Abstract
Quality control (QC) ranges for antimicrobial agents against QC strains for both dilution and disk diffusion testing are currently set by the Clinical and Laboratory Standards Institute (CLSI), using data gathered in predefined structured multilaboratory studies, so-called tier 2 studies. The ranges are finally selected by the relevant CLSI subcommittee, based largely on visual inspection and a few simple rules. We have developed statistical methods for analyzing the data from tier 2 studies and applied them to QC strain-antimicrobial agent combinations from 178 dilution testing data sets and 48 disk diffusion data sets, including a method for identifying possible outlier data from individual laboratories. The methods are based on the fact that dilution testing MIC data were log normally distributed and disk diffusion zone diameter data were normally distributed. For dilution testing, compared to QC ranges actually set by CLSI, calculated ranges were identical in 68% of cases, narrower in 7% of cases, and wider in 14% of cases. For disk diffusion testing, calculated ranges were identical to CLSI ranges in 33% of cases, narrower in 8% of cases, and 1 to 2 mm wider in 58% of cases. Possible outliers were detected in 8% of diffusion test data but none of the disk diffusion data. Application of statistical techniques to the analysis of QC tier 2 data and the setting of QC ranges is relatively simple to perform on spreadsheets, and the output enhances the current CLSI methods for setting of QC ranges.
Susceptibility testing in the diagnostic microbiology laboratory requires testing of standard quality control (QC) strains on a regular basis against the antimicrobial agents being used in order to ensure test performance. Unlike the procedures in biochemistry and hematology, where QC ranges for accurate test performance are generally established in each individual laboratory, susceptibility testing QC ranges are established by each authority responsible for developing and promulgating testing methods.
The Clinical and Laboratory Standards Institute (CLSI; formerly NCCLS) describes methods whereby these ranges are set for their susceptibility testing methods (8). Preliminary QC range studies, or tier 1 studies, are used principally for the purpose of controlling the performance of susceptibility tests during drug development. They are usually performed in a single laboratory with a limited number of replicates. For establishing published QC ranges for a new antimicrobial agent, a tier 2 study is recommended in the standard. A tier 2 study must involve at least seven independent laboratories, which are required to test the antimicrobial agent in or on three separate lots of medium from two different manufacturers at least 30 times (from 30 separately prepared inocula). In the case of disk testing, two separate disk lots from two manufacturers are tested. The choice of the number of laboratories, medium lots, disk lots, and replications has been determined by cumulative experience and with assistance over the years from statisticians employed in the susceptibility test manufacturing industry. Until now, QC ranges for MIC zone diameters (ZDs) have been determined largely by visual inspection of the histogram of the data generated, enhanced by “common sense” rules of thumb and, in the case of disk testing, by a statistical method involving medians which was developed in the early 1980s (4). In the latter method, a tentative QC range is calculated as the overall median of the ZDs observed in the study ± 0.5 times the range of the medians of ZDs of the individual laboratories, rounded up or down to the nearest whole millimeter. Current methods of setting QC ranges do not take advantage of the fact that the data generated follow statistical distributions, nor do they use any unbiased techniques to detect and reject outlier laboratories or results.
Here we show that relatively simple statistical techniques can be applied to data generated in CLSI QC studies and that these can be used as the primary output, to which few arbitrary rules need be applied, thereby reducing the risk of incorrectly setting QC ranges.
MATERIALS AND METHODS
Data sets.
Data sets were collected from presentations on QC studies made to the CLSI Subcommittee on Antimicrobial Susceptibility Testing and the Subcommittee of Veterinary Antimicrobial Susceptibility Testing between June 2004 and June 2006 for the purpose of establishing or revising QC ranges. In some cases, these data sets and the CLSI-determined ranges have been published (2, 3, 5, 7). The antimicrobial agents and the QC strains examined are listed in Table 1. All data were entered in raw and summarized formats into a spreadsheet (Microsoft Office Excel; Microsoft Corporation) which contained formulas for all the necessary calculations defined below.
TABLE 1.
QC strains and antimicrobial agents examined
| QC strain, medium, and methoda | Antimicrobials examined |
|---|---|
| Escherichia coli ATCC 25922 | |
| Mueller-Hinton medium, BMD | Cefquinome, ceftaroline, ceftobiprole, doripenem, faropenem, iclaprim, PTK-0796, tulathromycin |
| Brucella medium, BMDb | Azithromycin, chloramphenicol, ciprofloxacin, doxycycline, gentamicin, levofloxacin, rifampin, streptomycin, tetracycline, trimethoprim-sulfamethoxazole |
| Mueller-Hinton medium, DD | Cefquinome, ceftaroline, ceftobiprole, colistin, doripenem, faropenem, iclaprim, polymyxin B, tulathromycin |
| Staphylococcus aureus ATCC 29213 | |
| Mueller-Hinton medium, BMD | Cefquinome, ceftaroline, ceftobiprole, doripenem, faropenem, iclaprim, NXL-103, PTK-0796, tulathromycind |
| Brucella medium, BMDb | Azithromycin, chloramphenicol, ciprofloxacin, doxycycline, gentamicin, levofloxacin, rifampin, streptomycin, tetracycline, trimethoprim-sulfamethoxazole |
| Staphylococcus aureus ATCC 25923 | |
| Mueller-Hinton medium, DD | Cefquinome, ceftaroline, ceftobiprole, doripenem, faropenem, iclaprim, telavancin, tulathromycin |
| Mueller-Hinton medium plus sheep blood, DD | Ceftiofur, tulathromycin |
| Enterococcus faecalis ATCC 29212 | |
| Mueller-Hinton medium, BMD | Ceftobiprole, doripenem, NXL-103, PTK-0796, tulathromycind |
| Pseudomonas aeruginosa ATCC 27853 | |
| Mueller-Hinton medium, BMD | Ceftobiprole, doripenem |
| Mueller-Hinton medium, DD | Ceftobiprole, colistin, doripenem, polymyxin B |
| Streptococcus pneumoniae ATCC 49619 | |
| Mueller-Hinton medium plus lysed horse blood, BMD | Cefquinome, ceftaroline, ceftobiprole, doripenem, faropenem, iclaprim, NXL-103, PTK-0796, telavancin, tulathromycin |
| Brucella medium, BMDb | Azithromycin, chloramphenicol, ciprofloxacin, doxycycline, gentamicin, levofloxacin, rifampin, streptomycin, tetracycline, trimethoprim-sulfamethoxazole |
| Mueller-Hinton medium plus sheep blood, DD | Cefquinome, ceftaroline, ceftobiprole, doripenem, faropenem, iclaprim, telavancin, tulathromycin |
| Haemophilus influenzae ATCC 49247 | |
| Haemophilus test medium, BMD | Ceftaroline, ceftobiprole, doripenem, iclaprim, NXL-103, PTK-0796 |
| Haemophilus test medium, DD | Ceftaroline, ceftobiprole, doripenem, faropenem, iclaprim |
| Haemophilus influenzae ATCC 49766 | |
| Haemophilus test medium, BMD | Ceftaroline, ceftobiprole, doripenem, faropenem |
| Haemophilus test medium, DD | Ceftobiprole |
| Bacteroides fragilis ATCC 25286 | |
| Supplemented Brucella medium, BMD | Doripenem, faropenem |
| Supplemented Brucella medium plus lysed horse blood, AD | Faropenem |
| Bacteroides thetaiotaomicron ATCC 29241 | |
| Supplemented Brucella medium, BMD | Doripenem, faropenem |
| Supplemented Brucella medium plus lysed horse blood, AD | Faropenem |
| Eubacterium lentum ATCC 43055 | |
| Supplemented Brucella medium, BMD | Doripenem, faropenem |
| Supplemented Brucella medium plus lysed horse blood, AD | Faropenem |
| Campylobacter jejuni ATCC 33560 | |
| Mueller-Hinton medium plus lysed horse blood, BMDc | Azithromycin, chloramphenicol, ciprofloxacin, clarithromycin, clindamycin, doxycycline, erythromycin, florphenicol, gentamicin, levofloxacin, meropenem, nalidixic acid, telithromycin, tetracycline |
| Clostridium difficile ATCC 70057 | |
| Supplemented Brucella medium plus lysed sheep blood, AD | Amoxicillin-clavulanate, ampicillin, ampicillin-sulbactam, avilamycin, clindamycin, doripenem, garenoxacin, linezolid, meropenem, metronidazole, monensin, moxifloxacin, narasin, nitazoxanide, Opt 80, penicillin, piperacillin, piperacillin-tazobactam, ramoplanin, rifaximin, ticarcillin, ticarcillin-clavulanate, tigecycline, tilmicosin, tinidazole, tizoxanide, tylosin, vancomycin |
| Histophilus somni ATCC 700025 | |
| Veterinary fastidious medium, BMD | Cefquinome, tulathromycin |
| Chocolate Mueller-Hinton medium, DD | Cefquinome, ceftiofur, tulathromycin |
| Actinobacillus pleuropneumoniae ATCC 27090 | |
| Veterinary fastidious medium BMD | Cefquinome, tulathromycin |
| Chocolate Mueller-Hinton medium, DD | Cefquinome, ceftiofur, tulathromycin |
| Mannheimia haemolytica ATCC 33396 | |
| Mueller-Hinton medium, BMD | Tulathromycin |
| Mueller-Hinton medium plus sheep blood, DD | Ceftiofur, tulathromycin |
BMD, broth microdilution; AD, agar dilution; DD, disk diffusion.
Ranges were determined after 24 and 48 h of incubation.
Ranges were determined after incubation at 36°C for 48 h and 42°C for 24 h.
Evaluated with and without TES buffer [N-Tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid].
Calculated MIC ranges.
Because the distribution of MICs closely follows a log normal distribution, data were converted to logarithms to base 2 for ease of analysis. Means and standard deviations were calculated from these logarithms for each laboratory and for the pooled laboratory data. Using the pooled logarithms from all laboratories, MIC control ranges were calculated to encompass 95% of the values, that is, from the lower 2.5% of the distribution to the upper 97.5%. These ranges were adjusted downwards and upwards, respectively, to the nearest integer and then converted back to the relevant MICs on the conventional twofold dilution scale. This procedure ensured that at least 95%, and mostly more, of the predicted distribution of MICs in the range was included. In some cases, the range of dilutions included within the calculated range was only 1 or 2 dilutions. In line with current convention, these were adjusted to 3 dilutions, including 1 dilution above and below for ranges resulting in a single dilution and 1 dilution above or below the calculated modal dilution for ranges resulting in 2 dilutions.
Calculated ZD ranges.
Means and standard deviations were calculated for each laboratory and for the pooled laboratory data. From the pooled statistics, ZD ranges were calculated to encompass 95% of the values, that is, from the lower 2.5% of the distribution to the upper 97.5%. These values were adjusted downwards and upwards, respectively, to the nearest whole millimeter, thus ensuring that at least 95%, and mostly more, of the predicted distribution of MICs in the range was included.
Detection of possible outlier data.
Occasionally, visual inspection of the data suggested that data from some laboratories or individual values were substantially different from the others. This might be attributed to errors in test performance, including setup and reading, or to transcription errors or may indeed represent true variation in the test. In order to ensure that true variation in the data was not lost, a conservative approach was developed for the detection of possible outlier data. First, three central tendency statistics were calculated for each laboratory data set; these were the mean, the median, and the mode. Second, control ranges were set for each of these. For the mean, the control ranges were set to be within 1.645 standard deviations of the mean for the pooled data (90% of the data). For the median, the ranges were set at the 25th percentile of the pooled data minus 1.5 times the interquartile range to the 75th percentile of the pooled data plus 1.5 times the interquartile range. For the mode, the ranges were set at the mode of the pooled data ± 1 dilution for MIC tests and at 2 mm for disk diffusion tests. To be considered a possible outlier laboratory, at least two of the three central tendency statistics of an individual laboratory's data needed to be outside the control ranges.
Election to not set ranges.
CLSI has not formally established a set of rules for electing not to establish QC ranges. However, it has generally been agreed that ranges which result in an excessively broad range of MICs or ZDs are not acceptable. A twofold dilution range of ≥5 for MICs or a ZD range of >12 is thought to represent excessive scatter and/or interlaboratory variation, and usually ranges have not been established for such data sets by CLSI. Other reasons for not setting ranges have included (i) technical issues, e.g., the test dilution range did not go sufficiently high or low to accurately capture the variation in results; (ii) major differences in results between medium lots, i.e., usually one of the medium lots yielded significantly higher or lower results; and (iii) all results in MIC studies being at very low concentrations, where accuracy of preparation can be problematic.
In this study, ranges were not set when there were clearly identifiable technical, medium lot, or low-concentration issues as described above. Ranges were also not set if there was excessive variation between laboratories, defined as more than three laboratories with one central tendency statistic indicating them as possible outliers. In contrast, calculated ranges were accepted for MIC ranges of >4 dilutions and for all ZD ranges in order to allow comparison with those set by CLSI.
RESULTS
In total, 178 tier 2 broth microdilution MIC data sets and 48 disk diffusion data sets were examined. These included data on 55 different antimicrobial agents for the relevant CLSI QC strains, both aerobic and anaerobic. Both human and veterinary agents were included.
The QC ranges calculated by the statistical method were then compared to the actual QC ranges approved by the relevant CLSI subcommittee.
MIC ranges.
There were 178 MIC range comparisons (Table 2). In 15 instances (8.4%), one laboratory was identified as a possible outlier by the predefined rules. For 10 of these, all three criteria were met, and for the remaining 5, two criteria were met. The data from each of these laboratories were excluded before determination of the calculated QC ranges. In three instances, exclusion of one laboratory's data set led to a second becoming a possible outlier. Data from these second laboratories were also excluded, and ranges were then calculated using data from six laboratories. The relevant CLSI subcommittee elected not to set ranges for these particular QC strain-antimicrobial agent combinations.
TABLE 2.
Comparison of calculated MIC ranges with those set by CLSI
| Relationship of calculated and CLSI MIC ranges | No. of dilutions covered by range
|
No. (%) of ranges | |
|---|---|---|---|
| Calculated | CLSI | ||
| Identical | 3 | 3 | 77 (43.5) |
| 4 | 4 | 32 (18.0) | |
| 3 (after adjustment) | 3 | 12 (6.7) | |
| Subtotal | 121 (67.9) | ||
| Calculated ranges were narrower | 3 | 4 | 13 (7.3) |
| Calculated ranges were wider | 4 | 3 | 17 (9.6) |
| 5 | 4 | 5 (2.8) | |
| 5 | 3 | 1 (0.6) | |
| 6 | 3 | 1 (0.6) | |
| 6 | 4 | 1 (0.6) | |
| Subtotal | 25 (14.0) | ||
| CLSI ranges were not set | 3 (after adjustment) | No ranges set | 4 (2.2) |
| 4 | No ranges set | 3 (1.7) | |
| 3 | No ranges set | 1 (0.6) | |
| Subtotal | 8 (4.5) | ||
| Neither method set ranges | No ranges set | No ranges set | 11 (6.2) |
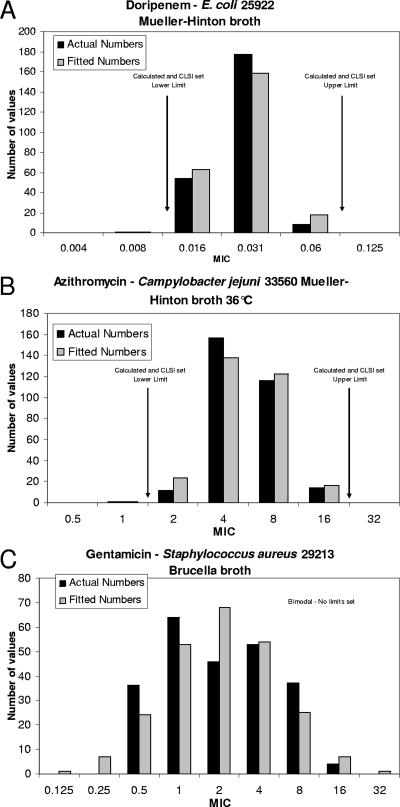
There were 11 cases where CSLI elected not to set ranges for the reasons noted in Materials and Methods. In one case (Haemophilus influenzae ATCC 49247 and doripenem), the reason was excessive variation between laboratories, which was readily captured by the rule defined in Materials and Methods. In four cases, there was a substantial difference in results with one medium lot compared to those with the other two lots. An example is presented in Fig. 1C, where the original data were obviously bimodal due to one medium lot giving significantly higher MICs than those seen with the other two medium lots. In four cases, the dilution series used did not go high enough, and in two further cases the dilution series did not go low enough, to capture all possible variation.
FIG. 1.
Comparison of calculated and observed MIC distributions for three QC strain-antimicrobial combinations. (A) Doripenem versus E. coli ATCC 25922 in Mueller-Hinton broth (2). (B) Azithromycin versus Campylobacter jejuni ATCC 33560 in Mueller-Hinton broth supplemented with lysed horse blood, incubated at 36°C for 49 h (7). (C) Gentamicin versus Staphylococcus aureus ATCC 29213 in Brucella broth.
In general, the fitting of the data to a log normal distribution worked well for the MIC data, with the number of strains at each dilution from the fitted data closely matching that actually observed (Fig. 1A and B).
Two-thirds (121/178 ranges) of the calculated MIC ranges were identical to those set by CLSI. In 12 of the 121 instances where they were identical, adjustments had been made to the calculated ranges as outlined in Materials and Methods. In all cases where the calculated ranges resulted in narrower ranges, the calculated ranges covered 3 dilutions while the ranges set by CLSI covered 4 dilutions. Frequently, in these cases, the CLSI ranges were extended to 4 dilutions because of the “shoulder” rule, which states that if the frequency of observations at an MIC above or below the modal MIC is about 65% that of the mode or greater, then the QC range should be extended 1 twofold dilution lower or higher than that concentration, respectively.
When calculated and CLSI ranges were different, the CLSI ranges were more likely to be narrower, mostly by a single dilution. There were six instances where the calculated ranges ran to 5 dilutions (e.g., gentamicin versus Escherichia coli ATCC 25922 in Brucella microdilution broth after 24 and 48 h of incubation) and two where the ranges included 6 dilutions (e.g., doripenem versus Bacteroides fragilis ATCC 25285 in supplemented Brucella microdilution broth). Closer inspection of the data raises the question of whether it was appropriate to set ranges at all because of considerable variation between laboratories, but without any standout individual laboratory. When there is that much statistical variation between laboratories, one questions the wisdom of trimming the ranges to include 3 or 4 dilutions, even if 95% of the observed values are captured, as it is likely that >5% of QC results will be out of control when the QC range is put into wide routine practice.
In eight instances, the new method calculated a range that was not set by CLSI. This suggests that the new method can give guidance on whether to set ranges, even if there are apparent difficulties with the data.
Sixteen sets (9%) of calculated ranges required adjustment to include 3 twofold dilutions, with 11 sets (6.2%) covering a single calculated dilution and 5 sets (2.8%) covering two calculated dilutions.
ZD ranges.
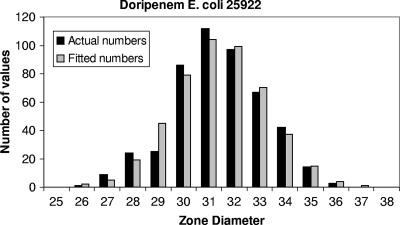
There were 48 ZD range comparisons (Table 3). No laboratory data were considered possible outliers, and all data were included in the calculation of the ranges. The majority of calculated ranges generated a wider range of ZDs than those determined by CLSI, but only by 1 or 2 mm. In some cases, the calculated ranges covered a narrower range, by 1 to 2 mm. In one-third of cases, the ranges were identical. By inspection, the fits to a normal distribution were generally very good (Fig. 2).
TABLE 3.
Comparison of calculated ZD ranges with those set by CLSI
| Relationship of calculated and CLSI ZD ranges | ZD (mm) covered by range
|
Difference between ranges (mm) | No. (%) of ranges | |
|---|---|---|---|---|
| Calculated | CLSI | |||
| Identical | 6 | 6 | 0 | 1 (2.1) |
| 7 | 7 | 0 | 5 (10.4) | |
| 8 | 8 | 0 | 1 (2.1) | |
| 9 | 9 | 0 | 6 (12.5) | |
| 10 | 10 | 0 | 1 (2.1) | |
| 11 | 11 | 0 | 2 (4.2) | |
| Subtotal | 16 (33.3) | |||
| Calculated ranges were narrower | 7 | 8 | 1 | 1 (2.1) |
| 8 | 9 | 1 | 2 (4.2) | |
| 9 | 11 | 2 | 1 (2.1) | |
| Subtotal | 4 (8.3) | |||
| Calculated ranges were wider | 6 | 5 | 1 | 2 (4.2) |
| 8 | 7 | 1 | 3 (6.3) | |
| 9 | 7 | 2 | 1 (2.1) | |
| 9 | 8 | 1 | 3 (6.3) | |
| 10 | 8 | 2 | 1 (2.1) | |
| 10 | 9 | 1 | 5 (10.4) | |
| 11 | 9 | 2 | 2 (4.2) | |
| 12 | 10 | 2 | 2 (4.2) | |
| 12 | 11 | 1 | 3 (6.3) | |
| 13 | 11 | 2 | 4 (8.3) | |
| 13 | 12 | 1 | 1 (2.1) | |
| 15 | 13 | 2 | 1 (2.1) | |
| Subtotal | 28 (58.3) | |||
| Ranges could not be set | Nil | Nil | ||
| Neither method set ranges | Nil | Nil | ||
FIG. 2.
Comparison of calculated and observed ZD distributions for doripenem versus E. coli ATCC 25922 (2).
DISCUSSION
Considerable time and thought have gone into the design and development of tier 2 studies for establishing CLSI QC ranges. The CLSI subcommittees and the Quality Control Working Group, in particular, have consciously tried to enhance the predictive value of their ranges by estimating the number of laboratories and replicate measurements required to ensure that the data are likely to be representative of the intra- and interlaboratory variation (M. Ullery, personal communication). While statistical methods have been applied to define the numbers of laboratories and replicates, up to now they have not been applied in a concerted way to the analysis of the data. Statistical methods lend themselves readily to the analysis of tier 2 QC study data because the data from an individual laboratory are approximately log normally (MIC) or normally (ZD) distributed, and data from multiple laboratories conform to the central limit theorem (which in this context implies that the mean of the individual laboratory means more closely approaches the true mean of the population, that is, the mean MIC of all QC tests that will be performed in the future).
In developing the statistical method, we have attempted to embrace the “rules of thumb” that are currently employed by the CLSI subcommittees, while enhancing them by (i) attempting to identify possible outlier data in a reproducible manner and (ii) using predominantly statistical values to define the ranges rather than having them defined by visual inspection plus capture of at least 95% of the observed data in the study.
Although participation of more laboratories will possibly generate ranges that have better predictive value, the costs of conducting these studies constrain the numbers, and it has been calculated that seven laboratories should provide sufficient data to allow estimates of ranges to be reasonably predictive of those likely to be observed in routine testing (Ullery, personal communication). On the basis that data from one laboratory might be nonrepresentative, it has therefore been common practice to use eight laboratories in tier 2 studies. However, CLSI has not established criteria that would detect nonrepresentative laboratories, and judgments are usually made “by committee.”
With regard to possible outlier detection, the statistical method proposed here has been designed to minimize the possibility of data rejection and to ensure that true variation is included. Indeed, while we excluded data from laboratories identified as possible outliers for the purposes of analysis and comparison, we would not recommend exclusion as a matter of course. Instead, we envision the identification of possible outlier data as a flag to investigate possible causes with the laboratory concerned before considering data exclusion. For instance, in one case where MIC ranges were being examined, it was clear that the possible outlier data from one laboratory for one QC organism were actually data from the same laboratory for another QC strain against the same agent, i.e., they were the result of a transcriptional error. On the other hand, we suggest that serious consideration be given to including such data when no reasonable technical or transcriptional cause can be found.
Calculated MIC ranges resulted in some QC ranges that were narrower than those set by the CLSI subcommittees. The calculated ranges merely reflect the amount of variation in MICs observed in the study. The current CLSI convention is to adjust the ranges to include at least 3 twofold dilutions, and this convention was applied to our calculated ranges for comparison purposes. Indeed, in some other susceptibility testing standards, this convention has been codified (1). However, the validity of doing this can be questioned. The fact that ranges calculated from eight-laboratory tier 2 studies can be only 1 or 2 twofold dilutions is a consequence of the relatively coarse grouping that the twofold dilution series imposes on the data. If finer grouping of MICs were to be used, such as that generated by the commercial Etest (Solna, Sweden) gradient diffusion method (6), it would be more obvious that the scatter in MICs for individual QC strain-antimicrobial agent combinations would vary significantly. It is therefore possible that the intra- and interlaboratory variation in MICs of QC strains could be quite small and consist of values within just 1 or 2 twofold dilutions. An example is the combination of Enterococcus faecalis ATCC 29212 and doripenem. Of 240 replicate measurements, 231 were at a single MIC (2 μg/ml), and in four of the eight laboratories this was the only value observed in 30 replicates. The calculated QC ranges were a single concentration, as this clearly captured much more than 95% of the data. According to the distribution of the data and the calculated statistical parameters, there is a 0.4% probability of observing a value of 1 μg/ml and a 0.6% probability of observing a value of 4 μg/ml. Adjusting to 3 dilutions is done to address the fear that a 1- or 2-twofold-dilution range will not be representative when applied to routine work. However, it seems inconsistent to apply this adjustment only to calculated 1- and 2-twofold-dilution ranges. This implies that these ranges are not predictive while those with 3 and 4 twofold dilutions are predictive when both are found in a single study. Further discussion will be required to address the problem of narrow MIC ranges.
In contrast, it can be argued more easily that calculated MIC ranges producing an excessively broad range of twofold dilutions are problematic. A calculated range of 5 or 6 twofold dilutions for a particular QC strain-antimicrobial agent combination means that there is significant interlaboratory variation, which suggests that the particular QC strain is not a reliable QC indicator for that antimicrobial agent. There were eight such ranges (4.5%) in the data sets we examined. At present, CLSI subcommittees are likely to accept such combinations if 95% of the observed data are within 4 twofold dilutions. However, it is the spread of variation with combinations that produce a 5- to 6-twofold-dilution range that should send a warning about the particular QC strain's reliability when applied in a routine context. We advocate caution in setting QC ranges for any QC strain where the calculated MIC ranges produce such broad ranges, because we can predict from the statistics that in routine practice, out-of-range values will occur more often than 5% of the time.
Far fewer issues arose in the comparison of ZD ranges. There were no problems with possible outliers, coarse grouping of data, or excessively broad or narrow calculated ranges. The tendency for calculated ranges to produce a slightly wider range of ZDs was expected, as the current CLSI method accommodates 95% of the data observed in the study while the calculated ranges are meant to apply to the indefinitely large number of QC tests that will be performed in routine laboratories.
Overall, we believe the statistical approach adds value to the current CLSI method of establishing QC ranges. It was easily set up in a spreadsheet which requires only entry of the raw data and has visual alerts to possible outlier data once entry is complete. The calculated ranges and other fields are automatically recalculated whenever the raw data are modified, such as exclusion of possible outlier data, allowing the group collating the tier 2 study data to examine the effects of data adjustment immediately. Ultimately, the proof of this concept will be in its application in the field. Unfortunately, CLSI does not currently have a direct system for measuring the performance of its published QC ranges. Rather, it relies on feedback from clients (e.g., laboratories and pharmaceutical sponsors) to raise concerns about “abnormal” rates of out-of-control data. We look forward to such data being collected in future, as it would allow direct comparison between the current range-setting methods and the proposed statistical method.
Acknowledgments
We thank all those individuals who conducted and presented the studies used in our analyses and the CLSI Subcommittee on Antimicrobial Susceptibility Testing, especially Steven Brown, Clyde Thornsberry, and David Hecht. We are extremely grateful to Sharon Cullen for providing important historical material from previous CLSI Quality Control Working Group papers and discussions.
Footnotes
Published ahead of print on 16 April 2007.
REFERENCES
- 1.British Society for Antimicrobial Chemotherapy. January 2006, posting date. BSAC methods for antimicrobial susceptibility testing, version 5. http://www.bsac.org.uk/_db/_documents/version_5_.pdf.
- 2.Brown, S. D., and M. M. Traczewski. 2005. Comparative in vitro antimicrobial activity of a new carbapenem, doripenem: disc diffusion criteria and quality control. J. Antimicrob. Chemother. 55944-949. [DOI] [PubMed] [Google Scholar]
- 3.Brown, S. D., M. M. Traczewski, and the Brucella QC Working Group. 2005. Broth microdilution susceptibility testing of Brucella species: quality control limits for ten antimicrobial agents against three standard quality control strains. J. Clin. Microbiol. 435804-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gavan, T. L., R. N. Jones, A. L. Barry, P. C. Fuchs, E. H. Gerlach, J. M. Matsen, L. B. Reller, C. Thornsberry, and L. D. Thrupp. 1981. Quality control limits for ampicillin, carbenicillin, mezlocillin and piperacillin disk diffusion susceptibility tests: a collaborative study. J. Clin. Microbiol. 1467-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones, R. N., T. M. Anderegg, J. M. Swenson, and the Quality Control Working Group. 2005. Quality control guidelines for testing gram-negative control strains with polymyxin B and colistin (polymyxin E) by standardized methods. J. Clin. Microbiol. 43925-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kronvall, G., I. Karlsson, M. Walder, M. Sörberg, and L. E. Nilsson. 2006. Epidemiological MIC cut-off values for tigecycline calculated from Etest MIC values using normalized resistance interpretation. J. Antimicrob. Chemother. 57498-505. [DOI] [PubMed] [Google Scholar]
- 7.McDermott, P. F., S. M. Bodeis-Jones, T. R. Fritsche, R. N. Jones, R. D. Walker, and the Campylobacter Susceptibility Testing Group. 2005. Broth microdilution susceptibility testing of Campylobacter jejuni and the determination of quality control ranges for fourteen antimicrobial agents. J. Clin. Microbiol. 436136-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.NCCLS. 2001. Development of in vitro susceptibility testing criteria and quality control parameters; approved guideline, 2nd ed. Document M23-A2. NCCLS, Wayne, PA.