665-9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neuman, B. W., D. A. Stein, A. D. Kroeker, A. D. Paulino, H. M. Moulton, P. L. Iversen, and M. J. Buchmeier. 2004. Antisense morpholino-oligomers directed against the 5′ end of the genome inhibit coronavirus proliferation and growth. J. Virol. 785891-5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puig-Basagoiti, F., T. S. Deas, P. Ren, M. Tilgner, D. M. Ferguson, and P.-Y. Shi. 2005. High-throughput assays using luciferase-expressing replicon, virus-like particle, and full-length virus for West Nile virus drug discovery. Antimicrob. Agent. Chemother. 494980-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puig-Basagoiti, F., M. Tilgner, C. Bennett, Y. Zhou, J. Munoz-Jordan, A. Garcia-Sastre, K. Bernard, and P. Shi. 18 December 2006, posting date. A mouse cell-adapted NS4B mutation attenuates West Nile virus RNA synthesis. Virology. doi: 10.1016/j.virol.2006.11.012. [DOI] [PMC free article] [PubMed]
- 39.Puig-Basagoiti, F., M. Tilgner, B. Forshey, S. Philpott, N. Espina, Wentworth, S. Goebel, P. S. Masters, B. Falgout, P. Ren, Ferguson, and P. Y. Shi. 2006. Triaryl pyrazoline compound inhibits flavivirus RNA replication. Antimicrob. Agents Chemother. 501320-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ray, D., A. Shah, M. Tilgner, Y. Guo, Y. Zhao, H. Dong, T. Deas, Y. Zhou, H. Li, and P. Shi. 2006. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 808362-8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rice, C. M., E. M. Lenches, S. R. Eddy, S. J. Shin, R. L. Sheets, and J. H. Strauss. 1985. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science 229726-733. [DOI] [PubMed] [Google Scholar]
- 42.Shi, P. Y., M. A. Brinton, J. M. Veal, Y. Y. Zhong, and W. D. Wilson. 1996. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry 354222-4230. [DOI] [PubMed] [Google Scholar]
- 43.Shi, P. Y., M. Tilgner, and M. K. Lo. 2002. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology 296219-233. [DOI] [PubMed] [Google Scholar]
- 44.Shi, P. Y., M. Tilgner, M. K. Lo, K. A. Kent, and K. A. Bernard. 2002. Infectious cDNA clone of the epidemic West Nile virus from New York City. J. Virol. 765847-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Summerton, J., and D. Weller. 1997. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 7187-195. [DOI] [PubMed] [Google Scholar]
- 46.Ta, M., and S. Vrati. 2000. Mov34 protein from mouse brain interacts with the 3′ noncoding region of Japanese encephalitis virus. J. Virol. 745108-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan, B. H., J. Fu, R. J. Sugrue, E. H. Yap, Y. C. Chan, and Y. H. Tan. 1996. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology 216317-325. [DOI] [PubMed] [Google Scholar]
- 48.Tilgner, M., T. S. Deas, and P.-Y. Shi. 2005. The flavivirus-conserved penta-nucleotide in the 3′ stem-loop of the West Nile virus genome requires a specific sequence and structure for RNA synthesis, but not for viral translation. Virology 331375-386. [DOI] [PubMed] [Google Scholar]
- 49.Tilgner, M., and P. Y. Shi. 2004. Structure and function of the 3′-terminal six nucleotides of the West Nile virus genome in viral replication. J. Virol. 788159-8171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van den Born, E., D. A. Stein, P. L. Iversen, and E. J. Snijder. 2005. Antiviral activity of morpholino oligomers designed to block various aspects of Equine arteritis virus amplification in cell culture. J. Gen. Virol. 863081-3090. [DOI] [PubMed] [Google Scholar]
- 51.Warfield, K., D. Swenson, G. Olinger, D. Nichols, W. Pratt, R. Blouch, D. Stein, M. Aman, P. Iversen, and S. Bavari. 2006. Gene-specific countermeasures against Ebola virus based on antisense phosphorodiamidate morpholino oligomers. PLoS Pathog. 2e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.World Health Organization. 2002. Fact sheet, no. 117. Dengue and dengue haemorrhagic fever. World Health Organization, Geneva, Switzerland. http://www.who.int/mediacentre/factsheets/fs117/en/.
- 53.World Health Organization. 2002. Immunization, vaccines and biologicals: Japanese encephalitis. World Health Organization, Geneva, Switzerland. http://www.wpro.who.int/health_topics/encephalitis_japanese/overview.htm.
- 54.World Health Organization. December 2001, revision date. Fact sheet, no. 100. Yellow fever. World Health Organization, Geneva, Switzerland. http://www.who.int/mediacentre/factsheets/fs100/en/.
- 55.You, S., and R. Padmanabhan. 1999. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 27433714-33722. [DOI] [PubMed] [Google Scholar]
- 56.Yuan, J., D. Stein, T. Lim, D. Qiu, S. Coughlin, Z. Liu, Y. Wang, R. Blouch, H. Moulton, P. Iversen, and D. Yang. 2006. Inhibition of coxsackievirus B3 in cell cultures and in mice by peptide-conjugated morpholino oligomers targeting the internal ribosome entry site. J. Virol. 8011510-11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng, L., B. Falgout, and L. Markoff. 1998. Identification of specific nucleotide sequences within the conserved 3′ stem-loop in the dengue type 2 virus genome required for replication. J. Virol. 727510-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou, Y., D. Ray, Y. Zhao, H. Dong, S. Ren, Z. Li, Y. Guo, K. Bernard, P. Shi, and H. Li. 31 January 2007. Structure and function of flavivirus NS5 methyltransferase. J. Virol. doi: 10.1128/JVI.02704-06. (Subsequently published, J. Virol. 813891-3903, 2007.) [DOI] [PMC free article] [PubMed] [Google Scholar]