Abstract
Nucleoside analogs are associated with various mitochondrial toxicities, and it is becoming increasingly difficult to accommodate these differences solely in the context of DNA polymerase gamma inhibition. Therefore, we examined the toxicities of zidovudine (AZT) (10 and 50 μM; 2.7 and 13.4 μg/ml), didanosine (ddI) (10 and 50 μM; 2.4 and 11.8 μg/ml), and zalcitabine (ddC) (1 and 5 μM; 0.21 and 1.1 μg/ml) in HepG2 and H9c2 cells without the presumption of mitochondrial DNA (mtDNA) depletion. Ethidium bromide (EtBr) (0.5 μg/ml; 1.3 μM) was used as a positive control. AZT treatment resulted in metabolic disruption (increased lactate and superoxide) and increased cell mortality with decreased proliferation, while mtDNA remained unchanged or increased (HepG2 cells; 50 μM AZT). ddC caused pronounced mtDNA depletion in HepG2 cells but not in H9c2 cells and increased mortality in HepG2 cells, but no significant metabolic disruption in either cell type. ddI caused a moderate depletion of mtDNA in both cell types but showed no other effects. EtBr exposure resulted in metabolic disruption, increased cell mortality with decreased cell proliferation, and mtDNA depletion in both cell types. We conclude that nucleoside analogs display unique toxicities within and between culture models, and therefore, care should be taken when generalizing about the mechanisms of nucleoside reverse transcriptase inhibitor toxicity. Additionally, mtDNA abundance does not necessarily correlate with metabolic disruption, especially in cell culture; careful discernment is recommended in this regard.
Nucleoside reverse transcriptase inhibitors (NRTIs) as a class represent one of the primary options for the treatment and prevention of human immunodeficiency virus infection. Despite the vast amount of clinical and experimental data demonstrating their toxicity, NRTIs remain at the forefront of human immunodeficiency virus chemotherapy. This dominance is due to several factors, namely, their efficacy, lack of effective alternatives, and an accepted but imperfect risk-benefit approach (i.e., toxicity versus viral inhibition). The future prospects for their use over extended periods inspire an urgency to better understand the toxic mechanisms of these drugs in order to minimize further patient risk.
By design, NRTIs share structural similarities with each other and with the endogenous nucleosides they mimic. Structure-activity assumptions would normally suggest that these compounds share similar mechanisms of toxicity; however, a survey of the literature involving NRTI research shows that the evidence paints a much more convoluted picture (34, 52). The distinctiveness and tissue specificities of NRTI toxicities suggests that unintended targets are varied among individual compounds within this family of drugs. Much of the variation in NRTI toxicity has been attributed to differing levels of prodrug activation in different tissues (reviewed in reference 21). The activities of NRTIs against viral reverse transcriptase and incorporation/termination of endogenous DNA by polymerases (i.e., mitochondrial DNA [mtDNA] polymerase gamma [Pol-γ]) requires their activation to the triphosphate nucleotide, which occurs through the endogenous deoxynucleotide phosphorylation processes. The activities of these phosphorylation pathways vary with the tissue and its respective mitotic state. For example, the first step in activation of thymidine analogs (zidovudine [3′-azido-3′-deoxythymidine; AZT] and stavudine [2′,3′-didehydro-3′-deoxythmidine; d4T]) preferentially occurs in mitotically active cells, because thymidine kinase 1 is expressed only during S phase (though mitochondrial thymidine kinase 2 may play a role in activation in quiescent cells). Activation pathways for other NRTIs (didanosine [2′3′-dideoxyinosine; ddI], zalcitabine [2′,3′-dideoxycytidine; ddC], and lamivudine) are constitutively expressed, and differences in toxicity could be attributed to differences in the rate-limiting steps specific to the analog. However, the apparent species-related difference in ddC activation suggests that other factors may play roles (2, 27). In terms of tissue culture cell lines in which the population of cells is intrinsically mitotically active, one would expect that all NRTIs would be activated according to their proposed pathways, which may or may not reflect the in vivo tissue state.
AZT is primarily associated with hematological toxicity and myopathy (skeletal and possibly cardiac); AZT-triphosphate is a weak inhibitor of Pol-γ (18, 32, 52). d4T is associated with lipdystrophy and an increased risk for lactic acidosis and is considered a moderate inhibitor of Pol-γ (18, 32, 52). ddI is associated with pancreatitis and peripheral neuropathy, while its active form is considered a strong inhibitor of Pol-γ (18, 32, 52). ddC is associated with a high risk for peripheral neuropathy, and its active form is considered the most potent against Pol-γ (18, 32, 52). These facts demonstrate NRTIs' dissimilar toxicological profiles, including tissue specificities and distinctive markers of metabolic disruption. At the center of these differences lies the mitochondrion, which has long been suspected to be the inadvertent target of NRTI toxicity. While there are some studies that provide mechanisms aside from mitochondria (31, 46, 54), based on the observed pathologies, the mitochondria remain the primary focus. Irrespective of this common focus, NRTIs display surprisingly variable toxicities, and it is becoming increasingly difficult to accommodate these differences within a single mechanism (i.e., the “DNA Pol-γ hypothesis” [23, 24]).
The Pol-γ hypothesis elegantly describes a mechanism that explains how nucleoside analogs could specifically cause mitochondrial dysfunction. However, it is becoming increasingly common to invoke the Pol-γ hypothesis in the absence of data demonstrating components of the complete mechanism (i.e., mtDNA depletion, mitochondrial mRNA depletion, respiratory complex dysfunction, etc.) or without consideration of data showing that these markers are not always directly related (3, 43, 47). Such considerations may be critical, especially when using cell culture models to study NRTI toxicity.
In this study, we investigated the effects of three commonly used NRTIs (AZT, ddI, and ddC) using two cell culture lines representing cardiac (H9c2-1) and hepatic (HepG2) tissue types. The purpose was to explore the distinct characteristics of cytotoxicity presented by these drugs without presupposing an obligatory role of mtDNA depletion as a primary determinant of cellular or metabolic dysfunction.
MATERIALS AND METHODS
Chemicals.
All reagents were of research or cell culture quality. AZT was purchased from Toronto Research Chemicals (North York, Ontario, Canada). Ethidium bromide (EtBr), ddI, and ddC were purchased from Sigma (St. Louis, MO). Tissue culture reagents were purchased from Invitrogen (Carlsbad, CA). MitoSOX Red, ethidium homodimer, and calcein AM were obtained from Molecular Probes (Eugene, OR). All other reagents were from Sigma unless otherwise specified.
Cell culture and treatment.
H9C2-1 cardiac myocytes obtained from the American Type Culture Collection (ATCC) (Manassas, VA) were cultured in Dulbecco's modified Eagle medium adjusted to contain 1.5 g/liter sodium bicarbonate and supplemented with 10 ml/liter of antibiotic-antimycotic solution (100×) and 10% fetal bovine serum (FBS). Cells were plated on 75-cm2 tissue culture flasks and grown in a 5% CO2 incubator at 37°C with saturating humidity with medium changes every 2 days. At ∼50% confluence, the cell growth medium was changed to Dulbecco's modified Eagle medium supplemented with 1% FBS and 10 nM all-trans-retinoic acid in order to maintain cardiac-like characteristics (36). Media were changed every 2 days, with daily additions of all-trans-retinoic acid. Cells were maintained under these conditions for 7 days before treatment began.
HepG2/C3A human hepatoma cells were also obtained from the ATCC. HepG2 cells were cultured in minimum essential medium supplemented with 1.5 g/liter sodium bicarbonate, 110 mg of sodium pyruvate, and 10% FBS. Cells were plated on 75-cm2 tissue culture flasks and grown in a 5% CO2 incubator at 37°C with saturating humidity with medium changes every 2 days. The cells were passaged by detachment with 0.05% trypsin and 0.5 mM EDTA.
Cells were cultured in the presence of 10 or 50 μM AZT, 10 or 50 μM ddI, or 1 or 5 μM ddC (control cells were cultured in the absence of compound); drug stocks were prepared in sterile phosphate-buffered saline (PBS) and replenished at medium changes. Drug stocks were stored at −20°C between doses, and new stocks were prepared before each experiment. The doses of AZT and ddI were selected to represent steady-state peak plasma levels of patients receiving NRTI therapy (17, 38), as well as a supratherapeutic dose to observe concentration-related effects; supratherapeutic doses of ddC were used after preliminary experiments showed minimal toxicity with ddC in H9c2 cells. Additionally, another treatment group containing 0.5 μg/ml (1.3 μM) EtBr was included as a positive control.
Cell viability.
The number of dead cells was determined using the LIVE/DEAD viability assay kit (Molecular Probes). Briefly, harvested cells were resuspended in 1 ml PBS containing 100 nM calcein AM and 8 μM ethidium homodimer 1 and incubated in the dark at 37°C for 30 min. The cells were then analyzed on a FACSCaliber flow cytometer using 488-nm excitation, and calcein AM emission was measured with a 530/30 bandpass filter and ethidium homodimer 1 emission with a 661/16 bandpass filter. After gating was performed to exclude debris, dead cells were considered to be those that exhibited high ethidium homodimer 1 fluorescence and low calcein AM fluorescence. The values reported are out of 10,000 events.
Cell proliferation.
A measure of cell proliferation was determined using sulforhodamine B (SRB) as described previously (19). Briefly, cells were seeded in a 12-well culture plate at approximately 2,500 cells per well. At day 6, the medium was removed from the wells, and the adhered cells were left to air dry and were fixed overnight at −10°C with methanol containing 1% acetic acid. The methanol was then aspirated, and the plates were air dried before 2 ml of 0.5% SRB (in 1% acetic acid) was added to each well. The plates were incubated at room temperature for 1 h, the dye was removed, and each well was rinsed with 1% acetic acid before being air dried one more time. The bound dye was eluted with 2 ml of 10 mM Tris buffer (pH 10), and the absorbance was measured in a spectrophotometer at 540 nm. Cell density restrictions prevented the measurement of proliferation beyond day 6.
Extracellular lactate.
Lactate was measured in media drawn from the 12-well culture plates used in the cell proliferation experiment. Briefly, 0.5 ml medium was mixed with 1 ml ice-cold 10% trichloroacetic acid and placed on ice for 30 min. Samples were then centrifuged at 5,000 × g for 3 min, and the clarified supernatant was used for lactate determination. Lactate was measured through the catalytic generation of NADH on a per-mole basis; briefly, 290 μl of a 0.2 M glycine-hydrazine (pH 9.2) reaction buffer containing 0.6 mg/ml NAD+ and 17 U/ml lactate dehydrogenase was added to each well of a 96-well plate. Samples (10 μl) were added to the appropriate wells, and the reaction mixtures were incubated for 30 min at room temperature before being read at 340 nm on a microplate reader. Lactate is reported as nmol/well and was normalized by cell number using the respective SRB absorption values.
Mitochondrial superoxide.
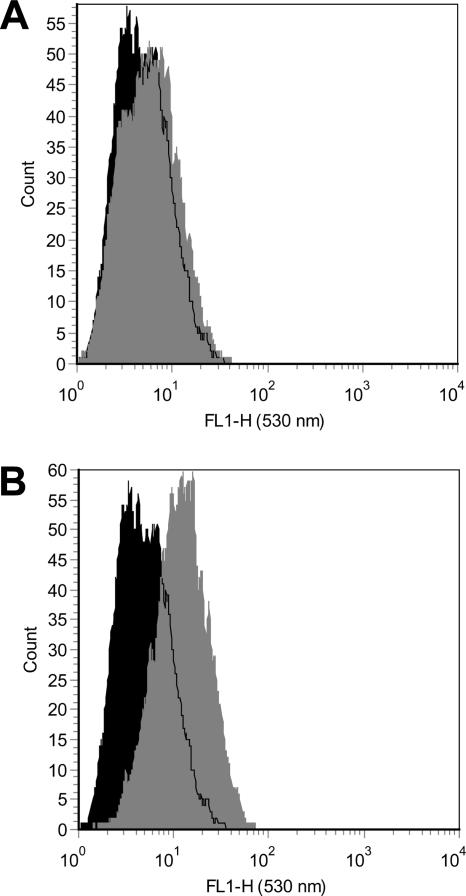
Intramitochondrial superoxide production was measured using the live-cell permeable and mitochondrial localizing MitoSOX Red fluorescent probe (hydroethidine analog) (M. S. Janes, D. M. Hill, C. M. Cardon, K. M. Robinson, J. R. Walls, W.-Y. Leung, J. S. Beckman, and M. J. Ignatius, presented at the American Society for Cell Biology 44th Annual Meeting, Washington, DC, 2004). To our knowledge, this is the first time this probe has been used to measure superoxide by flow cytometry. Mitochondrial localization was confirmed by fluorescence microscopy showing co-staining of the mitochondria by MitoTracker Green and MitoSOX Red in both HepG2 and H9c2 cells (data not shown). Medium from each 75-cm2 flask was aspirated, and the attached cells were rinsed with PBS before adding 3 ml of a 5 μM MitoSOX solution in Hank's balanced salt solution with calcium and magnesium. Coated cells were incubated for 10 min at 37°C in the dark before being harvested by trypsinization. The cells were resuspended in Hank's balanced salt solution with calcium and magnesium for analysis on the FACSCaliber flow cytometer, using 488-nm excitation and a 530/30-bp emission filter; use of this filter allowed the observation of MitoSOX fluorescence while minimizing interference from EtBr, as well as non-superoxide hydroethidine oxidation products (55). Values are reported as the geometric mean fluorescence of a population of 10,000 cells; representative histograms demonstrating the effect of AZT on MitoSOX fluorescence in HepG2 mitochondrial superoxide production after 14 days of exposure are shown in Fig. 1.
FIG. 1.
Representative flow cytometry histograms of MitoSOX Red fluorescence (530-nm emission) in HepG2 cells (10,000 cells). Control (black) and treated (gray) cells were prepared as described in Materials and Methods. Treated cells were exposed to 10 μM (A) and 50 μM (B) AZT for 14 days.
mtDNA abundance.
Cells for DNA isolation were harvested by trypsinization and resuspended in 200 μl of PBS. DNA was extracted from the cells using a DNeasy Tissue Kit (QIAGEN, Valencia, CA). To assess the mtDNA content per cell, the number of copies of well-conserved single-copy genes was measured (6). Pyruvate kinase (forward primer, ACTGGCCGGTGTCATAGTGA; reverse primer, TGTTGACCAGCCGTATGGATA; 347-bp product) was used as a marker for nuclear DNA, while cytochrome b (forward primer, CACCCACATCTGCCGAGAC; reverse primer, GGAAATGCGAAGAAGCGTGTTA; 348-bp product) was used as a marker for mtDNA (IDT, Coralville, IA). Real-time PCR mixtures were prepared with LightCycler FastStart DNA Master SYBR Green 1 reagents according to the kit suggestions, with 0.5 μM concentrations of each primer and 2 mM MgCl2, in a LightCycler 1.5 (Roche Diagnostics, Indianapolis, IN). Quantification of mtDNA was accomplished by calculating the ratio of the mitochondrially encoded gene (cytochrome b) to the nuclear-encoded gene (pyruvate kinase) and expressing it as mtDNA copy number per nuclear DNA copy number. Copy numbers for each gene were calculated based on standard curves. The standards were generated by PCR of DNA with specific primers for each gene using QIAGEN HotStartTaq Master Mix. PCR was conducted in a Perkin-Elmer thermocycler. Agarose gel electrophoresis was performed to verify the products. The products were then purified using a QIAquick PCR purification kit according to the manufacturer's instructions (QIAGEN) and quantitated by measuring absorbance at 260 nm. The DNA copy number was calculated using the resulting concentration of DNA and the following equation: MW = (product length × 607.4) + 157.9 (Ambion Inc., Austin, TX).
Statistics.
Data, grouped by days of treatment, were analyzed using GraphPad Prism 4 software by one-way analysis of variance, followed by the Tukey post hoc test for comparison between treatment groups. Unless otherwise stated, values represent the means of three separate experiments plus the mean standard error; significance was assumed at 95% confidence.
RESULTS
This study was designed to contrast the possible mitochondrial toxicities of several NRTIs with that of EtBr, which is known to deplete mtDNA and mitochondrial RNA (16, 37, 56), in order to determine whether mtDNA abundance necessarily correlates with toxicity. Although NRTIs are now commonly administered in pairs, single NRTI exposures were used in this study to better differentiate their activities.
Preliminary time course experiments using H9c2 cells revealed that this cell line is generally nonresponsive to NRTI treatment. In consideration of this observation, data for the cell line were collected only after 12 days of treatment.
AZT.
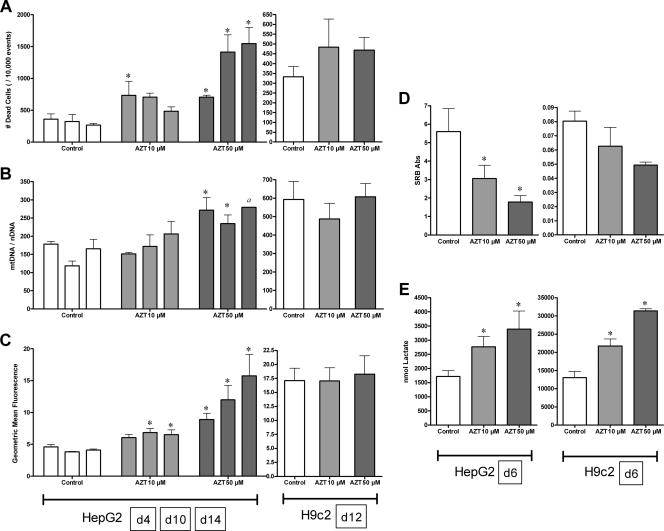
In this study, AZT treatment caused metabolic disruption without mtDNA depletion; these effects were more pronounced in HepG2 cells than in H9c2 cells.
While all cell populations were consistently above 80% viable, AZT caused an increase in HepG2 cell death on day 4 at 10 μM and days on 4, 10, and 14 at 50 μM (Fig. 2A). This effect was not observed in H9c2 cells at day 12. Similarly, HepG2 proliferation was inhibited by AZT (50 μM) at day 6; a similar trend was observed in H9c2 cells (Fig. 2D).
FIG. 2.
Effects of AZT (10 [light gray] and 50 [dark gray] μM) on HepG2 and H9c2 cells. HepG2 cells were treated for 4, 10, and 14 days (A to C) and 6 days (D and E); H9c2 cells were treated for 12 days (A to C) and 6 days (D and E). (A) Cell mortality determined using the LIVE/DEAD assay for flow cytometry and reported as the number of dead cells/10,000 events. (B) mtDNA abundance determined by quantitative real-time PCR reported as the ratio of cytochrome b gene copy number/pyruvate kinase gene copy number (mtDNA/nuclear DNA[nDNA]). (C) Mitochondrial superoxide generation determined using the MitoSOX Red fluorescent probe by flow cytometry, reported as the geometric mean fluorescence of 10,000 events. (D) Cell proliferation determined using the SRB binding assay, reported as the dilution-adjusted absorbance at 540 nm. Abs, antibodies. (E) Extracellular lactate determined using NAD+ reduction by lactate dehydrogenase, reported as nmol lactate per culture well normalized by cell number using the respective SRB absorption values. The data reported are the means of three separate experiments plus mean standard errors unless otherwise indicated. *, statistically different from time-matched control (P < 0.05). a, n = 2 due to insufficient material for analysis.
Metabolic fidelity was measured using two end points, extracellular lactate and mitochondrial superoxide. AZT caused a significant elevation in medium lactate by day 6 at both 10 and 50 μM and in both culture models (Fig. 2E). Mitochondrial superoxide was increased in HepG2 cells at all time points with AZT (50 μM); in contrast, H9c2 cells showed no change in mitochondrial superoxide with AZT (Fig. 2C).
AZT treatment did not result in mtDNA depletion (Fig. 2B). In fact, in HepG2 cells, 50 μM AZT caused an increase in the mtDNA copy numbers on days 4 and 10 (there was insufficient material for day 14 due to inhibited cell proliferation). No change in mtDNA was observed in H9c2 cells treated with AZT through day 12.
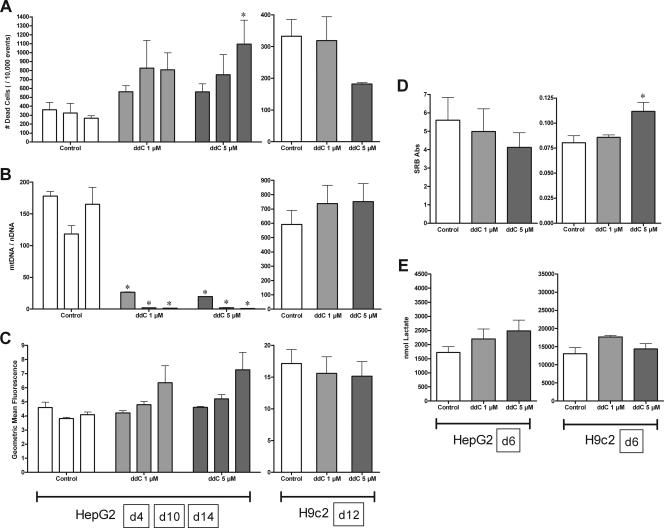
ddC.
Treatment with ddC caused rapid and severe depletion of mtDNA in HepG2 cells only; no significant metabolic effects were observed in either cell type.
Cells were generally viable with ddC, though a significant increase in dead cells was observed with 5 μM ddC on day 14 with HepG2 cells (Fig. 3A); HepG2 cell proliferation was not affected (Fig. 3D). H9c2 cell viability was unaffected by ddC treatment (Fig. 3A), and 5 μM ddC caused an increase in cell proliferation by day 6 (Fig. 3D). Neither extracellular lactate nor mitochondrial superoxide was significantly affected by ddC treatment in either culture model, though a time-dependent increasing trend might be suggested for mitochondrial superoxide in HepG2 cells (Fig. 2C and 3E).
FIG. 3.
Effects of ddC (1 [light gray] and 5 [dark gray] μM) on HepG2 and H9c2 cells. HepG2 cells were treated for 4, 10, and 14 days (A to C) and 6 days (D and E); H9c2 cells were treated for 12 days (A to C) and 6 days (D and E). See the legend to Fig. 1 for panel descriptions. The data reported are the means of three separate experiments plus mean standard errors. *, statistically different from time-matched control (P < 0.05).
Most striking with ddC were the differential responses of the culture models with respect to mtDNA depletion (Fig. 3B). HepG2 cells showed a dramatic drop in mtDNA abundance: 15% and 11% of control for 1 and 5 μM ddC, respectively, by day 4. This continued through day 14, when mtDNA abundance dropped to less than 1% that of the control for both 1 and 5 μM ddC. In contrast, H9c2 cells showed no change in mtDNA abundance through day 12.
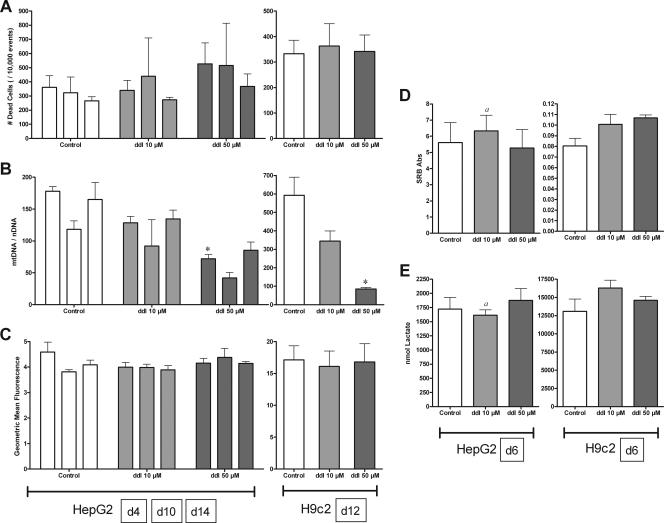
ddI.
Cells treated with ddI showed moderate depletion in mtDNA in both culture models without metabolic disruption.
No increase in cell death was observed for HepG2 or H9c2 cells exposed to ddI (Fig. 4A). Although observation of ddI-treated H9c2 cells grown in 75-cm2 flasks suggested a higher density on day 12, this effect was not measured with the SRB assay on day 6; no change in proliferation was seen in HepG2 cells (Fig. 4D). Neither cell line demonstrated metabolic alterations with ddI treatment (Fig. 3E and 4C).
FIG. 4.
Effects of ddI (10 [light gray] and 50 [dark gray] μM) on HepG2 and H9c2 cells. HepG2 cells were treated for 4, 10, and 14 days (A to C) and 6 days (D and E); H9c2 cells were treated for 12 days (A to C) and 6 days (D and E). See the legend to Fig. 1 for panel descriptions. The data reported are the means of three separate experiments plus mean standard errors unless otherwise indicated. *, statistically different from time-matched control (P < 0.05). a, n = 2 due to insufficient material for analysis.
In contrast to ddC, both HepG2 and H9c2 showed depletion of mtDNA with ddI exposure (Fig. 4B). HepG2 cells grown with 10 μM ddI consistently had mtDNA levels at ∼80% those of controls, while those grown in 50 μM ddI had about 40 to 50% of control levels at all time points. H9c2 cells treated with ddI for 12 days had mtDNA values ∼60% and 15% those of controls for 10 and 50 μM, respectively.
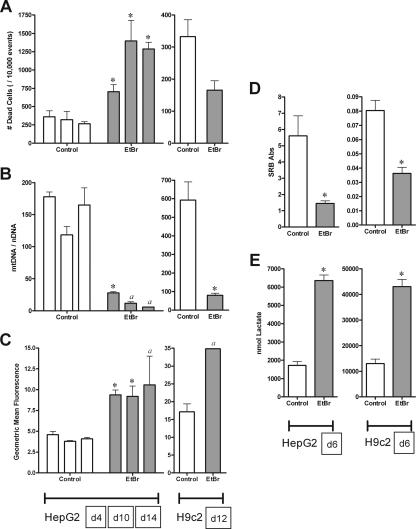
EtBr.
EtBr, shown to be effective at generating ρ0 cells (cells lacking mtDNA) (37), was used as a positive control for mtDNA depletion. Range-finding experiments (data not shown) showed that treatment with 0.5 μg/ml EtBr was effective at depleting mtDNA in both HepG2 (16% of control by day 6) and H9c2 (21% of control by day 6) cell lines.
Increased cell death was seen in EtBr-treated HepG2 cells on days 10 and 14, though overall viability was still above 80% (Fig. 5A). H9c2 cell viability was unaffected by EtBr treatment (Fig. 5A). EtBr inhibited cell proliferation for both HepG2 and H9c2 cells through day 6 (Fig. 5D).
FIG. 5.
Effects of EtBr (0.5 μg/ml [light gray]) on HepG2 and H9c2 cells. HepG2 cells were treated for 4, 10, and 14 days (A to C) and 6 days (D and E); H9c2 cells were treated for 12 days (A to C) and 6 days (D and E). See the legend to Fig. 1 for panel descriptions. The data reported are the means of three separate experiments plus mean standard errors unless otherwise indicated. *, statistically different from time-matched control (P < 0.05). a, n = 2 due to insufficient material for analysis.
In terms of metabolic disruption, EtBr treatment increased extracellular lactate in HepG2 and H9c2 cells by day 6 (Fig. 5E). Additionally, the measure of mitochondrial superoxide was increased in both cell lines at all time points (Fig. 5C). Though we tried to avoid the nonspecific ethidium fluorescence, the increased signal for mitochondrial superoxide could still represent nonspecific ethidium reduction products, due to the higher concentration of ethidium in these cells.
As expected, EtBr diminished the mtDNA content in both cell lines (Fig. 5B). The inhibited rate of HepG2 cell proliferation prevented the collection of a full data set (n = 3) by days 10 and 14, though a trend for depletion was observed: ∼16%, 10%, and 4% of control for days 4, 10, and 14, respectively. H9c2 cells grown in 0.5 μg/ml EtBr for 12 days had ∼14% of the mtDNA content of the controls.
DISCUSSION
The mitochondrion is surely a target for NRTI toxicity, as evidenced by the vast amount of literature showing morphological and biochemical alterations of the organelle with NRTI exposure in vivo and in vitro (12, 21, 29, 52). However, this shared locus of toxicity belies the apparent complexity of NRTI toxicity. Suggested mechanisms of toxicity include, but are not limited to, the inhibition of DNA Pol-γ (23, 24, 32), inhibition of endogenous nucleotide kinases (30, 35, 49), direct inhibition of oxidative phosphorylation (4, 5, 29, 40), reactive oxygen species (ROS) generation (31, 44, 46), and mtDNA and nuclear DNA mutagenesis (1, 10, 45), yet mtDNA abundance is commonly employed as the hallmark of NRTI toxicity. The results of this investigation demonstrate that inhibition of DNA Pol-γ is but one of many possible cytotoxic mechanisms of the different NRTIs to alter the metabolic status of the cell and that the essences of mtDNA depletion in the mechanism of cytotoxicity differ among the different cell lines.
In this study, we demonstrated that AZT, ddC, and ddI exhibit distinct toxicity profiles both within a single cell culture model and between two different cell types. Our results suggest that HepG2 cells are more responsive to NRTI exposure while H9c2 cells are generally refractory. This is in agreement with other authors, who have suggested that HepG2 cells are perhaps the “ideal” cell culture line to study NRTI-induced mitochondrial toxicity (20, 41). However, one should be careful to consider the clinical relevance of choosing a model solely on the basis of its responsiveness to the test compounds without regard to its clinical surrogate.
It should be emphasized that there is a danger in extrapolating toxicity results obtained from tissue culture to their related organ system or whole organism, especially with regard to the mitochondria. Transformed cells are highly glycolytic, and it could be argued that, in terms of energy requirements, the mitochondria are at least secondary and possibly accessory in importance. Indeed, our laboratory has exposed H9c2 cells grown in 4.5 g/liter glucose to 20 μM rotenone over 48 h without affecting viability or ATP concentration (unpublished results). In this light, the observation that in HepG2 cells ddC caused 99% depletion of mtDNA without pronounced downstream effects may be understandable. In a tissue heavily dependent on mitochondria, this extent of depletion is far above predicted threshold effects (43) and could be devastating. It is interesting, however, that despite the minimal amount of mtDNA present, the cells were still able to provide sufficient NAD+ to support glycolysis without having to resort to lactate production.
This observation provides evidence to show that mitochondrial metabolic fidelity does not always correlate with mtDNA abundance (especially in cell culture models); indeed, in this study, the two factors appeared unrelated. The compound most effective at increasing lactate and mitochondrial superoxide (AZT) was the least effective at depleting mtDNA, while ddC and ddI both depleted mtDNA without significantly altering lactate or superoxide production. The only compound that seemed to demonstrate components of the Pol-γ hypothesis—mtDNA depletion, which results in downstream metabolic disruption and oxidative stress—was the positive control, EtBr. EtBr intercalates into DNA and at low concentrations specifically inhibits both replication and transcription of mtDNA irrespective of Pol-γ activity (16, 56). The fact that EtBr can directly inhibit transcription may explain the differences observed between ddC and EtBr in HepG2 cells. While ddC was more effective at depleting mtDNA, transcription is not believed to be directly affected by NRTIs, and perhaps the remaining mitochondrial genomes were sufficient to maintain adequate mitochondrial functionality in culture in this time frame. The time-dependent increasing trend in cell mortality and mitochondrial superoxide observed with ddC (Fig. 2A and C) may suggest that mtDNA depletion was beginning to affect mitochondrial fidelity; longer exposures may have exacerbated this effect, as demonstrated previously (50, 51). If this was the case, ddC would appear to verify elements of the Pol-γ hypothesis in HepG2 cells, but not in H9c2 cells.
In general, for the respective doses and time frames, the HepG2 data presented here are in agreement with previous studies using this cell line (7, 39, 50) and confirm the observation that AZT is cytotoxic without depleting mtDNA. Oxidative damage and ROS have been observed to increase with AZT treatment (14, 15, 46, 53), and both AZT and AZT-monophosphate have been shown to be pro-oxidant in vitro (31). In this study, we demonstrate that AZT exposure increases intramitochondrial superoxide production in HepG2 cells. Though the mechanism for this increase is uncertain, it suggests that mitochondrial sources for superoxide generation may be targets for AZT or its phosphorylated forms. Indeed, AZT has been shown to alter the functionality of respiratory complexes I and III (28, 29, 40, 48). Mitochondrion-localized elevation of ROS by AZT may also provide a mechanism for the morphological alteration and mtDNA disruption observed with long-term exposure (13, 25, 33) without involving Pol-γ, which has a low affinity for AZT-triphosphate (26, 32).
In the context of glycolytically active cells, the differences evoked by AZT and ddC are suggestive of the mechanisms of action. Others have reported early metabolic and functional disruption with ddC in the rat heart (44); however, our study suggests that ddC has no direct effect on HepG2 or H9c2 mitochondrial function but instead primarily depletes mtDNA (HepG2), presumably through the inhibition of Pol-γ. With HepG2 cells, in spite of a dramatic loss in mtDNA with ddC, extracellular lactate remained stable through day 6, suggesting that for these cultured cells, mtDNA is not required in substantial quantities. In contrast, AZT showed an increase in lactate, which suggests a significant restriction in the availability of NAD+. Whether this is due to poly-ADP-ribose polymerase activation (46) potentially caused by the increase in superoxide/ROS, the direct inhibition of NADH oxidation in the mitochondria, or a combination of these factors is unclear. It would be convenient if the source of superoxide and the target for NADH oxidation were the same, which would suggest that AZT interacts with complex I to exert its early metabolic effects.
The H9c2 cell line, derived from embryonic rat heart tissue, represents one of a few cardiac-derived cell lines that maintain some cardiac-like characteristics under reasonably simple culture conditions (22). The use of low-serum (1% FBS) media supplemented with all-trans-retinoic acid has been shown to maintain the cardiac phenotype and inhibit the spontaneous differentiation to skeletal myotubes (36). H9c2 cells grown under these conditions are not terminally differentiated, though they have decreased proliferation rates and will form large multinucleate cells. With that understanding, H9c2 cells represent a prospective culture model for cardiac tissue, which is the context in which they were used in this study.
Compared to HepG2 cells, the effect of NRTI treatment on H9c2 cells is markedly subdued. Whether this difference is reflective of tissue-, species-, or cell-line-specific characteristics (or some combination thereof) is unclear. The clinically observed tissue-specific toxicities (and efficacies) of NRTIs have been attributed to differences in prodrug activation (phosphorylation) in the various tissues (21). In this scheme, AZT and d4T are preferentially phosphorylated in active cells while ddC and ddI are activated in postmitotic/resting cells. However, in the context of actively proliferating cultured cells, this scheme cannot explain the difference observed in mtDNA depletion with ddC in this study. Considering that the H9c2 cells were proliferating and the mtDNA content was being maintained, it must be assumed that dCTP was being produced and transported into the mitochondria. However, there is evidence to suggest that ddC activation (and subsequent toxicity) is dependent on the cell species and that rat and mouse cells have a significantly smaller ability to phosphorylate ddC than human, monkey, and rabbit cells (2, 27). Whether this difference is due to transport barriers in rat cells, such as H9c2, or other differences in nucleotide metabolism (i.e., production of ddC liponucleotide adducts (42) is open to speculation and warrants investigation, especially if this is an adequate model for cardiac-type cells. The capacity of HepG2 or H9c2 cells to phosphorylate/activate the various NRTIs has not been reported in the open literature and might be an area for future study, especially if HepG2 cells represent an attractive model for NRTI-induced mitochondrial dysfunction (20, 41). Regardless, the cell-specific differences observed with ddC between H9c2 and HepG2 cells underscore the danger of assuming that NRTI toxicity is a result of mtDNA depletion in all experimental models.
The Pol-γ hypothesis provides a valuable model to understand the toxicity of several NRTIs in many tissues; however, this study confirms that the inhibition of Pol-γ is unlikely to be the exclusive mechanism by which an NRTI exerts its toxic effects and that demonstration of mtDNA depletion is insufficient to explain metabolic dysfunction in cell culture. In the current study, it required an almost complete depletion of mtDNA over an extended period before mitochondrial dysfunction was suggested with ddC. Given enough time, ddI may have also caused sufficient mtDNA depletion to result in metabolic disruption, as observed previously (50, 51). In light of these observations, it is important, especially in cell culture models, to distinguish between mitochondrial dysfunction and mtDNA abundance. Conclusions based on moderate depletion of mtDNA should be accompanied by evidence to show that the depletion is sufficient to disrupt metabolism in that model. Likewise, observations of NRTI-related metabolic disruption should not be attributed to mtDNA depletion without evidence demonstrating the latter. It is generally accepted that AZT toxicity may result from several alternate pathways (4, 5, 8, 29, 35); therefore, we should assume that the other NRTIs may also display a diverse range of effects in the context of different tissues/models. The clinical and experimental data reflect the complexity of this problem (9, 11, 34) and suggest that it should be examined from several perspectives. A more comprehensive and useful understanding of nucleoside metabolism, signaling, regulation, and NRTI toxicity/activity would result from this more discriminating approach.
Acknowledgments
This research was supported by the NIH (HL072715).
Footnotes
Published ahead of print on 30 April 2007.
REFERENCES
- 1.Agarwal, R. P., and O. A. Olivero. 1997. Genotoxicity and mitochondrial damage in human lymphocytic cells chronically exposed to 3′-azido-2′,3′-dideoxythymidine. Mutat. Res. 390223-231. [DOI] [PubMed] [Google Scholar]
- 2.Balzarini, J., R. Pauwels, M. Baba, P. Herdewijn, E. de Clercq, S. Broder, and D. G. Johns. 1988. The in vitro and in vivo anti-retrovirus activity and intracellular metabolism of 3′-azido-2′,3′-dideoxythymidine and 2′,3′-dideoxycytidine are highly dependent on the cell species. Biochem.l Pharmacol. 37897-903. [DOI] [PubMed] [Google Scholar]
- 3.Barazzoni, R., K. R. Short, and K. S. Nair. 2000. Effects of aging on mitochondrial DNA copy number and cytochrome c oxidase gene expression in rat skeletal muscle, liver, and heart. J. Biol. Chem. 2753343-3347. [DOI] [PubMed] [Google Scholar]
- 4.Barile, M., D. Valenti, G. A. Hobbs, M. F. Abruzzese, S. A. Keilbaugh, E. Quadgliariello, S. Passarella, and M. V. Simpson. 1994. Mechanisms of toxicity of 3′-azido-3′-deoxythymidine: its interaction with adenylate kinase. Biochem. Pharmacol. 481405-1412. [DOI] [PubMed] [Google Scholar]
- 5.Barile, M., D. Valenti, S. Passarella, and E. Quagliariello. 1997. 3′-Azido-3′-deoxythymidine uptake into isolated rat liver mitochondria and impairment of ADP/ATP translocator. Biochem. Pharmacol. 53913-920. [DOI] [PubMed] [Google Scholar]
- 6.Berthiaume, J., and K. B. Wallace. 2002. Perfluorooctanoate, perfluorooctanesulfonate, and N-ethyl perfluorooctanesulfonamido ethanol; peroxisome proliferation and mitochondrial biogenesis. Toxicol. Lett. 12923-32. [DOI] [PubMed] [Google Scholar]
- 7.Birkus, G., M. J. M. Hitchcock, and T. Cihlar. 2002. Assessment of mitochondrial toxicity in human cells treated with Tenofovir: comparison with other nucleoside reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 46716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carnicelli, V., A. D. Giulio, A. Bozzi, R. Strom, and A. Oratore. 2006. Zidovudine inhibitis protein kinase C activity in human chronic myeloid (K562) cells. Pharmacol. Toxicol. 99317-322. [DOI] [PubMed] [Google Scholar]
- 9.Chiappini, F., E. Teicher, R. Saffroy, P. Pham, B. Falissard, A. Barrier, S. Chevalier, B. Debuire, D. Vittecoq, and A. Lemoine. 2004. Prospective evaluation of blood concentration of mitochondrial DNA as a marker of toxicity in 157 consecutively recruited untreated or HAART-treated HIV-positive patients. Lab. Investig. 84908-914. [DOI] [PubMed] [Google Scholar]
- 10.Copeland, W. C., M. S. Chen, and T. S. Wang. 1992. Human DNA polymerases alpha and beta are able to incorporate anti-HIV deoxynucleotides into DNA. J. Biol. Chem. 26721459-21464. [PubMed] [Google Scholar]
- 11.Cote, H. C. F. 2005. Possible ways nucleoside analogues can affect mitochondrial DNA content and gene expression during HIV therapy. Antivir. Ther. 10M3-M11. [PubMed] [Google Scholar]
- 12.Dagan, T., C. Sable, J. Bray, and M. Gerschenson. 2002. Mitochondrial dysfunction and antiretroviral nucleoside analog toxicities: what is the evidence? Mitochondrion 1397-412. [DOI] [PubMed] [Google Scholar]
- 13.Dalakas, M. C., I. Illa, G. H. Pezeshkpour, J. P. Laukaitis, B. Cohen, and J. L. Griffin. 1990. Mitochondrial myopathy caused by long-term zidovudine therapy. N. Engl. J. Med. 3221098-1105. [DOI] [PubMed] [Google Scholar]
- 14.de la Asuncion, J. G., M. L. del Olmo, J. Sastre, A. Millan, A. Pellin, F. V. Pallardo, and J. Vina. 1998. AZT treatment induces molecular and ultrastructural oxidative damage to muscle mitochondria (prevention by antioxidant vitamins). J. Clin. Investig. 1024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Asuncion, J. G., M. L. del Olmo, J. Sastre, F. V. Pallardo, and J. Vina. 1999. Zidovudine (AZT) causes an oxidation of mitochondrial DNA in mouse liver. Hepatology 29985-987. [DOI] [PubMed] [Google Scholar]
- 16.Desjardins, P., E. Frost, and R. Morais. 1985. Ethidium bromide-induced loss of mitochondrial DNA from primary chicken embryo fibroblasts. Mol. Cell. Biol. 51163-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estrela, R. D. C. E., M. C. Salvadori, and G. Suarez-Kurtz. 2004. A rapid and sensitive method for simultaneous determination of lamivudine and zidovudine in human serum by on-line solid-phase extraction coupled to liquid chromatography/tandem mass spectrometry detection. Rapid Commun. Mass Spectrom. 181147-1155. [DOI] [PubMed] [Google Scholar]
- 18.Flint, O. P. 1994. In vitro studies of the toxicity of nucleoside analogues used in the treatment of HIV infection. Toxicol. in Vitro 8677-683. [DOI] [PubMed] [Google Scholar]
- 19.Holy, J., G. Lamont, and E. Perkins. 2006. Disruption of nucleocytoplasmic trafficking of cyclin D1 and topoisomerase II by sanguinarine. BMC Cell Biol. 713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoschele, D. 2006. Cell culture models for the investigation of NRTI-induced mitochondrial toxicity. Relevance for the prediction of clinical toxicity. Toxicol. in Vitro 20535-546. [DOI] [PubMed] [Google Scholar]
- 21.Kakuda, T. N. 2000. Pharmacology of nucleoside and nucleotide reverse transcriptase inhibitor-induced mitochondrial toxicity. Clin. Therapeutics 22685-708. [DOI] [PubMed] [Google Scholar]
- 22.Kimes, B. W., and B. L. Brandt. 1976. Properties of a clonal muscle cell line from rat heart. Exp. Cell Res. 98376-381. [DOI] [PubMed] [Google Scholar]
- 23.Kohler, J. J., and W. Lewis. 2007. A brief overview of mechanisms of mitochondrial toxicity from NRTIs. Environ. Mol. Mutagen. 48166-172. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, W., and M. C. Dalakas. 1995. Mitochondrial toxicity of antiviral drugs. Nat. Med. 1417-422. [DOI] [PubMed] [Google Scholar]
- 25.Lewis, W., B. Gonzalez, A. Chomyn, and T. Papoian. 1992. Zidovudine induces molecular, biochemical, and ultrastructural changes in rat skeletal muscle mitochondria. J. Clin. Investig. 891354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim, S. E., and W. C. Copeland. 2001. Differential incorporation and removal of antiviral deoxynucleotides by human DNA polymerase γ. J. Biol. Chem. 27623616-23623. [DOI] [PubMed] [Google Scholar]
- 27.Lipman, J. M., J. A. Reichert, A. Davidovich, and T. D. Anderson. 1993. Species differences in nucleotide pool levels of 2′,3′-dideoxycytidine: a possible explanation for species-specific toxicity. Toxicol. Appl. Pharmacol. 123137-143. [DOI] [PubMed] [Google Scholar]
- 28.Lund, K. C., and K. B. Wallace. 2004. Direct effects of nucleoside reverse transcriptase inhibitors on rat cardiac mitochondrial bioenergetics. Mitochondrion 4193-202. [DOI] [PubMed] [Google Scholar]
- 29.Lund, K. C., and K. B. Wallace. 2004. Direct, DNA pol-γ-independent effects of nucleoside reverse transcriptase inhibitors on mitochondrial bioenergetics. Cardiovasc. Toxicol. 4217-228. [DOI] [PubMed] [Google Scholar]
- 30.Lynx, M. D., A. T. Bentley, and E. E. McKee. 2006. 3′-Azido-3′-deoxothymidine (AZT) inhibits thymidine phosphorylation in isolated rat liver mitochondria: a possible mechanism of AZT hepatotoxicity. Biochem. Pharmacol. 711342-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mak, I. T., L. F. Nedelec, and W. B. Weglicki. 2004. Pro-oxidant properties and cytotoxicity of AZT-monophosphate and AZT. Cardiovasc. Toxicol. 4109-115. [DOI] [PubMed] [Google Scholar]
- 32.Martin, J. L., C. E. Brown, N. Matthews-Davis, and J. E. Reardon. 1994. Effects of antiviral nucleoside analogs on human DNA polymerases and mitochondrial DNA synthesis. Antimicrob. Agents Chemother. 382743-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masini, A., C. Scotti, A. Caligaro, O. Cazzalini, L. A. Stivala, L. Bianchi, F. Giovannini, D. Ceccarelli, U. Muscatello, A. Tomasi, and V. Vannini. 1999. Zidovudine-induced experimental myopathy: dual mechanism of mitochondrial damage. J. Neurol. Sci. 166131-140. [DOI] [PubMed] [Google Scholar]
- 34.McComsey, G., R. K. Bai, J. F. Maa, D. Seekins, and L. J. Wong. 2005. Extensive investigations of mitochondrial DNA genome in treated HIV-infected subjects: beyond mitochondrial DNA depletion. J. Acquire. Immune Defic. Syndr. 39181-188. [PubMed] [Google Scholar]
- 35.McKee, E. E., A. T. Bentley, M. Hatch, J. Gingerich, and D. Susan-Resiga. 2004. Phosphorylation of thymidine and AZT in heart mitochondria: elucidation of a novel mechanism of AZT cardiotoxicity. Cardiovasc. Toxicol. 4155-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menard, C., S. Pupier, D. Mornet, M. Kitzmann, J. Nargeot, and P. Lory. 1999. Modulation of L-type calcium channel expression during retinoic acid-induced differentiation of H9c2 cardiac cells. J. Biol. Chem. 27429063-29070. [DOI] [PubMed] [Google Scholar]
- 37.Moraes, C. T., R. Dey, and A. Barrientos. 2001. Transmitochondrial technology in animal cells, p. 397-412. In L. A. Pon and E. A. Schon (ed.), Mitochondria, vol. 65. Academic Press, San Diego, CA. [DOI] [PubMed] [Google Scholar]
- 38.Moyer, T. P., Z. Temesgen, R. Enger, L. Estes, J. Charlson, L. Oliver, and A. Wright. 1999. Drug monitoring of antiretroviral therapy for HIV-1 infection: method validation and results of a pilot study. Clin. Chem. 451465-1476. [PubMed] [Google Scholar]
- 39.Pan-Zhou, X.-R., L. Cui, X.-J. Zhou, J.-P. Sommadossi, and V. M. Darley-Usmar. 2000. Differential effects of antiretroviral nucleoside analogs on mitochondrial function in HepG2 cells. Antimicrob. Agents Chemother. 44496-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pereira, L. F., M. B. M. Oliveira, and E. G. S. Carnieri. 1998. Mitochondrial sensitivity to AZT. Cell Biochem. Funct. 16173-181. [DOI] [PubMed] [Google Scholar]
- 41.Pinti, M., L. Troiano, M. Nasi, R. Ferraresi, J. Dobrucki, and A. Cossarizza. 2003. Hepatoma HepG2 cells as a model for in vitro studies on mitochondrial toxicity of antiretroviral drugs: which correlation with the patient? J. Biol. Regul. Homeostat. Agents 17166-171. [PubMed] [Google Scholar]
- 42.Rossi, L., S. Serafini, G. F. Schiavano, A. Casabianca, G. Vallanti, L. Chiarantini, and M. Magnani. 1999. Metabolism, mitochondrial uptake and toxicity of 2′,3′-dideoxycytidine. Biochem. J. 344915-920. [PMC free article] [PubMed] [Google Scholar]
- 43.Rossignol, R., B. Faustin, C. Rocher, M. Malgat, J.-P. Mazat, and T. Letellier. 2003. Mitochondrial threshold effects. Biochem. J. 370751-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skuta, G., G. M. Fischer, T. Janaky, Z. Kele, P. Szabo, J. Tozser, and B. Sumegi. 1999. Molecular mechanism of the short-term cardiotoxicity caused by 2′,3′-dideoxycytidine (ddC): modulation of reactive oxygen species levels and ADP-ribosylation reactions. Biochem. Pharmacol. 581915-1925. [DOI] [PubMed] [Google Scholar]
- 45.Sussman, H. E., O. A. Olivero, Q. Meng, S. M. Pietras, M. C. Poirier, J. P. O'Neill, B. A. Finette, M. J. Bauer, and V. E. Walker. 1999. Genotoxicity of 3′-azido-3′-deoxythymidine in the human lymphoblastoid cell line, TK6: relationships between DNA incorporation, mutant frequency, and spectrum of deletion mutations in HPRT. Mutat. Res. 429249-259. [DOI] [PubMed] [Google Scholar]
- 46.Szabados, E., G. M. Fischer, K. Toth, B. Csete, B. Nemeti, K. Trombitas, T. Habon, D. Endrei, and B. Sumegi. 1999. Role of reactive oxygen species and poly-ADP-ribose polymerase in the development of AZT-induced cardiomyopathy in rat. Free Radic. Biol. Med. 26309-317. [DOI] [PubMed] [Google Scholar]
- 47.Tang, Y., E. A. Schon, E. Wilichowski, M. E. Vasquez-Memije, E. Davidson, and M. P. King. 2000. Rearrangements of human mitochondrial DNA (mtDNA): new insights into the regulation of mtDNA copy number and gene expression. Mol. Biol. Cell 111471-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valenti, D., A. Atlante, M. Barile, and S. Passarella. 2002. Inhibition of phosphate transport in rat heart mitochondria by 3′-azido-3′-deoxythymidine due to stimulation of superoixide anion mitochondrial production. Biochem. Pharmacol. 64201-206. [DOI] [PubMed] [Google Scholar]
- 49.Valenti, D., M. Barile, E. Quagliariello, and S. Passarella. 1999. Inhibition of nucleoside diphosphate kinase in rat liver mitochondria by added 3′-azido-3′-deoxythymidine. FEBS Lett. 444291-295. [DOI] [PubMed] [Google Scholar]
- 50.Walker, U. A., B. Setzer, and N. Venhoff. 2002. Increased long-term mitochondrial toxicity in combinations of nucleoside analog reverse-transcriptase inhibitors. AIDS 162165-2173. [DOI] [PubMed] [Google Scholar]
- 51.Walker, U. A., N. Venhoff, E. C. Koch, M. Olschewski, J. Schneider, and B. Setzer. 2003. Uridine abrogates mitochondrial toxicity related to nucleoside analogue reverse transcriptase inhibitors in HepG2 cells. Antivir. Ther. 8463-470. [PubMed] [Google Scholar]
- 52.White, A. J. 2001. Mitochondrial toxicity and HIV therapy. Sex. Transm. Infect. 77158-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamaguchi, T., I. Katoh, and S.-I. Kurata. 2002. Azidothymidine causes functional and structural destruction of mitochondria, glutathione deficiency and HIV-1 promoter sensitization. Eur. J. Biochem. 2692782-2788. [DOI] [PubMed] [Google Scholar]
- 54.Yan, J.-P., D. D. Ilsley, C. Frohlick, R. Steet, E. T. Hall, R. K. Kuchta, and P. Melacon. 1995. 3′-Azidothymidine (Zidovudine) inhibits glycosylation and dramatically alters glycosphingolipid synthesis in whole cells at clinically relevant concentrations. J. Biol. Chem. 27022836-22841. [DOI] [PubMed] [Google Scholar]
- 55.Zhao, H., J. Joseph, H. M. Fales, E. A. Sokoloski, R. L. Levine, J. Vasquez-Vivar, and B. Kalyanaraman. 2005. Detection and characterization of the products of hydroethdine and intracellular superoxide by HPLC and limitations of fluorescence. Proc. Natl. Acad. Sci. USA 1025727-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zylber, E., C. Vesco, and S. Penman. 1969. Selective inhibition of the synthesis of mitochondria-associated RNA by ethidium bromide. J. Mol. Biol. 44195-204. [DOI] [PubMed] [Google Scholar]