Abstract
Long-term lamivudine (LMV) treatment of chronic hepatitis B almost inevitably engenders viral resistance. Mutations that result in the replacement of the methionine at position 204 of the deoxynucleoside triphosphate-binding site of the hepatitis B virus (HBV) reverse transcriptase (rt) by isoleucine, valine, or (rarely) serine (rtM204I/V/S) confer high-level resistance to LMV but reduce replication efficiency. The subsequent selection or coselection of secondary mutations that partially restore replication efficiency is common and may influence drug resistance. Genotyping has shown that LMV treatment can select for HBV rtL80V/I mutants, but their prevalence and phenotype have not been documented. Analysis of a large sequence database revealed that rtL80V/I occurred almost exclusively in association with LMV resistance, and 85% of these isolates encoded rtL80I. Coselection of rtL80V/I occurred in 46% of isolates in which LMV resistance was attributable to rtM204I but only 9% of those in which resistance was attributable to rtM204V. Moreover, rtL80V/I did not occur in HBV genotype A isolates but occurred at similar frequencies in genotype B, C, and D isolates. In vitro phenotyping showed that although the rtL80I mutant by itself replicated less efficiently and was hypersensitive to LMV compared to the replication efficiency and sensitivity of its wild-type parent, the presence of rtL80I enhanced the replication efficiency of rt204I/V mutants without significantly affecting LMV resistance. Molecular modeling revealed that rt80 does not interact directly with the enzyme's substrates. Collectively, these results suggest that coselection of rtL80V/I and rtM204I/V occurs because the former compensates for the loss of replication efficiency associated with the acquisition of LMV resistance, particularly in the case of rtM204I.
The introduction in 1998 of lamivudine (LMV), the first safe and efficacious orally available inhibitor of hepatitis B virus (HBV) replication, revolutionized the treatment of chronic hepatitis B (CHB). LMV is a synthetic deoxycytidine analogue that is phosphorylated intracellularly by host cell enzymes which generate its 5′-triphosphate (LMV-TP). LMV-TP inhibits replication by competing with dCTP for incorporation into nascent viral DNA. Since LMV lacks a 3′-hydroxyl homologue, which is required for chain extension, incorporation of an LMV monophosphate residue causes immediate replication arrest. Treatment with LMV rapidly and significantly decreases viremia in a majority of patients with CHB and arrests or reverses liver disease in many, as indicated by the normalization of the results of liver function tests and of liver histology (18). Unfortunately, the long-term effectiveness of LMV is reduced by the development of viral resistance (1), which increases cumulatively at an annual rate of approximately 14 to 20% and occurs most frequently in individuals who are coinfected with human immunodeficiency virus (HIV) type 1 (20). Provided that the viral load is less than 6 log10 copies/ml, most cases of viral breakthrough while patients are receiving LMV can be rescued by treatment with adefovir dipivoxil (ADV), an acyclic dAMP analogue approved for use for the treatment of CHB in 2002 (17, 24).
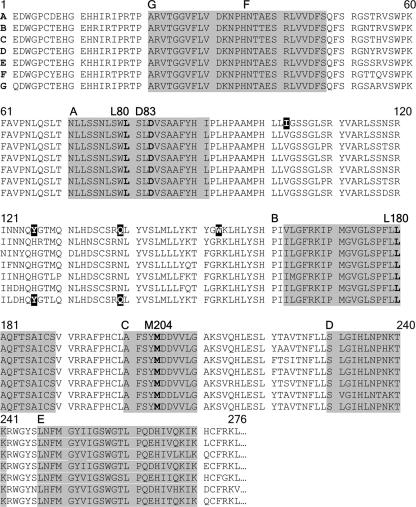
Unlike most other DNA viruses, HBV uses an unusual replication strategy that entails an essential reverse transcription step catalyzed by the HBV polymerase (P) gene product. The HBV polymerase is a multifunctional protein that, besides having RNA- and DNA-dependent DNA polymerase activity, self-primes reverse transcription, acts as an RNase H, and coordinates the intracellular assembly of viral nucleocapsids (for reviews, see references 32 and 14). The protein comprises four distinct structural/functional domains, namely (in order from the N terminus), (i) the terminal (reverse transcriptase [rt]-priming) domain, (ii) a spacer domain, (iii) the rt domain, and (iv) an RNase H domain. The HBV P gene is homologous to retroviral pol genes, and the HBV polymerase's rt domain shows close sequence homology with the HIV rt (4). Clusters of highly conserved amino acid residues in the rt domain that are common to all RNA-dependent polymerases have been ascribed specific functions that are essential for enzymatic activity and have been designated motifs A to G (Fig. 1) (3, 9, 26).
FIG. 1.
Alignment of rt reference sequences for genotypes A to G. Highly conserved sequence motifs A to G are shaded. L80, D83, and M204 in the dNTP-binding tetrad are emphasized in boldface type. Three amino acid residues that occur rarely or not at all in genotypes other than genotype A and that never occur in conjunction with rtL80V/I are identified by white type on a black background. Reference sequences (276 N-terminal residues) are from Stuyver et al. (27), and the conserved sequence motifs were identified as described by Bartholomeusz et al. (4).
Mutations that result in the replacement of methionine in the tyrosine-methionine-aspartate-aspartate (YMDD) deoxynucleoside triphosphate (dNTP)-binding tetrapeptide in motif C by valine, leucine, or, rarely, serine (designated rtM204V, rtM204I, and rtM204S, respectively, by using the genotype-independent nomenclature proposed in 2001 [27]) are necessary and sufficient to confer high-level LMV resistance. The rtM204I substitution has been detected in isolation, but rtM204V and rtM204S are found only in association with other changes, notably, rtL180M in the B motif (Fig. 1) (1, 3, 27). Early studies that used in vitro assays established that rtM204I and rtM204V mutants replicate less efficiently than wild-type HBV (19, 21). It was subsequently observed that the replication impairment imparted by rtM204V/I can be partially or completely overcome not only by further rt changes (particularly rtL180M [12, 23], rtV173L [10], and/or rtV207I [36]) but also by the presence of the precore (PC) G1896A mutation, which creates a premature stop codon (pcW28*) and prevents HBeAg synthesis (5, 31). Mutations that affect the core protein have also been shown to confer a replication advantage (29). Numerous amino acid substitutions in rt besides rtM204I/V/S, rtL180M, and rtV173L have since been detected by genotyping of clinical HBV isolates from patients undergoing long-term LMV treatment. The clinical histories of these patients, together with limited in vitro phenotyping data, suggested that the acquisition of some secondary mutations might accelerate treatment failure. Mutations that cause the lysine at residue 80 of the HBV rt to be replaced by valine or isoleucine (rtL80V/I) were first reported by Ogata and colleagues (22). They found a high prevalence of rtL80V/I mutants in clinical isolates recovered from five Japanese patients who had experienced viral breakthrough and disease exacerbation while undergoing long-term LMV therapy. Although rtL80V/I was clearly implicated in treatment failure, whether and how it may contribute to drug resistance have not been established. We used in vitro assays to characterize the phenotypic consequences of rtL80I for viral replication as well as LMV and ADV resistance, analyzed a large sequence database to estimate its prevalence, and used molecular modeling to suggest possible explanations for our observations.
MATERIALS AND METHODS
Determination of mutation frequencies.
SEQHEPB is a computer software program and database that is specifically designed for rapid computer-assisted virtual phenotyping of HBV and that accepts either genome (nucleic acid) or protein (amino acid) sequences as input. SEQHEPB searches for homology between input sequences and others already stored in its database, which also stores relevant clinical data. Details about SEQHEPB have recently been published elsewhere (35). The data accumulated by SEQHEPB were analyzed to determine the frequencies of occurrence of mutations of interest.
Generation of HBV mutants by site-directed mutagenesis.
The point mutations required to produce the selected changes in the amino acid sequence of the HBV polymerase were introduced by site-directed mutagenesis into a 1.3-times-genome-length wild-type HBV genotype D isolate, which was inserted into the plasmid vector pBlueBac4.5 (8). Mutagenesis was carried out according to the instructions provided with the Quickchange mutagenesis kit (Stratagene, La Jolla, CA). The primers used for mutagenesis (Table 1) were synthesized by GeneWorks (Adelaide, SA, Australia) and were purified to 99.9% homogeneity by high-performance liquid chromatography. The decision to create a series of rtL80I (rather than rtL80V) variants was influenced by logistic constraints and the predominance of the former in clinical isolates (see below). Mutants that encoded multiple changes were generated by consecutive site-directed mutagenesis reactions. Automated DNA sequencing in both directions (BigDye Terminator cycle sequencing; PE Applied Biosystems, Foster City, CA) was used to verify that each mutant encoded only the desired sequence changes. Polymerase mutants were created in both HBeAg-positive and HBeAg-negative backgrounds. HBeAg negativity was produced by site-directed mutagenesis to generate the G1896A mutation (pcW28*) (5).
TABLE 1.
Primers used for site-directed mutagenesis
| Primer | Primer sequencea |
|---|---|
| rtL80I | 5′-CCAACTTGTCCTGG ATATCGCTGGATGTGTCTGCG-3′ |
| rtL180M | 5′-CCTCAGTCCGTTTCTCATGGCTCAGTTTACTAG-3′ |
| rRtM204I | 5′-GGCTTTCAGTTATATCGATGATGTGGTATTGGGGG-3′ |
| PC | 5′-GCCTTGGGTGGCTTTAGGGCATGGACATCGACC-3′ |
Nucleotide changes are in boldface and underlined.
Cell culture and transfection.
Huh7 cells were grown in Dulbecco's modified eagle medium (Gibco BRL) supplemented with 10% fetal bovine serum at 37°C in a humidified environment containing 5% CO2. For transfection, cells were seeded into 60-mm dishes and allowed to adhere overnight. On the following day, when the cells were 60 to 80% confluent, the culture medium was replaced with fresh medium with or without different concentrations of either LMV or ADV. Sets of replicate cultures were transiently transfected (6) with 5 μM each HBV construct by using 8 μl of Fugene6 reagent (Roche Diagnostics, Mannheim, Germany), according to the manufacturer's instructions. The culture medium was changed on day 2 or day 3, and the cells were harvested on day 5 posttransfection.
Relative replication yield assays.
After 5 days of culture in drug-free medium, intracellular core-associated HBV DNA was harvested from the transfectants and quantified by Southern blotting, autoradiography, and densitometry, as described previously (6). The mean replication yield for each mutant was standardized relative to the mean yield for the wild-type replicates, defined as 1.0. Transfection efficiency was standardized in terms of HBsAg secretion by using the IMX enzyme immunoassay (Abbott Laboratories, North Chicago, IL). Data from three independent assays were pooled for analysis (5). Since mutations that encode rtL80I and rtL180M do not affect coding by the overlapping envelope reading frame sequence, they should not affect standardization of the transduction efficiency relative to the level of HBsAg secretion. The change in the envelope protein (sW196S) caused by mutations that produce rtM204I does not alter the antigenicity of HBsAg (31).
Antiviral drug sensitivity assays.
LMV was purchased from Moravek Biochemicals (Brea, CA). A 50 mM stock solution was prepared in distilled water and stored frozen at −20°C. ADV (provided by Gilead Sciences, Foster City, CA) was also prepared as a 50 mM stock solution, adjusted to pH 7.4 with NaOH, and stored at −20°C. All other chemicals and reagents were purchased from local suppliers and were analytical grade. Replicate cultures of Huh7 transfectants, prepared as described above, were continuously exposed to different concentrations of LMV (0, 0.001, 0.01, 0.1, 1, or 10 μM) or ADV (0, 0.1,0.5, 1, 5, or 10 μM). After 5 days in culture, intracellular core-associated HBV DNA was harvested and quantified by Southern blotting, autoradiography, and densitometry, as described previously (6).
Data analysis and statistics.
The mean amount of HBV DNA in the drug-treated cultures was standardized relative to the mean amount (defined as 1.0) in untreated replicate controls for each virus. Data from three independent assays were pooled for analysis. Dose-dependent inhibition of HBV DNA replication, when it occurred, could be described by a three-parameter logistic dose-response equation: y = a/[1 +(x/b)c]. The best-fit values for individual dose-response equations were determined by using TableCurve2D software (SPSS Inc., Chicago, IL), which was also used to estimate the 50% effective concentrations (EC50s). SigmaStat software (version 3.0; SPSS Inc.) was used to test for statistical significance. Differences with P values of <0.05 were regarded as significant.
Molecular modeling.
A three-dimensional model of HBV rt was previously constructed, based on homology with the HIV rt, the structure of which has been resolved by X-ray crystallography (4) (also see http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid = pfam00078). Both Sybyl software (version 6.9; Tripos Associates) and Swiss PDB Deepview software (http://au.expasy.org/spdb) were used to model the effects of amino acid substitutions in the HBV rt domain (15).
RESULTS
Prevalence of rtL80V/I in clinical isolates.
The SEQHEPB program was used to determine the prevalence of rtL80V/I in the 2,085 complete polymerase sequences of HBV isolates from 1,376 patients with CHB. The presence of rtM204I/V in sequences from 503 (37%) of the 1,376 patients indicated that they were infected with LMV-resistant HBV. Eleven of these isolates contained mixed populations of different mutants (such as rtM204I/V with or without rtL180M), and these were excluded from further analysis. Two hundred fifty-nine (53%) of the remaining 492 patients were infected with rtM204I mutants and 233 (47%) were infected with rt204V variants. In cases in which two or more HBV isolates were obtained from an individual patient during the course of LMV treatment, only the most recent rt sequence (corresponding to the longest LMV exposure) was included in the final analysis. Sequence evolution during treatment is summarized with other relevant details in Table 2, which confirms that in the majority of cases, rtL80V/I and rtL180M were acquired after the acquisition of primary LMV resistance. The rtL80V/I substitutions were much more frequent in conjunction with rtM204I than with rtM204V (frequencies, 119/259 [46%] and 21/233 [9%], respectively; P < 0.005) (Table 3). Of the subpopulation of isolates harboring rtL80V/I, rtL80I was predominant: the frequencies were 119/140 (85%) and 21/140 (15%) for rtL80I and rtL80V, respectively (Table 3). The occurrences of rtL80V/I in isolation were rare, being present in only 5 of the 2,085 rt sequences in the SEQHEPB database, and all were from HBV isolates from LMV-naïve patients. The four most common types of LMV-resistant HBV mutants accounted for >80% of the population. In order of decreasing frequency they were rtL180M/M204V, rM204I, rtL80I/M204I, and rtL80I/L180M/M204I, with frequencies of 43%, 20%, 13%, and 6%, respectively (Table 3). Further grouping by genotype revealed that rtL80V/I occurred in isolates of genotypes B, C, and D at similar frequencies (37%, 36%, and 32% respectively) but was undetected in the sequences of 84 LMV-resistant isolates of genotype A (Table 3).
TABLE 2.
Changes in amino acid sequences of LMV-resistant rt during treatment
| Change and case no. | Genotype | Initial change(s) | Subsequent change(s)a | Elapsed time (mo)b | HBV DNA changec |
|---|---|---|---|---|---|
| Gain of L80I or L80V | |||||
| 52 | B | M204I | +L80V+L180M; M204I→V | 22 | 40 |
| 59 | D | M204I | +L80V+L180M | 31 | 0.4 |
| 225 | C | M204V | +L80V+L180M | 11 | 0.03 |
| 301 | C | M204I | +L80I+L180M | 4 | 0.5 |
| 375 | C | M204I | +L80V | 16 | 3.6 |
| 550 | D | L180M/M204V | +L80V | 13 | 2.0 |
| Loss of L80I | |||||
| 333 | D | L80I/M204V | −L80I; M204V→I | 4 | 0.05 |
| 953 | C | L80I/M204I | −L80I | 15 | NA |
| Change L80V→L80I | |||||
| 267 | B | L80V/L180M/M204V | L80V→I | 8 | 2.2 |
| 872 | D | L80V/L180M/M204I | L80V→I | 2 | 1.8 |
| Gain of L180M | |||||
| 57 | B | M204I | +L180M; M204I→V | 14 | 4.8 |
| 120 | A | M204I | +L180M | 5 | NC |
| 172 | A | M204I | +L180M; M204I→V | 7 | NA |
| 187 | A | M204I | +L180M; M204I→V | 58 | 1.2 |
| 192 | D | M204I | +L180M | 6 | NC |
| 214 | A | M204I | +L180M; M204I→V | 7 | 3.6 |
| 277 | D | L80I/M204I | +L180M | 2 | 1,164 |
| 332 | C | L80I/M204I | +L180M; M204I→V | 39 | 0.7 |
| 1316 | B | L80I/M204I | +L180M | 5 | 29.1 |
| Change M204V→M204I | |||||
| 5 | D | L80I/L180M/M204V | M204V→I | 11 | 6.6 |
| 555 | C | L180M/M204V | M204V→I | 19 | NA |
Changes listed in the “initial change(s)” column were maintained unless indicated otherwise in the “subsequent change(s)” column.
Elapsed time between sampling for isolates in which changes were detected.
Fold change in viral load (as serum HBV DNA load) associated with rt change; NC, no change (low, stable viremia); NA, not available. Note that L80I/V appears after the acquisition of primary LMV resistance.
TABLE 3.
Prevalence and frequency distribution of specific rt variants in LMVr clinical HBV isolatesa
| Variant | Amino acid substitutions in rt
|
No. (%) of isolates of the following genotype with the indicated changes:
|
Fraction (%) of all LMVr isolates | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M204 | L180 | L80 | A | B | C | D | E | F | G | All | ||
| Resistance conferred by rtM204Ib | I | 15 | 27 | 23 | 30 | 0 | 0 | 1 | 96 | 20 | ||
| I | M | 3 | 1 | 23 | 17 | 0 | 0 | 0 | 44 | 9 | ||
| I | I | 0 | 20 | 30 | 15 | 0 | 0 | 0 | 65 | 13 | ||
| I | M | I | 0 | 4 | 14 | 10 | 0 | 0 | 0 | 28 | 6 | |
| I | V | 0 | 6 | 6 | 4 | 0 | 0 | 0 | 16 | 3 | ||
| I | M | V | 0 | 2 | 4 | 4 | 0 | 0 | 0 | 10 | 2 | |
| Genotype total | 18 | 60 | 100 | 80 | 0 | 0 | 1 | 259 | 53 | |||
| Resistance conferred by rtM204Vc | V | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | <1 | ||
| V | M | 65 | 37 | 63 | 37 | 1 | 0 | 7 | 210 | 43 | ||
| V | M | I | 0 | 0 | 5 | 2 | 0 | 0 | 0 | 7 | 1 | |
| V | M | V | 0 | 6 | 4 | 4 | 0 | 0 | 0 | 14 | 3 | |
| Genotype total | 66 | 43 | 73 | 43 | 1 | 0 | 7 | 233 | 47 | |||
| Genotype total (no.) for rtM204II/V | 84 | 103 | 173 | 123 | 1 | 0 | 8 | 492 | 100 | |||
| Frequency (%) of rtL80V/I | 0 | 37 | 36 | 32 | 0 | 0 | 28 | |||||
Data are from the SEQHEPB database. LMVr, lamividine resistant. Only the most recent LMV-resistant isolate from each patient was included.
A total of 119/259 (46%) isolates coselected rtL80V/I.
A total of 21/233 (9%) isolates coselected rtL80V/I.
Influence of rtL80I on replication efficiency of HBeAg-positive and HBeAg-negative HBV.
Different combinations of point mutations were introduced by site-directed mutagenesis into a pair of infectious HBV clones of genotype D which were genetically identical, except that one contained the G1896A PC mutation, which confers an HBeAg-negative phenotype. The wild-type parent clone and the entire panel of mutant derivatives were subjected to parallel sets of assays to determine their relative replication efficiencies and sensitivities to LMV and ADV. The contribution of the rtL80I amino acid substitution to the viral phenotype was deduced by parallel comparisons of the phenotype of the wild-type HBV clone with the phenotypes of its mutant derivatives in both HBeAg-positive and -negative backgrounds.
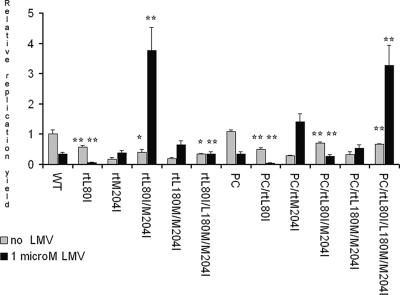
All LMV-resistant mutants were found to be replication deficient compared to the replication efficiency of the wild type, as observed previously (19, 21) (Table 4 and Fig. 2). The presence of the PC stop codon (G1896A) mutation, which confers HBeAg negativity, enhanced the replication efficiency, whereas the rtL180M substitution in isolation did not significantly affect the replication yield, also confirming previous findings (5, 12, 30). In isolation, rtL80I alone significantly reduced the replication yield but, paradoxically, increased it by up to 150% when it was present in conjunction with rtM204V or rtM204I. The replication yields were slightly greater in an HBeAg-negative background, with the enhancing effects of rtL80I and the PC mutation being independent and approximately additive (Fig. 2 and Table 4). In the presence of 1 μM LMV, a concentration sufficient to inhibit the replication of wild-type virus by approximately two-thirds, the replication yield was very dramatically increased by the presence of rtL80I in only two cases, suggesting that either the sequence in which mutations are acquired may be important or that the conditions in vitro cannot accurately mimic those in vivo.
TABLE 4.
LMV sensitivities and relative replication yields of wild-type HBV and its mutant derivativesa
| HBV variant | LMV EC50 (μM)b | Relative replication yieldc
|
Effect of L80I on replication efficiencyd
|
||
|---|---|---|---|---|---|
| Without LMV | With 1 μM LMV | Without LMV | With 1 μM LMV | ||
| Wild type | 0.41 ± 0.19 | 1.00 ± 0.15 | 0.33 ± 0.04 | ||
| rtL80I | 0.03 ± 0.01 | 0.57 ± 0.05 | 0.06 ± 0.01 | 43% decrease** | 82% decrease** |
| rtM204I | R | 0.18 ± 0.00 | 0.38 ± 0.04 | ||
| rtL80I/M204I | R | 0.39 ± 0.06 | 3.76 ± 0.43 | 117% increase* | 902% increase** |
| rtL180M/M204I | R | 0.19 ± 0.04 | 0.65 ± 0.08 | ||
| rtL80I/L180M/M204I | R | 0.35 ± 0.01 | 0.34 ± 0.04 | 84% increase* | 48% decrease** |
| PC | 0.46 ± 0.12 | 1.08 ± 0.06 | 0.35 ± 0.04 | ||
| PC/rtL80I | 0.03 ± 0.02 | 0.5 ± 0.06 | 0.04 ± 0.00 | 54% decrease** | 88% decrease** |
| PC/rtM204I | R | 0.28 ± 0.03 | 1.40 ± 0.16 | ||
| PC/rtL80I/M204i | R | 0.70 ± 0.08 | 0.27 ± 0.03 | 150% increase** | 80% decrease** |
| PCrtL180M/M204I | R | 0.32 ± 0.09 | 0.54 ± 0.06 | ||
| PC/rtL80I/L180M/M204I | R | 0.67 ± 0.02 | 3.27 ± 0.38 | 109% increase** | 506% increase** |
The wild type is genotype D; PC signifies that the G1896A mutation in the PC region (which prevents HBeAg synthesis) is present.
EC50s are means ± standard deviations of the pooled results from three independent assays. R, resistant (i.e., no dose-dependent inhibition of viral replication observed at LMV concentrations up to 10 μM).
Relative replication yields are the means ± standard deviations from at least three independent assays.
Percent changes in relative replication yield for each pair with and without LMV, with statistically significant differences of P < 0.05 (*) and P < 0.01(**) indicated.
FIG. 2.
Relative replication yields following transfection of Huh7 cells with wild-type HBV and each of its mutant derivatives and culture for 5 days in the presence or absence of 1 μM LMV, a concentration sufficient to reduce the replication of wild-type (WT) HBV to approximately one-third of that for the uninhibited control. The concentration of HBsAg in the culture supernatants was used to correct for variations in transfection efficiencies, and the amount of replication in each case has been standardized relative to the mean value for the uninhibited controls, defined as 1.0. Each column represents data from at least three experiments; error bars show standard deviations. The significance of differences in replication associated with the acquisition of rtL80I are indicated by asterisks above each of the “+rtL80I” pair of columns: *, P < 0.05; **, P < 0.01.
Influence of rtL80I on sensitivity to LMV and ADV.
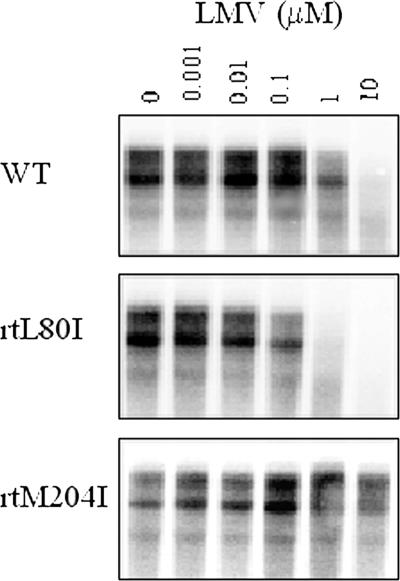
Despite the replication advantage conferred by the presence of the PC mutation, it did not significantly affect LMV resistance, confirming earlier observations (5). Estimated EC50s (in μM; means ± standard deviations) were 0.41 ± 0.19 and 0.46 ± 0.12 for the wild type and the PC mutant, respectively (Fig. 3 and Table 4). The rtL80I change in isolation was never observed in HBV isolates from patients treated with LMV and caused hypersensitivity to LMV in vitro, decreasing the EC50 estimates more than 10-fold, but it failed to alter the resistance of LMV-resistant (i.e., rtM204I/V) mutants sufficiently to permit estimation of EC50 values, which remained ≫10 μM. This implies that the level of LMV resistance conferred by rt204I/V is so large that it precludes the in vitro measurement of the relatively smaller effects conferred by additional mutations (Fig. 3 and Table 4). The presence of rtL80I had no significant effect on the susceptibilities of any of the isolates in the HBV panel to ADV: none of the estimated EC50s for ADV was significantly different from that for the wild type (results not shown).
FIG. 3.
Assays for LMV resistance. Replicate cultures of Huh7 cells were transiently transfected with a laboratory clone of wild-type (WT) HBeAg-negative HBV or one of its mutant derivatives. After 5 days of continuous exposure to LMV at the concentrations indicated, intracellular core-associated HBV DNA was isolated, separated by agarose gel electrophoresis, and detected by Southern blotting and autoradiography.
Molecular modeling of effects of rtL80I on enzyme structure.
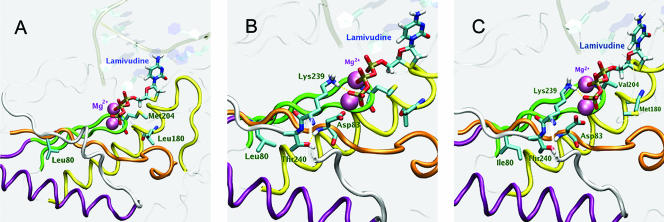
The location and spatial orientation of leucine at HBV rtL80 and the effects of its replacement by isoleucine or valine (rtL80V/I) were determined by computer simulation and by use of a previously constructed three-dimensional homology model of HBV rt (4). This model located both rtL180 and rtM204 in the enzyme's dNTP-binding pocket, where they can interact with incoming dNTPs, but places rtL80 in a location where a direct interaction with incoming dNTPs or their analogues is not possible (Fig. 4A). The model places rtL80 in a hydrophobic part of the enzyme's core, near α helix F (residues 216 to 231), adjacent to the β sheets that encompass motif D (residues 234 to 245) and within 2.6 Å of the methyl group of an essential threonine residue (rtT240) within motif D. In addition, rtL80 is separated from an essential aspartate residue (rtD83) by only 2 amino acid residues (Fig. 4B).
FIG. 4.
(A) Model of the wild-type HBV rt showing the location of residue L80 in relation to residues L180 and M204, which are located in the dNTP-binding pocket. Conserved sequence motifs are colored: A, orange; B, yellow; C, green; D, white. The conserved alpha helix and the chelated magnesium ions are purple. LMV-TP is shown occupying the dNTP-binding pocket. (B) Model of the wild-type rt active site showing the spatial relationships between residues L80, T240, D83, and K239. (C) Model of the dNTP-binding pocket of the active site of LMV-resistant rt showing the location of rtL80I in relation to rtM204V and rtL180M. Note the alteration in the spatial relationship between rtL80I and rtT240 compared to that in panel B.
DISCUSSION
Coselection of rtL80V/I and LMV resistance was first observed by Ogata and colleagues (22) during their investigation into the reasons for the failure of LMV treatment of five Japanese patients with CHB, all of whom were infected with HBeAg-positive HBV strains of serotype adr and genotype C. Drug resistance was attributable to selection for rtL80I/M204I/V in three cases and selection for rtL80I/180M/M204V and rtL82M/M204V in the remaining two cases, respectively. The emergence of resistant mutants was associated with increases in viral load to greater than baseline levels, accompanied by elevations in serum aminotransferase activity and exacerbation of liver disease in every case. Although they speculated that rtL80V/I may have contributed to clinical deterioration, Ogata's team did not determine the prevalence of rtL80V/I in other, independent clinical isolates, nor did they investigate how the rtL80V/I change might affect the viral phenotype. We have analyzed a large cohort of clinical HBV isolates for the prevalence of rtL80V/I and characterized its effects on in vitro viral replication and sensitivity to LMV and ADV.
Interrogation of the SEQHEPB database revealed that the rtL80V/I substitution occurs almost exclusively in HBV isolates that are LMV resistant and, furthermore, that it is found significantly more frequently in association with rtM204I than in association with rtM204V (Table 3). The mutations responsible for the rtL80V/I change in the HBV polymerase protein are silent in the overlapping surface gene reading frame and should not affect either the assembly and release of virions or their subsequent infectivity and immunogenicity. Positive selection for rtL80V/I mutants during LMV therapy and their eclipse by wild-type virus posttherapy implies that the rtL80V/I substitution may enhance LMV resistance or viral replication, or both, possibilities that were investigated by the use of in vitro assays. In addition, we found that rtL80V/I occurred only in HBV isolates of genotypes B, C, and D and not in genotype A isolates, which could be interpreted as implying that certain mutational patterns are restricted by structural/functional constraints to particular genotypes (3, 27). Indeed, comparison of reference sequences representative of each genotype (25, 27) shows that even rt sequences outside the most conserved motifs are remarkably invariant, although only a single amino acid residue, the isoleucine at position 103 (rtI103), appears to be unique to genotype A, as it is valine (V) in all other genotypes (Fig. 1). Other residues common to only one or two other genotypes besides genotype A include the glutamine at position 139 (rtQ139) and the tryptophan at position 153 (rtW153), which are replaced by asparagine (N) and arginine (R) in other genotypes except variants of genotypes G and B, respectively (Fig. 1). This suggests that the presence of rtI103, rtQ139, and/or rtW153 mutually excludes rtL80V/I, a notion supported by the absence of evidence of their coexistence in any of the 2,085 rt sequences in the SEQHEPB database. An additional possibility is that whatever advantage is conferred on LMV-resistant HBV isolates of other genotypes by the acquisition of rtL80V/I is already available in genotype A. Considering this possibility, it is interesting that recent evidence implicates inherent genotype-dependent differences in replication efficiency and gene expression as contributors to disease severity (28).
Several independent studies have established that rtM204I/V, which is necessary and sufficient to confer LMV resistance, also reduces viral replication efficiency (1, 19, 21). The replication defects associated with the acquisition of LMV resistance can partially be compensated for by selection for secondary mutations that are not necessarily located in genetic sequences that encode catalytically essential amino acid residues. Selection for these “compensatory” mutations appears to be almost inevitable during prolonged treatment with LMV, but the phenotypic effects of the majority of the many documented secondary mutations are unknown. Since rtM204I/V confers high-level resistance to LMV, exemplified by large (>100-fold) increases in EC50s in vitro, most secondary mutations associated with LMV resistance probably contribute more to restoring replication efficiency than to enhancing drug resistance. Indeed, a few have been shown to enhance the replication of rtM204I and/or rtM204V mutants in vitro. For example, rtL180M (which was originally observed in patients treated with famciclovir [2], the prodrug of penciclovir, to which it confers low-level resistance) partially compensates for the replication defect imparted by rtM204V and, to a lesser extent, rtM204I without contributing significantly to LMV resistance (7, 11, 12, 23). Similarly, rtA181T contributes minimally to LMV resistance but can partially compensate for the replication deficiency of particular mutants (34).
Molecular modeling of the HBV rt-substrate complexes shows that rtL180 and rtM204 form part of the enzyme's dNTP-binding pocket, where they are closely associated with each other and can interact directly with incoming dNTPs and their analogues, whereas rtL80 is located too far from this site to be able to contribute directly to substrate binding. Despite the remoteness of rtL80 from the enzyme's active sites, substitution of isoleucine for leucine is clearly sufficient to affect replication, causing a >10-fold decrease in replication efficiency. Its proximity to rtD83 and to the methyl group of rtT240 may be significant. Earlier studies of the homologous HIV rt by Wrobel and colleagues (33) show that juxtaposition of the α F helix and the two β sheets that contain the three catalytically essential aspartate residues (corresponding to rtD83, rtD205, and rtD206, respectively, in the HBV enzyme) is crucial for conformational stability. The structure of this region is highly conserved in other polymerases as distantly related as the Klenow fragment (16). The highly conserved threonine residue (rtT240) in HBV rt is part of a bent structure formed by residues 235 to 240 that is required to stabilize binding of the TP group of the incoming dNTP by direct interaction with rtK239 (Fig. 4B). In addition, the hydroxyl group of rtT240 interacts with the amide group of rtD83, which is essential for catalysis. We propose that substitution of either I or V for rt80L alters the relative position of rtD83, which is only two residues away, and also changes the spatial orientation of rtT240, affecting the overall conformation of the TP-binding site as a consequence (Fig. 4C). Considering that rtL80V/I in isolation occurs rarely in vivo and significantly decreases the replication yield in vitro, it is probable not only that the conformational changes that it causes decrease the enzyme's dNTP-binding (and dNTP analogue-binding) affinity but also that the decrease in affinity is greater for LMV-TP than for natural dNTPs, in particular, competing dCTP. Several previous studies have established that although rtM204V imparts a more severe replication defect than rtM204I (19, 21), the compensation provided by rtL180M (which by itself imparts a small replication defect) is much greater for rtM204V than for rtM204I, an observation supported by cell-free in vitro kinetic studies with mutant HBV polymerases (13). The replication disadvantage imparted by rtL180M alone has been reported to be associated with an increase in the Km (relative to that for the wild type) for dCTP of only 1.3-fold. The fold increases in the Km values for dCTP of other variants were estimated to be 3.3 and 2.7 for rtM204I and rtL180M/M204I, respectively, compared to values of 3.6 and 1.8 for rtM204V and rtL180M/M204V, respectively (13). These decreases correspond to efficiency gains of 18% and 50% for rtM204I and rtM204V, respectively. Our results suggest that the acquisition of rtL80V/I (which alone confers a relatively large replication disadvantage) causes the converse compensatory changes, i.e., that it imparts a significant and useful gain in efficiency to rtM204I, especially in the presence of LMV, but not to rtM204V. Kinetic studies could establish the mechanism for this gain and whether (as the results presented here imply) the sequence in which mutations are acquired is important.
In conclusion, we have determined the prevalence or rtL80V/I and characterized the in vitro phenotype conferred by rtL80I, which occurs almost exclusively in LMV-resistant HBV isolates, partially compensating for their otherwise low replication efficiency. Although rtL80I in isolation increases sensitivity to LMV and imparts a replication defect, it enhances the in vitro replication of LMV-resistant HBV. Molecular models of HBV rt show that although rt residue 80 is located distal to the enzyme's dNTP-binding pocket, substitution of isoleucine for leucine at this site sufficiently changes the overall spatial alignment of other residues that are important for catalysis to partially restore replication efficiency. These results imply that the presence of rtL80I decreases the enzyme's affinity for both dNTPs and LMV-TP and that the decrease in affinity for LMV-TP is greater than the decrease in affinity for competing dCTP.
Acknowledgments
We thank David Chalmers for assistance with molecular modeling and Scott Bowden for reviewing the manuscript.
This work was supported by a grant from the National Institutes of Health, Bethesda, MD (R01 grant AIO60449).
Footnotes
Published ahead of print on 16 April 2007.
REFERENCES
- 1.Allen, M. I., M. Deslauriers, C. W. Andrews, G. A. Tipples, K. A. Walters, D. L. Tyrrell, N. Brown, L. D. Condreay, et al. 1998. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Hepatology 271670-1677. [DOI] [PubMed] [Google Scholar]
- 2.Aye, T. T., A. Bartholomeusz, T. Shaw, S. Bowden, A. Breschkin, J. McMillan, P. Angus, and S. Locarnini. 1997. Hepatitis B virus polymerase mutations during antiviral therapy in a patient following liver transplantation. J. Hepatol. 261148-1153. [DOI] [PubMed] [Google Scholar]
- 3.Bartholomeusz, A., and S. Schaefer. 2004. Hepatitis B virus genotypes: comparison of genotyping methods. Rev. Med. Virol. 143-16. [DOI] [PubMed] [Google Scholar]
- 4.Bartholomeusz, A., B. G. Tehan, and D. K. Chalmers. 2004. Comparisons of the HBV and HIV polymerase, and antiviral resistance mutations. Antivir. Ther. 9149-160. [PubMed] [Google Scholar]
- 5.Chen, R. Y., R. Edwards, T. Shaw, D. Colledge, W. E. T. Delaney, H. Isom, S. Bowden, P. Desmond, and S. A. Locarnini. 2003. Effect of the G1896A precore mutation on drug sensitivity and replication yield of lamivudine-resistant HBV in vitro. Hepatology 3727-35. [DOI] [PubMed] [Google Scholar]
- 6.Chin, R., T. Shaw, J. Torresi, V. Sozzi, C. Trautwein, T. Bock, M. Manns, H. Isom, P. Furman, and S. Locarnini. 2001. In vitro susceptibilities of wild-type or drug-resistant hepatitis B virus to (−)-beta-d-2,6-diaminopurine dioxolane and 2′-fluoro-5-methyl-beta-l-arabinofuranosyluracil. Antimicrob. Agents Chemother. 452495-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delaney, W. E., IV, R. Edwards, D. Colledge, T. Shaw, J. Torresi, T. G. Miller, H. C. Isom, C. T. Bock, M. P. Manns, C. Trautwein, and S. Locarnini. 2001. Cross-resistance testing of antihepadnaviral compounds using novel recombinant baculoviruses which encode drug-resistant strains of hepatitis B virus. Antimicrob. Agents Chemother. 451705-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delaney, W. E., IV, and H. C. Isom. 1998. Hepatitis B virus replication in human HepG2 cells mediated by hepatitis B virus recombinant baculovirus. Hepatology 281134-1146. [DOI] [PubMed] [Google Scholar]
- 9.Delaney, W. E., IV, S. Locarnini, and T. Shaw. 2001. Resistance of hepatitis B virus to antiviral drugs: current aspects and directions for future investigation. Antivir. Chem. Chemother. 121-35. [DOI] [PubMed] [Google Scholar]
- 10.Delaney, W. E., IV, H. Yang, C. E. Westland, K. Das, E. Arnold, C. S. Gibbs, M. D. Miller, and S. Xiong. 2003. The hepatitis B virus polymerase mutation rtV173L is selected during lamivudine therapy and enhances viral replication in vitro. J. Virol. 7711833-11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Man, R. A., A. I. Bartholomeusz, H. G. Niesters, P. E. Zondervan, and S. A. Locarnini. 1998. The sequential occurrence of viral mutations in a liver transplant recipient re-infected with hepatitis B: hepatitis B immune globulin escape, famciclovir non-response, followed by lamivudine resistance resulting in graft loss. J. Hepatol. 29669-675. [DOI] [PubMed] [Google Scholar]
- 12.Fu, L., and Y. C. Cheng. 1998. Role of additional mutations outside the YMDD motif of hepatitis B virus polymerase in L(−)SddC (3TC) resistance. Biochem. Pharmacol. 551567-1572. [DOI] [PubMed] [Google Scholar]
- 13.Gaillard, R. K., J. Barnard, V. Lopez, P. Hodges, E. Bourne, L. Johnson, M. I. Allen, P. Condreay, W. H. Miller, and L. D. Condreay. 2002. Kinetic analysis of wild-type and YMDD mutant hepatitis B virus polymerases and effects of deoxyribonucleotide concentrations on polymerase activity. Antimicrob. Agents Chemother. 461005-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganem, D., J. R. Pollack, and J. Tavis. 1994. Hepatitis B virus reverse transcriptase and its many roles in hepadnaviral genomic replication. Infect. Agents Dis. 385-93. [PubMed] [Google Scholar]
- 15.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 182714-2723. [DOI] [PubMed] [Google Scholar]
- 16.Jacobo-Molina, A., J. Ding, R. G. Nanni, A. D. Clark, Jr., X. Lu, C. Tantillo, R. L. Williams, G. Kamer, A. L. Ferris, P. Clark, et al. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 906320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lampertico, P., M. Vigano, E. Manenti, M. Iavarone, G. Lunghi, and M. Colombo. 2005. Adefovir rapidly suppresses hepatitis B in HBeAg-negative patients developing genotypic resistance to lamivudine. Hepatology 421414-1419. [DOI] [PubMed] [Google Scholar]
- 18.Lau, D. T., M. F. Khokhar, E. Doo, M. G. Ghany, D. Herion, Y. Park, D. E. Kleiner, P. Schmid, L. D. Condreay, J. Gauthier, M. C. Kuhns, T. J. Liang, and J. H. Hoofnagle. 2000. Long-term therapy of chronic hepatitis B with lamivudine. Hepatology 32828-834. [DOI] [PubMed] [Google Scholar]
- 19.Ling, R., and T. J. Harrison. 1999. Functional analysis of mutations conferring lamivudine resistance on hepatitis B virus. J. Gen. Virol. 80(Pt 3)601-606. [DOI] [PubMed] [Google Scholar]
- 20.Matthews, G. V., A. Bartholomeusz, S. Locarnini, A. Ayres, J. Sasaduesz, E. Seaberg, D. A. Cooper, S. Lewin, G. J. Dore, and C. L. Thio. 2006. Characteristics of drug resistant HBV in an international collaborative study of HIV-HBV-infected individuals on extended lamivudine therapy. AIDS 20863-870. [DOI] [PubMed] [Google Scholar]
- 21.Melegari, M., P. P. Scaglioni, and J. R. Wands. 1998. Hepatitis B virus mutants associated with 3TC and famciclovir administration are replication defective. Hepatology 27628-633. [DOI] [PubMed] [Google Scholar]
- 22.Ogata, N., K. Fujii, S. Takigawa, M. Nomoto, T. Ichida, and H. Asakura. 1999. Novel patterns of amino acid mutations in the hepatitis B virus polymerase in association with resistance to lamivudine therapy in Japanese patients with chronic hepatitis B. J. Med. Virol. 59270-276. [PubMed] [Google Scholar]
- 23.Ono, S. K., N. Kato, Y. Shiratori, J. Kato, T. Goto, R. F. Schinazi, F. J. Carrilho, and M. Omata. 2001. The polymerase L528M mutation cooperates with nucleotide binding-site mutations, increasing hepatitis B virus replication and drug resistance. J. Clin. Investig. 107449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schiff, E. R., C. L. Lai, S. Hadziyannis, P. Neuhaus, N. Terrault, M. Colombo, H. L. Tillmann, D. Samuel, S. Zeuzem, L. Lilly, M. Rendina, J. P. Villeneuve, N. Lama, C. James, M. S. Wulfsohn, H. Namini, C. Westland, S. Xiong, G. S. Choy, S. Van Doren, J. Fry, and C. L. Brosgart. 2003. Adefovir dipivoxil therapy for lamivudine-resistant hepatitis B in pre- and post-liver transplantation patients. Hepatology 381419-1427. [DOI] [PubMed] [Google Scholar]
- 25.Simmonds, P., and S. Midgley. 2005. Recombination in the genesis and evolution of hepatitis B virus genotypes. J. Virol. 7915467-15476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh, K., and M. J. Modak. 1998. A unified DNA- and dNTP-binding mode for DNA polymerases. Trends Biochem. Sci. 23277-281. [DOI] [PubMed] [Google Scholar]
- 27.Stuyver, L. J., S. A. Locarnini, A. Lok, D. D. Richman, W. F. Carman, J. L. Dienstag, and R. F. Schinazi. 2001. Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology 33751-757. [DOI] [PubMed] [Google Scholar]
- 28.Sugiyama, M., Y. Tanaka, T. Kato, E. Orito, K. Ito, S. K. Acharya, R. G. Gish, A. Kramvis, T. Shimada, N. Izumi, M. Kaito, Y. Miyakawa, and M. Mizokami. 2006. Influence of hepatitis B virus genotypes on the intra- and extracellular expression of viral DNA and antigens. Hepatology 44915-924. [DOI] [PubMed] [Google Scholar]
- 29.Suk, F. M., M. H. Lin, M. Newman, S. Pan, S. H. Chen, J. D. Liu, and C. Shih. 2002. Replication advantage and host factor-independent phenotypes attributable to a common naturally occurring capsid mutation (I97L) in human hepatitis B virus. J. Virol. 7612069-12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tillmann, H. L., C. Trautwein, T. Bock, K. H. Boker, E. Jackel, M. Glowienka, K. Oldhafer, I. Bruns, J. Gauthier, L. D. Condreay, H. R. Raab, and M. P. Manns. 1999. Mutational pattern of hepatitis B virus on sequential therapy with famciclovir and lamivudine in patients with hepatitis B virus reinfection occurring under HBIg immunoglobulin after liver transplantation. Hepatology 30244-256. [DOI] [PubMed] [Google Scholar]
- 31.Torresi, J., L. Earnest-Silveira, G. Civitico, T. E. Walters, S. R. Lewin, J. Fyfe, S. A. Locarnini, M. Manns, C. Trautwein, and T. C. Bock. 2002. Restoration of replication phenotype of lamivudine-resistant hepatitis B virus mutants by compensatory changes in the “fingers” subdomain of the viral polymerase selected as a consequence of mutations in the overlapping S gene. Virology 29988-99. [DOI] [PubMed] [Google Scholar]
- 32.Will, H., W. Reiser, T. Weimer, E. Pfaff, M. Buscher, R. Sprengel, R. Cattaneo, and H. Schaller. 1987. Replication strategy of human hepatitis B virus. J. Virol. 61904-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wrobel, J. A., S. F. Chao, M. J. Conrad, J. D. Merker, R. Swanstrom, G. J. Pielak, and C. A. Hutchison III. 1998. A genetic approach for identifying critical residues in the fingers and palm subdomains of HIV-1 reverse transcriptase. Proc. Natl. Acad. Sci. USA 95638-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yatsuji, H., C. Noguchi, N. Hiraga, N. Mori, M. Tsuge, M. Imamura, S. Takahashi, E. Iwao, Y. Fujimoto, H. Ochi, H. Abe, T. Maekawa, C. Tateno, K. Yoshizato, F. Suzuki, H. Kumada, and K. Chayama. 2006. Emergence of a novel lamivudine-resistant hepatitis B virus variant with a substitution outside the YMDD motif. Antimicrob. Agents Chemother. 503867-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuen, L. K., A. Ayres, M. Littlejohn, D. Colledge, A. Edgely, W. J. Maskill, S. A. Locarnini, and A. Bartholomeusz. 2006. SEQHEPB: a sequence analysis program and relational database system for chronic hepatitis B. Antivir. Res. 7564-74. [DOI] [PubMed] [Google Scholar]
- 36.Zollner, B., M. Sterneck, K. Wursthorn, J. Petersen, M. Schroter, R. Laufs, and H. H. Feucht. 2005. Prevalence, incidence, and clinical relevance of the reverse transcriptase V207I mutation outside the YMDD motif of the hepatitis B virus polymerase during lamivudine therapy. J. Clin. Microbiol. 432503-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]