Abstract
Methicillin resistance in Staphylococcus aureus is primarily mediated by the acquired penicillin-binding protein PBP 2a, which is encoded by mecA. PBP 2a acts together with native PBP 2 to mediate oxacillin resistance by contributing complementary transpeptidase and transglycosylase activities, respectively. In this study, we have investigated a phenotype of β-lactam dependence in a clinical methicillin-resistant S. aureus strain (strain 2884D) obtained by in vitro selection with ceftobiprole. 28884D, which grew very poorly in blood agar, required the presence of the β-lactam antibiotics to grow. On the basis of this observation, we hypothesized that a gene or genes essential for growth were dependent on oxacillin induction. Identification and analysis of genes regulated by oxacillin were performed by both real-time reverse transcription-PCR and spotted microarray analysis. We found that mecA was constitutively expressed in strain 2884D and that the constitutive expression resulted from perturbations in the two systems involved in its regulation, i.e., MecI/MecR1 (staphylococcal chromosome cassette mec type I) and BlaI/BlaR1 (nonfunctional penicillinase operon). PBP 2 appeared to be poorly induced by oxacillin in 2884D. Further analysis of the PBP 2 two-component VraSR regulatory system showed that it was nonfunctional, accounting for the lack of response to oxacillin. Together, these results support the notion that limited PBP 2 availability may have led 2884D to become dependent on oxacillin-mediated mecA induction as a required survival mechanism.
Methicillin-resistant Staphylococcus aureus (MRSA) is an increasingly common cause of nosocomial infections and now is also appearing in community populations (25). The therapeutic agents available for the treatment of staphylococcal infections have been limited as a result of acquired resistance to the actions of the most active antimicrobials, especially β-lactams. Methicillin resistance in S. aureus is mediated by the acquisition of a new drug-resistant target, a penicillin-binding protein (PBP), PBP 2a, which has a decreased affinity for β-lactam antibiotics, but it can continue to cross-link the cell wall once the native PBPs (i.e., PBPs 1 to 4) have been inactivated. PBP 2a is encoded by the mecA gene, which is located on a 21- to 67-kb genomic island called staphylococcal chromosome cassette mec (SCCmec) (18). The transcription of mecA can be regulated by two distinct but related sets of regulatory genes. One of them is mecI/mecR1, which is located upstream of mecA and which is divergently transcribed from mecA. The second set is the homologous regulatory element of staphylococcal penicillinase, blaI/blaR1 (23). MecI and BlaI are repressors that can each repress mecA transcription as well as blaZ transcription (10, 23, 33). In contrast, MecR1 and BlaR1 are the corresponding sensor transducers which, in contrast to MecI and BlaI, are specific for the cognate repressors and cannot substitute for each other (23). Sensing of the signals of these transmembrane repressors occurs by binding of one inducer to the extracellular sensor domain, which results in the activation of the intracellular metalloprotease domain. This protease cleaves the repressor, enabling the transcription of either mecA through mecR1/mecI or blaZ through blaR1/blaI (1, 41).
Among other factors, one of the native staphylococcal PBPs, PBP 2, is also required for the full expression of methicillin resistance (29). PBP 2 is a high-molecular-weight class A PBP with both transpeptidase and transglycosylase domains (29). In the presence of high concentrations of methicillin, PBP 2 is completely acylated; but its β-lactam-insensitive transglycosylase domain is still required for resistance, implying that PBP 2a and PBP 2 functionally cooperate during growth in the presence of β-lactams (27, 28). The cell wall of transglycosylase mutants is enriched for shorter glycan strands. Therefore, the cooperation between PBP 2a and PBP 2 has been proposed to be a mechanism by which MRSA strains compensate for the decreased cell wall cross-linking that results from the presence of β-lactams by generating longer glycan strands (27, 28).
In addition to these observations, a β-lactam-inducible two-component regulatory system (VraSR) consisting of a sensor kinase (VraS) and a response regulator (VraR) has been found to control β-lactam-dependent induction of PBP 2 transcription as well as that of other cell wall biosynthesis genes (21). In MRSA clinical strains, the disruption of VraSR results not only in reductions of β-lactam and/or vancomycin MICs but also in the reduction of PBP 2 transcription (7, 14, 21).
In the present report we describe a clinical oxacillin-dependent MRSA strain (strain 2884D) that was selected in vitro with ceftobiprole, a novel broad-spectrum cephalosporin that binds with a high affinity to PBP 2a and that is active against MRSA and vancomycin-resistant S. aureus strains (8). Ceftobiprole is stable in the presence of the class A penicillinases produced by S. aureus and enteric gram-negative organisms (16) and is relatively stable in the presence of some class C β-lactamases produced by some enteric gram-negative organisms (16). Oxacillin-dependent strain 2884D requires the presence of β-lactam antibiotics for growth, suggesting that the expression of a gene or genes essential for growth is dependent on induction by oxacillin. The purpose of this study was to investigate the phenotype of β-lactam dependence and to determine the potential involvement of oxacillin-dependent responsive genes in that phenotype.
MATERIALS AND METHODS
Materials and media.
Trypticase soy agar (TSA) II 5% sheep blood (BBL, Sparks, MD) and Mueller-Hinton agar (BBL Microbiology Systems, Cockeysville, MD) with and without additives (Sigma, St. Louis, MO; United States Biochemicals, Cleveland, OH) were used for the subculture and maintenance of the S. aureus strains.
Bacterial strains.
S. aureus 2884 was isolated from the sputum and nose of an 83-year-old woman on the day of hospitalization for a lower respiratory tract infection. S. aureus was identified by use of the Pastorex Staph Plus kit (Bio-Rad, Marnes la Coquette, France). The strain showed resistance to β-lactam and fluoroquinolone antibiotics. Mutant strain S. aureus 2884D was selected as a one-step mutant in the presence of ceftobiprole with a mutation frequency of 1.4 × 10−10 Briefly, 0.1 ml of a heavy inoculum (1 × 1010) was streaked and selected on a Mueller-Hinton agar (Difco) plate containing 4% NaCl and 5 μg of ceftobiprole. After 48 h of incubation at 37°C in air, the colonies were tested for their susceptibilities to β-lactams.
Antibiotics.
Standard reference powders and disks were obtained from Basilea Pharmaceutica, Basilea, Switzerland (ceftobiprole); Aventis, France (penicillin G and cefoxitin); Bristol-Myers Squibb, France (oxacillin and cloxacillin); Merck Sharp & Dohme, Chibret, France (imipenem); and Glaxo SmithKline, France (amoxicillin and amoxicillin clavulanate). Resistance to oxacillin, cefoxitin, penicillin, ceftobiprole, amoxicillin clavulanate, and imipenem was determined according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI; formerly NCCLS) (24) and CASFM (12). Disk diffusion tests were performed with disks obtained from Bio-Rad (Marnes la Coquette, France) for all antibiotics except ceftobiprole; ceftobiprole disks were obtained from Basilea Pharmaceutica. A 0.5 McFarland standard suspension was used as the inoculum for a 100-mm-diameter Mueller-Hinton agar plate, upon which disks of oxacillin (5 μg), cefoxitin (30 μg), ceftobiprole (30 μg), amoxicillin-clavulanic acid (20 μg/10 μg), ofloxacin (5 μg), and imipenem (10 μg) were placed as they would be for a routine disk diffusion test. The ranges of inhibition zone diameters were determined according to the guidelines of the CLSI and CASFM.
Population analysis.
Population analysis profiles were determined as described previously (36). Overnight cultures were plated at various dilutions on TSA plates containing a series of concentrations of oxacillin (Sigma-Aldrich), and the bacterial colonies were counted after incubation of the plates at 37°C for 48 h.
Determination of SCCmec types.
Chromosomal DNA was prepared by using a QIAGEN (Valencia, CA) genomic DNA preparation kit according to the manufacturer's directions. SCCmec types were determined by the use of specific primers for amplification of the key genetic elements mecA (primers mecA1 and mecA2), mecI (primers 214 and 215), and IS1272 (primers F3 and mA2 and primers F1 and B3) (38) and also by using those described by Oliveira et al. (26). To detect the ccr gene complex, primers α1, α2, α3, and β2 were used as described previously (25). PCR was performed with a Taq PCR MasterMix kit (QIAGEN, Valencia, CA) with a 50-μl reaction volume in a MiniCycler thermocycler (MJ Research, Boston, MA). As SCCmec standards, we used S. aureus strains COL (SCCmec type I), N315 (SCCmec type II), ANS46 (SCCmec type III), and MW2 (SCCmec type IV).
Sequences analysis.
Chromosomal DNA was prepared by using a QIAGEN genomic DNA preparation kit, according to the manufacturer's directions. PCR amplification of the DNA sequences was performed to confirm the presence of mecA, mecA-mecR1 promoter operator sequences, blaZ-blaR1 promoter operator sequences, and blaR1 and blaI regulators by use of the corresponding primers shown in Table 1. A QIAGEN Taq PCR master mix kit was used for the amplification reaction, and the thermocycling conditions were those recommended by the manufacturer. Sequence analysis was performed by using the automated laser fluorescence technique with fluorescein-labeled oligonucleotides (Applied Biosystems, Foster City, CA). Consensus sequences were assembled from both orientations with Vector NTI Advance 10 software for Windows (InforMax, Bethesda, MD). S. aureus N315 and S. aureus N315 plasmid pN315 (GenBank accession no. NC_003140) were used as controls.
TABLE 1.
Primers used for SCCmec typing and real-time RT-PCR
| Procedure and primer | Sequence |
|---|---|
| SCCmec typing | |
| F3 | TTGGGTTTCACTCGGATG |
| mA2 | AACGTTGTAACCACCCCAAGA |
| F1 | CACAATCTGTATTCTCAGGTCG |
| B3 | ATTAGTGCTCGTCTCCACG |
| α2 | AACCTATATCATCAATCAGTACGT |
| α3 | TAAACGCATCAATGCACAAACACT |
| α4 | AGCTCAAAAGCAAGCAATAGAAT |
| β2 | ATTGCCTTGATAATAGCCTCT |
| mecA1 | GGAGGATATTGATGAAAAAG |
| mecA2 | GCTTCACTGTTTTGTTATTC |
| 214 | CGGATCCGAAATGGAATTAATATAATG |
| 215 | CGGAATTCGACTTGATTGTTTCCTC |
| BlaR1-501 | TCAATCTCCAATAACTTTTTGG |
| BlaR1-1100 | GGATCTTTCGTTATGTACAG |
| BLAI-11463 | ACAATCGTATCTCCTGACCTCGCA |
| Real-time RT-PCR | |
| 16S | 562F-TCCGGAATTATTGGGCGTAA |
| 682R-CCACTTTCCTCTTCTGCACTCA | |
| 613T-TGAAAGCCCACGGCTCAACCGa | |
| BlaZ | 647F-CCTGCTGCTTTCGGTAAGACTT |
| 783R-TCTTTTGGAACACCGTCTTTAATTAA | |
| 682T-TTCTTGTTTTCTTTGCTTAATTTTCCATTTGCGa | |
| BlaR | 232F-AACCATAACATAAATACCACCAAACCTAT |
| 342R-AACTAAAACTATCCAAATAACTGTGCAGAT | |
| 264T-AATCCCAATTAAACTTATGGATATCTGTTGCGAACTCTa | |
| mecA | 512F-GTTAGATTGGGATCATAGCGTCATT |
| 657R-TGCCTAATCTCATATGTGTTCCTGTAT | |
| 538T-TTCCAGGAATGCAGAAAGACCAAAGCATGAa | |
| PBP2 | 165F-CGGTGCTGAAACTTATTCACAATATAAT |
| 249R-AGTGTATTGAGGTGTAAAGCCGTTAA | |
| 196T-CCACACGTCTTTCGCTGCATTATCAGGa | |
| VraR | 93F-GAATCCTCCTTATTTAAAGGTGCTTTC |
| 174R-TTGGTGCAACGTTCCATATTG | |
| 121T-CCTCGATACGTGTACCTGAATCTGGCAATGa | |
| BlaI | 265F-TAAACTGTATGGAGGGGATATGAAA |
| 356R-TGTCTCGCAATTCTTCAATTTCTT | |
| 307T-GCGAAAAATGAAGAATTAAATAACa |
Probe sequence.
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed as described previously (4, 22). The genomic DNA was prepared (2, 18) and digested with 10 U of SmaI enzyme (New England, Biolabs, Beverly, MA). Separation was performed in a CHEF-DRIII system (Bio-Rad Laboratories, Hercules, CA). The resolved macrorestriction patterns were compared on the basis of the recommendations of Tenover et al. (35). The relatedness of the strains was judged by visual comparison of the banding patterns of samples run together in the same gel, according to previously described criteria (35). Strains were considered identical when their PFGE patterns contained the same numbers and sizes of fragments.
Microarray transcriptional profiling of parental and oxacillin-dependent 2884 strains.
Microarray transcriptional profiling was carried out with a spotted DNA microarray containing 4,546 oligonucleotides (70-mer) covering the genomes of S. aureus COL (2,654 open reading frames [ORFs]), S. aureus N315 (2,623 ORFs), S. aureus Mu50 (2,748 ORFs), MRSA 252 (2,744 ORFs), methicillin-susceptible S. aureus MSSA 476 (2,619 ORFs), and pLW043 (62 ORFs). The entire protocol for the printing of the DNA microarray slides, probe preparation, and hybridization is described in detail at htpp//www.tigr.org/microarray/protocolsTIGR.shtml. RNA was isolated from parental (strain 2884) and oxacillin-dependent (strain 2884D) cells harvested at exponential phase of growth and resuspended in 500 μl QIAGEN RNeasy kit buffer RLT. The resuspended cells were then transferred to Q-Biogene FastPrep Lysing Matrix B tubes. The cells were disrupted in a FastPrep FP120 cell disrupter for 40 s at setting 6.0 and placed on ice for 5 min. The disrupted cells were then centrifuged at maximum speed (13,000 × g) for 15 min at 4°C. The aqueous phase was transferred to a fresh 1.5-ml microcentrifuge tube, and 350 μl RLT buffer was added per 100 μl sample. After centrifugation for 15 s at 8,000 × g, the supernatant was transferred to a fresh tube and 250 μl 100% ethanol was added per 100 μl sample. The samples were then applied to a QIAGEN RNeasy mini column and processed according to the manufacturer's instructions. All RNA samples were analyzed by spectrophotometry for determination of A260/A280 and gel electrophoresis to assess their concentrations and integrities. The samples were then treated with DNase (Ambion Inc., Austin, TX). cDNA probes were produced by reverse transcription (RT) of RNA (2 μg) and were indirectly labeled with either Cy3 or Cy5 dye (Amersham Biosciences, Piscataway, NJ). All hybridizations were done with cDNA probes into which a minimum of 170 pmol of dye molecule per microgram of cDNA produced was incorporated. TIFF images of the hybridized arrays were analyzed by using TIGR Spotfinder software (http://www.tigr.org/software/); the data set was normalized by applying the LOWESS algorithm (block mode; smooth parameter, 0.33) and using TIGR MIDAS software (http://www.tigr.org/software/), and significant changes were identified with SAM (significance analysis of microarrays) software (http://www-stat.stanford.edu/∼tibs/SAM/index.html). Several controls were used to ensure that the data obtained were of good quality. First, each ORF was present in duplicate on the array. Second, three independent RNA batches per strain were used. Third, the quality of the RNA samples was checked in self-hybridization experiments. Finally, each RNA preparation was used to make probes for at least two separate arrays for which the incorporated dye was reversed (dye flip). Differential expression was defined as a change of more then twofold in the amount of transcript compared with that of the comparator strain.
Real-time RT-PCR.
Oligonucleotide primers and probes (Table 1) were designed with Primer Express 1.0 software from ABI Prism (Perkin-Elmer Applied Biosystems, Foster City, CA) and purchased from Megabase Inc. (Evanston, IL). The probes consisted of an oligonucleotide labeled with the reporter dye 6-carboxyfluorescein at the 5′ end and with the quencher dye N,N′,N′-tetramethyl-6 carboxytetramethylrhodamine at the 3′ end. RT-PCR was done with the TaqMan One-Step RT-PCR Master Mix Reagents kit, as described by the manufacturer (Perkin-Elmer Applied Biosystems). The RT-PCR mixture (25 μl) contained 6.25 U of Multiscribe reverse transcriptase, 10.0 U of RNase inhibitor, 500 nM (each) gene-specific primer, 100 nM (each) probe, and 25 ng of total RNA template. Amplification and detection of the specific products were performed with an ABI Prism 7700 sequence detection system (Perkin-Elmer Applied Biosystems) with the following cycle profile: 1 cycle at 48°C for 30 min, 1 cycle at 95°C for 10 min, and 40 cycles at 95°C for 15 s and 60°C for 1 min. The critical threshold cycle (CT) was defined as the cycle at which the fluorescence became detectable above the background level and was inversely proportional to the logarithm of the initial number of template molecules. For each primer-probe set, a standard curve was plotted with CT values obtained from the amplification of known quantities of RNA isolated from strain S. aureus N315. The standard curves were used to transform the CT values to the relative number of RNA molecules. The amount of contaminating chromosomal DNA in each sample was determined from control reactions in which the reaction mixtures did not contain reverse transcriptase. The quantity of cDNA for each experimental gene was normalized to the quantity of 16S cDNA in each sample. Each RNA sample was run in triplicate.
RESULTS
Initial characterization of parental and oxacillin-dependent S. aureus 2884.
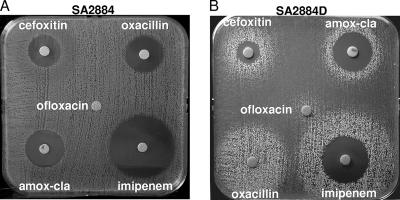
To determine whether strain 2884D, the oxacillin-dependent mutant, represented an isogenic derivative of parental S. aureus 2884, we performed PFGE. Both the parental 2884 and the oxacillin-dependent 2884D strains displayed identical PFGE profiles (data not shown). However, they differed in their susceptibilities to antibiotics. In fact, parental strain 2884 showed ranges of inhibition zone diameters for oxacillin (15 mm), cefoxitin (13 mm), and amoxicillin-clavulanic acid (16 mm) compatible with a heterogeneous MRSA phenotype; but it was still susceptible to ceftobiprole (disk zone diameter, 34 mm) (data not shown) (Fig. 1A), while oxacillin-dependent strain 2884D grew around the β-lactam disks only as satellite colonies; a very light growth appeared in areas between the growth around the disks, demonstrating its dependence on β-lactams for growth (Fig. 1B). Nevertheless, both 2884 and 2884D were resistant to ofloxacin, indicating that the antibiotic dependence of 2884D was observed only with β-lactam antibiotics. These results were confirmed by determination of MICs for parental strain 2884 (ceftobiprole MIC, 2 μg/ml; oxacillin MIC, 64 μg/ml; cefoxitin MIC, 32 μg/ml; amoxicillin-clavulanic acid [Augmentin] MIC, 16 μg/ml; and imipenem MIC, 8 μg/ml). Due to the growth limitations of the dependent strain, which grew only in the presence of 0.5 μg/ml of oxacillin, only the oxacillin MIC was determined, resulting in values that were ≥256 μg/ml.
FIG. 1.
Disk diffusion tests for resistance to amoxicillin-clavulanic acid (amox-cla), imipenem, oxacillin, and ceftobiprole in S. aureus 2884 (A) and S. aureus 2884D (B). A 0.5 McFarland standard suspension was used as the inoculum for a 100-mm-diameter Mueller-Hinton agar plate upon which oxacillin (5 μg), amoxicillin-clavulanic acid (20/10 μg), cefoxitin (30 μg), ofloxacin (5 μg), and imipenem (10 μg) disks were placed as they would be for a routine disk diffusion test. Resistance to these antimicrobials was determined according to the guidelines of the CLSI and CASFM.
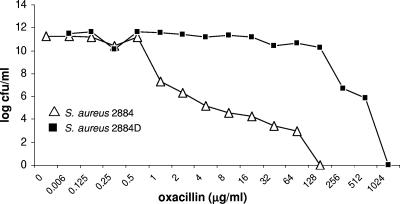
The phenotypes of methicillin resistance expressed by parental strain 2884 and oxacillin-dependent strain 2884D were investigated by population analysis in Mueller-Hinton agar, as described in Materials and Methods. As shown in Fig. 2, parental strain 2884 expressed a curve typical of that for heterogeneous methicillin resistance, with a more than 4-log-fold decrease in the number of CFU/ml at oxacillin concentrations of 1 μg/ml. Strain 2884D displayed a homogeneous expression profile, with the first 3-log-fold reduction in CFU/ml observed only at concentrations of oxacillin ≥256 μg/ml (Fig. 2).
FIG. 2.
Phenotypic expression of methicillin resistance. Overnight cultures from parental S. aureus 2884 (open triangles) and oxacillin-dependent strain S. aureus (filled squares) strains were plated at various dilutions on TSA plates containing a series of concentrations of oxacillin and incubated at 37°C for 48 h, after which the bacterial colonies were counted. The results of a representative study are shown. Two other experiments gave similar results. +oxa, growth in the presence of oxacillin.
SCCmec types.
The SCCmec type (type I) and key SCCmec genetic elements were identical between strains 2884 and 2884D. SCCmec type I contains a deletion of the entire mecI gene and 45% of the mecR1 gene, including the penicillin-binding domain, with insertion of a truncated copy of IS1272 (2, 3). Analysis of the mecA and the mecA-mecR1 promoter operator sequences of parental strain 2884 and oxacillin-dependent strain 2884D showed that they were identical to those of wild-type strain S. aureus N315 (GenBank accession no. NC_002745).
Transcription analysis of mecA.
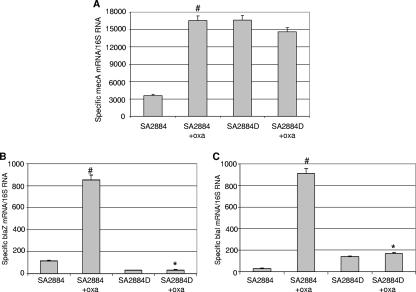
Among the genes for which transcription is induced directly by oxacillin are pbp2 and mecA, with pbp2 being essential for growth, while mecA is essential for growth in the presence of oxacillin (5, 6, 21). Therefore, we first investigated mecA expression levels by using real-time RT-PCR both in 2884 grown either in the absence or in the presence of oxacillin and in oxacillin-dependent strain 2884D (Fig. 3A). Despite the difficulty of doing definitive studies because the strain cannot be grown without oxacillin, we attempted to grow 2884D in blood agar. Even though growth was poorly observed, enough RNA could be extracted to perform RT-PCR analysis. A marked fourfold increase in the mecA mRNA content was observed in parental strain 2884 grown in the presence of 0.5 μg/ml oxacillin, while mecA expression in oxacillin-dependent strain 2884D grown either in blood agar (without oxacillin) or in the presence of 0.5 μg/ml oxacillin showed no significant changes compared to that detected in strain 2884 grown in the presence of oxacillin (Fig. 3A). Since 2884D grown in the absence of oxacillin (blood agar) displayed values similar to those observed in the presence of oxacillin, these results suggested that mecA expression in 2884D may be constitutive. Moreover, these results were consistent with the structural analysis of SCCmec, showing that in 2884D both mecI and mecR1 were disrupted and, therefore, could not regulate mecA transcription.
FIG. 3.
Determination of levels of expression of mecA (A), blaZ (B), and blaI (C) by TaqMan real-time RT-PCR in parental S. aureus strain 2884 and oxacillin-dependent strain 2884D. Relative values of the amount of specific mRNAs over 16S mRNA are shown on the vertical axis. #, transcription was significantly greater for strain 2884 grown in the presence of oxacillin compared with that seen for the same strain grown in the absence of oxacillin (SA2884+oxa and SA2884, respectively; P < 0.001); *, transcription levels were significantly lower in strain 2884D than in strain 2884D induced with oxacillin (SA2884D and SA2884+oxa, respectively; P < 0.001).
MecA expression, in addition to mecRI/mecRI, may also be regulated by the blaI/blaR1 system, which also controls the expression of the structural gene blaZ (10, 23, 33). We analyzed the penicillinase operon in both strain 2884 and strain 2884D in the presence or absence of 0.5 μg/ml of oxacillin. As shown in Fig. 3B, a 7.4-fold increase in the blaZ mRNA content in oxacillin-induced parental strain 2884 compared to that in the corresponding uninduced strain grown without oxacillin was observed (i.e., 851.8 ± 45.2 and 114.6 ± 4.5, respectively). In contrast, no induction of blaZ expression was observed in 2884D grown in 0.5 μg/ml oxacillin (35.4 ± 4.1), and similar values were observed for 2884D grown in blood agar (31.2 ± 3.9) (Fig. 3B). The same results were observed when the cells were grown and induced with 1 μg/ml ampicillin (data not shown). Furthermore, and in close correlation with the transcription data, a colorimetric β-lactamase assay revealed barely detectable activity in 2884D (grown in the presence of oxacillin) compared with that observed for strain 2884 induced with oxacillin (i.e., 0.060 ± 0.003 and 0.360 ± 0.018, respectively). The altered expression of β-lactamase observed in 2884D led us to predict the existence of some alteration in either the structure or the function of the BlaI/BlaR1 regulators. Therefore, blaI, blaR1, as well as the promoter operator of blaR1/blaZ in both isolate 2884 and isolate 2884D were amplified and sequenced. The corresponding set of primers was designed on the basis of the published sequences of the S. aureus N315 plasmid pN315 (GenBank accession no. NC_003140). The sequences of the blaR1/blaZ promoter operator in both 2884 and 2884D were identical, although the sequences of both strains differed from the sequence of wild-type S. aureus N315 plasmid pN315 at 5 nucleotides (data not shown). Similarly, analysis of the blaI sequences of 2884 and 2884D showed that they were identical and differed from the sequence of the wild-type strain at only one amino acid (T2A). On the other hand, analysis of the sensor transducer BlaR1 revealed important changes (Fig. 4). Both parental strain 2884 and oxacillin-dependent strain 2884D harbored the same 7 amino acid substitutions compared with the sequence of S. aureus N315 plasmid pN315. However, 2884D displayed the insertion of an extra T at nucleotide position 453 (ACG→ATC; Thr151Ile), causing a frame shift and the introduction of a premature stop codon at amino acid position 156 (Fig. 4).
FIG. 4.
Alignment of BlaR1 sensor transducer protein amino acid sequences. The differences in amino acids between parental strain 2884, oxacillin-dependent strain 2884D, and wild-type S.aureus/N315 plasmid pN315 (GenBank accession no. NC_003140) are highlighted in boldface. The premature stop codon in 2884D is represented by an asterisk.
Transcription analysis of blaI/blaR1 regulators by real-time RT-PCR revealed undetectable levels of blaR1 in strain 2884D compared to those determined in strain 2884 induced with oxacillin (0.05 ± 0.002 and 211.1 ± 15.2 specific BlaR1/16S mRNAs, respectively). Similarly, analysis of the blaI mRNA revealed a pronounced increase in expression levels in 2884 exposed to oxacillin (Fig. 3C), while no changes in expression were determined in 2884D, for which the values remained low under all conditions (Fig. 3C). Together, these results indicate that the penicillinase operon (BlaR1/BlaI-BlaZ) is impaired in 2884D, and given the constitutive expression of mecA, it does not appear to be involved in its regulation.
Transcription analysis of PBP 2.
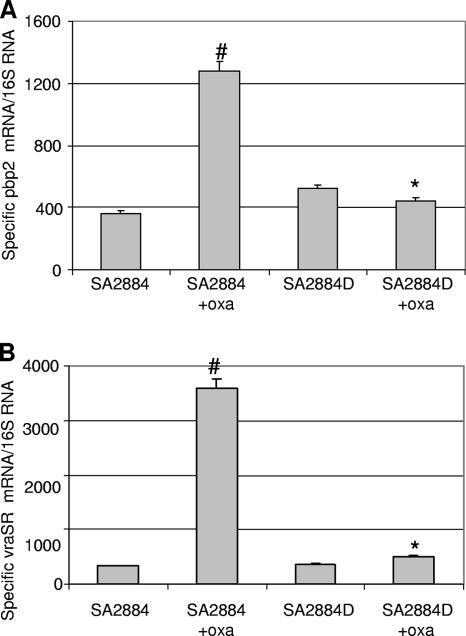
Recent studies have shown that the expression of high-level resistance requires the cooperative functioning of PBP 2a (mecA) and the penicillin-insensitive transglycosylase domain of PBP 2 (28). These observations brought back into focus the possibility that a functional or structural alteration of the native PBP 2 may be linked to the dependence mechanism observed in 2884D. The level of expression of PBP 2 was then determined by real-time RT-PCR with RNAs from both parental strain 2884 (with or without oxacillin) and 2884D (grown in blood agar either without oxacillin or with oxacillin). As shown in Fig. 5A, a 3.5-fold increase in the level of transcription of PBP 2 was observed in oxacillin-induced strain 2884 compared to that observed in the corresponding uninduced strain 2884 (1,278.4 ± 63.9 and 363 ± 18.15, respectively). Surprisingly, independently of the growing conditions, the expression of PBP 2 in oxacillin-dependent strain 2884D was significantly reduced in comparison to that in induced parental 2884 (i.e., 442.25 ± 22.11 and 1278.43 ± 6.39, respectively).
FIG. 5.
(A) Analysis of pbp2 mRNA expression by TaqMan real-time RT-PCR. Relative values of the amount of pbp2 mRNA compared with the amount of 16S mRNA are shown on the vertical axis. #, the transcription of pbp2 with oxacillin induction is significantly greater than that seen in the absence of oxacillin (SA2884+oxa and SA2884, respectively; P < 0.001); *, the transcription of pbp2 is significantly lower in strain 2884D induced with oxacillin than in strain 2884 induced with oxacillin (SA2884D+oxa and SA2884+oxa, respectively; P < 0.001). (B) Quantitation of vraSR mRNA by TaqMan real-time RT-PCR in S. aureus isolates. Relative values of mecA mRNA over 16S mRNA are shown on the vertical axis. #, the transcription of vraSR is significantly in the presence of oxacillin greater than in the absence of oxacillin (2884+oxa and 2884, respectively; P < 0.001); *, the transcription of vraSR is significantly lower in 2884D induced with oxacillin than in 2884 induced with oxacillin (2884D+oxa and 2884+oxa, respectively; P < 0.001).
In order to determine the mechanism associated with the lack of oxacillin-mediated PBP 2 induction in 2884D, we sequenced a 3.696-kb fragment that included the full-length sequence of PBP 2 and the upstream region, i.e., the P1-P2 promoters and the prfa gene. The corresponding set of primers was designed on the basis of the published sequence of S. aureus N315 (GenBank accession no. NC_002745). Alignment of the amino acid sequences of 2884 and 2884D and the amino acid sequence of wild-type strain S. aureus N315 showed a point mutation located at the transglycosylase domain of PBP 2 (nucleotide 588; Cys197Tyr). Since this mutation was present in both 2884 and 2884D, we inferred that these changes could not account for the differences in the levels of expression of PBP 2 observed.
Transcription of VraSR two-component regulatory system.
The transcription of PBP 2 as well as that of other cell wall biosynthesis genes is under the control of a β-lactam-inducible two-component regulatory system (VraSR) consisting of a sensor kinase (VraS) and a response regulator (VraR) (21). We examined the regulation of PBP 2 by the VraSR two-component regulatory system by transcriptional analysis performed with the same RNAs described above, i.e., RNAs from 2884 (grown with or without oxacillin at 0.5 μg/ml) and 2884D (grown in blood agar either without oxacillin or in the presence of 0.5 μg/ml oxacillin). As shown in Fig. 5B, the level of expression of vraSR was markedly induced (∼10-fold) by oxacillin in parental strain 2884 (i.e., from 328.8 ± 12.9 to 3,579 ± 178.9 in the absence and in the presence of oxacillin, respectively). By contrast, the levels of VraSR remained low in 2884D (i.e., 744.5 ± 32.7 and 3,579 ± 178.9 for 2884D and 2884 grown in the presence of oxacillin, respectively). Thus, these results indicate that the PBP 2 regulator VraSR is not responsive to the induction of oxacillin in 2884D and, therefore, may account for the lack of PBP 2 induction observed in this strain.
Transcriptional profiling between parental and oxacillin-dependent 2884.
To complement our observations on the functional role of the regulatory pathways related to oxacillin resistance or dependence, we compared the gene expression of parental strain 2884 and oxacillin-dependent strain 2884D, both of which were grown in the presence of oxacillin at 0.5 μg/ml. Pairwise comparisons, performed in triplicate, between these two isogenic isolates were made (Table 2). The results are based on a series of statistical analyses (filtering) in which the ratios of Cy3 and Cy5 were converted to log2 values and the cutoff was set at above 1 (present) or below −1 (absent). Table 2 shows the most relevant genes that were found to be differentially expressed between strains 2884 and 2884D. The most significant changes in oxacillin-dependent strain 2884D were those related to the penicillinase operon, which included BlaI/BlaR1-BlaZ. Marked decrease in the levels of expression of genes encoding β-lactamase (blaZ SAP012_N315) and the corresponding repressor (blaI SAP010_N315) were observed. In addition, the two-component sensor histidine kinase VraSR, represented by ORFs SA1667_N315 and 2390_COL, also appeared to be downregulated (Table 2). These results were consistent with the reduction in the levels of expression of blaZ and vraSR, determined by real-time RT-PCR analysis (Fig. 3B and 5B, respectively). Among the genes with increased transcription, mecA was found to be upregulated (SA0038_N315), as determined by real-time RT-PCR analysis (Fig. 3A). Other genes showing significant changes were related to stress and metabolic pathways (Table 2).
TABLE 2.
Differential gene expression in parental S. aureus strain 2884 and oxacillin-dependent S. aureus strain 2884
| ORF | Gene | Product or putative function | Fold change |
|---|---|---|---|
| SA1518_N315 | citZ | SA1518 N315, citrate synthase II | +3.64 |
| SA1557_COL | ccpA | SA1557 COL, catabolite control protein | +4.79 |
| SA2098_N315 | SA2098 N315, “ORFID:SA2098, hypothetical protein, similar to glycerate dehydrogenase” | +6.12 | |
| SAV1002_Mu50 | SAV1002 Mu50, conserved hypothetical protein | +3.34 | |
| SA2740_COL | SA2740 COL, ribosomal protein L34 | +3.99 | |
| SA0229_N315 | SA0229 N315, hypothetical protein, similar to nickel ABC transporter | +9.03 | |
| SA0958_COL | SA0958 COL, general stress protein 13 | +9.60 | |
| SA2150_COL | fmtB | SA2150 COL, FmtB protein | +6.82 |
| SA1585_N315 | SA1585 N315, ORFID:SA1585, proline dehydrohenase homolog | +4.36 | |
| SA1437_COL | SA1437 COL, “cold shock protein, CSD family” | +4.48 | |
| SA0807_N315 | mnhG | SA0807 N315, Na+/H+ antiporter subunit | +9.61 |
| NTL08SA0562 | MRSA 252, putative glycosyltransferase; NTL08SA0562_MRSA252 | +2.99 | |
| SA1941_N315 | dps | 67559 NTORF2007 SA1941 N315, general stress protein 20U | +4.32 |
| SA0038_N315 | mecA | 67921 NTORF0040 SA0038 N315, PBP 2′ | +3.4 |
| SAP012_N315 | blaI | SAP012 N315, penicillinase repressor | −8.88 |
| SAP010_N315 | blaZ | SAP010 N315, β-lactamase; SAP010_N315 | −4.26 |
| SA1667_N315 | vraR | Two-component sensor histidine kinase homolog | −2.71 |
| SA2390_COL | vraR | SA2390 COL, “sensory box histidine kinase, putative” | −2.49 |
| SA0249_N315 | SA0249 N315, cell division and morphogenesis-related protein | −3.20 | |
| SA2273_N315 | SA2273 N315, hypothetical protein | −2.89 | |
| SA2257_N315 | SA2257 N315, conserved hypothetical protein | −3.93 | |
| SA1462_COL | SA1462 COL, thymidylate synthase | −3.36 | |
| SAV1176_Mu50 | SAV1176 Mu50, conserved hypothetical protein | −6.83 | |
| SA0105_N315 | SA0105 N315, hypothetical protein | −3.68 | |
| SA1686_COL | SA1686 COL, histidyl-tRNA synthetase | −3.14 | |
| SA1259_N315 | dfrA | SA1259 N315, dihydrofolate reductase | −4.99 |
| SA1478_COL | 67658 ORF01035 SA1478 COL, alanine dehydrogenase | −20.43 |
DISCUSSION
In the present study we have investigated the phenotype of β-lactam-dependent strain 2884D, a clinical MRSA strain that was obtained by in vitro selection with ceftobiprole, a novel cephalosporin with a broad spectrum of activity against MRSA (8, 9, 13, 17, 19). The phenotypes of antibiotic dependence have previously been described in vancomycin-dependent enterococci (i.e., Enterococcus avium [30, 34], Enterococcus faecalis [37], and Enterococcus faecium [20]). In Staphylococcus, an earlier report described the emergence of a β-lactam-dependent coagulase-negative strain that displayed a phenotype similar to the one that we have described in the present study (39). However, the previous studies did not analyze the potential mechanisms responsible for the antimicrobial dependence. This is the first report describing a MRSA strain in which its growth improved remarkably in the presence of β-lactam antibiotics.
We hypothesized that a gene or group of genes responsive to oxacillin induction should be required for growth in this strain. Therefore, the experiments delineated in this study were designed to investigate the involvement of known oxacillin-dependent elements. We found, first, that mecA was expressed in both parental strain 2884 and oxacillin-dependent strain 2884D and, second, that 2884D was a type I SCCmec strain; i.e., it contained a combination of a type I ccr and a class B mec element (IS1272-ΔmecR1-mecA). On the basis of these structural studies of SCCmec, it was predicted that regulation through MecR1/MecI in 2884D was nonfunctional. MecA induction involves cleavage of the cognate repressor (MecI for MecR1 and BlaI for BlaR1), which is specific for each inducer's homologous repressor (23, 33). In previous studies, we have shown that in some clinical isolates MecI is inactivated or modified by deletion or mutation, leaving mecA regulated solely by the BlaI/BlaR1 system (31, 32). However, in the present case, our results suggested that BlaR1 and BlaI were not involved in the regulation of mecA in 2484D and were active only in parental strain 2884, as demonstrated by the oxacillin-induced expression of blaZ by this strain and the β-lactamase production by this strain. Moreover, our data suggested that the penicillinase operon, including the BlaI repressor, which, as mentioned above, can also regulate MecA expression (1, 41), was not transcriptionally active in 2884D. Therefore, in a context determined by the absence of both the MecI/MecR1 and the BlaI/BlaR1 inhibitory systems, mecA expression becomes constitutively expressed. As was the case in 2884D, unregulated mecA expression has also been observed previously in S. aureus 450 M, in which no mec or bla regulatory sequence (ΔmecI/ΔblaI) is present (31). An interesting feature in 2884D was that increased mecA transcription appeared to be associated with a slight production of BlaZ in the context of low levels of the BlaI repressor. This observation opens the possibility to further studies investigating the role and/or existence of other unknown regulators of the Bla system.
It is well established that the resident PBPs (i.e., PBP 1 to PBP 4) are involved in the assembly of the bacterial cell wall peptidoglycan (5). Once these PBPs are inactivated by β-lactam antibiotics, the strategy of β-lactam resistance requires the addition of the new acquired PBP 2a which has a decreased affinity for β-lactam antibiotics but which can still continue to cross-link the cell wall (15). However, recent observations have demonstrated that inactivation of the transglycosylase domain, but not the transpeptidase domain, of PBP 2 in S. aureus prevents the expression of β-lactam resistance, despite the presence of mecA (28). The finding that PBP 2 activity is needed to express resistance may suggest that PBP 2a has a protective effect on cell wall biosynthesis. In our case, the level of expression of PBP 2 in oxacillin-dependent strain 2884D was significantly decreased compared to that in induced parental strain 2884. Thus, these results raise the possibility that a minimal level of expression of PBP 2 is still required to complement the constitutively expressed PBP 2a.
Previous studies have demonstrated that the two-component regulatory system VraSR controls the induction of PBP 2 by β-lactam antibiotics (14, 21, 40). In fact, disruption of VraSR leads to a reduction in the level of β-lactam resistance of MRSA strains because of an inhibited transcriptional response (14, 21, 40). Our findings showed a dramatic reduction in the level of VraSR induction in oxacillin-dependent strain 2884D, which was grown in the presence of oxacillin, compared to that in oxacillin-induced parental strain 2884. These observations led us to conclude that the VraSR regulator is not responsive to the induction of oxacillin in 2884D, and therefore, we may assume that the lack of PBP 2 induction observed in this strain reflects a failure in its regulation. Our results also point to the fact that since PBP 2 is poorly regulated by the VraSR two-component regulator, in the presence of oxacillin, PBP 2a (mecA) becomes important for survival. In this case, given that mecA is not being regulated through its known regulators (i.e., MecI/MecR1 and BlaI/BlaR1), it is plausible that unregulated MecA production is necessary as a compensatory mechanism for survival. Recently, Boyle-Vavra et al. have proposed that the vraS/vraR regulatory system may play a critical role in the response to β-lactams by MRSA strains (7). Those authors found, similar to our observations for strain 2884D, increased mecA expression in a vraS mutant with SCCmec type IV (i.e., the strain harbored deleted portions of mecI/mecR1). This effect did not appear to be mediated solely by BlaI/BlaR1, which, as they and others have suggested, may support the notion that a factor other than BlaR1 or MecR1 may be involved in the induction of blaZ and mecA, respectively (11).
Finally, the role of genes related to stress response and metabolic pathways whose transcription appeared to be altered in oxacillin-dependant strain 2884D may also be important to compensate for survival. However, the involvement of these genes in the mechanism of dependence in 2884D was at this point beyond of the scope of the present study. In summary, the present study provides evidence of the existence of a particular phenotype of an S. aureus strain dependent on oxacillin for survival.
Acknowledgments
This work was supported by award 1K22-AI057724-01A1 from the National Institute of Allergy and Infectious Diseases (to A.E.R.).
We thank Brigitte Berger-Bachi and Mariana Pinho for helpful discussions and critical reading of the manuscript.
Footnotes
Published ahead of print on 30 April 2007.
REFERENCES
- 1.Archer, G. L., and J. M. Bosilevac. 2001. Signaling antibiotic resistance in staphylococci. Science 2911915-1916. [DOI] [PubMed] [Google Scholar]
- 2.Archer, G. L., D. M. Niemeyer, J. A. Thanassi, and M. J. Pucci. 1994. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob. Agents Chemother. 38447-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer, G. L., J. A. Thanassi, D. M. Niemeyer, and M. J. Pucci. 1996. Characterization of IS1272, an insertion sequence-like element from Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 40924-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger-Bachi, B., and S. Rohrer. 2002. Factors influencing methicillin resistance in staphylococci. Arch. Microbiol. 178165-171. [DOI] [PubMed] [Google Scholar]
- 6.Boyle-Vavra, S., S. Yin, M. Challapalli, and R. S. Daum. 2003. Transcriptional induction of the penicillin-binding protein 2 gene in Staphylococcus aureus by cell wall-active antibiotics oxacillin and vancomycin. Antimicrob. Agents Chemother. 471028-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyle-Vavra, S., S. Yin, and R. S. Daum. 2006. The VraS/VraR two-component regulatory system required for oxacillin resistance in community-acquired methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 262163-171. [DOI] [PubMed] [Google Scholar]
- 8.Bozdogan, B., D. Esel, C. Whitener, F. A. Browne, and P. C. Appelbaum. 2003. Antibacterial susceptibility of a vancomycin-resistant Staphylococcus aureus strain isolated at the Hershey Medical Center. J. Antimicrob. Chemother. 52864-868. [DOI] [PubMed] [Google Scholar]
- 9.Chambers, H. F. 1995. In vitro and in vivo antistaphylococcal activities of L-695,256, a carbapenem with high affinity for the penicillin-binding protein PBP 2a. Antimicrob. Agents Chemother. 39462-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke, S. R., and K. G. Dyke. 2001. Studies of the operator region of the Staphylococcus aureus beta-lactamase operon. J. Antimicrob. Chemother. 47377-389. [DOI] [PubMed] [Google Scholar]
- 11.Cohen, S., and H. M. Sweeney. 1968. Constitutive penicillinase formation in Staphylococcus aureus owing to a mutation unlinked to the penicillinase plasmid. J. Bacteriol. 951368-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comite de l'Antibiogramme de la Societe Francaise. 2003. Comite de l'Antibiogramme de la Societe Francaise de Microbiologie report. Int. J. Antimicrob. Agents 21364-391. [DOI] [PubMed] [Google Scholar]
- 13.Fung-Tomc, J. C., J. Clark, B. Minassian, M. Pucci, Y. H. Tsai, E. Gradelski, L. Lamb, I. Medina, E. Huczko, B. Kolek, S. Chaniewski, C. Ferraro, T. Washo, and D. P. Bonner. 2002. In vitro and in vivo activities of a novel cephalosporin, BMS-247243, against methicillin-resistant and -susceptible staphylococci. Antimicrob. Agents Chemother. 46971-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardete, S., S. W. Wu, S. Gill, and A. Tomasz. 2006. Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob. Agents Chemother. 503424-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartman, B. J., and A. Tomasz. 1984. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158513-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hebeisen, P., I. Heinze-Krauss, P. Angehrn, P. Hohl, M. G. Page, and R. L. Then. 2001. In vitro and in vivo properties of Ro 63-9141, a novel broad-spectrum cephalosporin with activity against methicillin-resistant staphylococci. Antimicrob. Agents Chemother. 45825-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, V., W. J. Brown, and M. J. Rybak. 2004. In vitro activities of a novel cephalosporin, CB-181963 (CAB-175), against methicillin-susceptible or -resistant Staphylococcus aureus and glycopeptide-intermediate susceptible staphylococci. Antimicrob. Agents Chemother. 482719-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 451323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, A. P., M. Warner, M. Carter, and D. M. Livermore. 2002. In vitro activity of cephalosporin RWJ-54428 (MC-02479) against multidrug-resistant gram-positive cocci. Antimicrob. Agents Chemother. 46321-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirkpatrick, B. D., S. M. Harrington, D. Smith, D. Marcellus, C. Miller, J. Dick, L. Karanfil, and T. M. Perl. 1999. An outbreak of vancomycin-dependent Enterococcus faecium in a bone marrow transplant unit. Clin. Infect. Dis. 291268-1273. [DOI] [PubMed] [Google Scholar]
- 21.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49807-821. [DOI] [PubMed] [Google Scholar]
- 22.Matushek, M. G., M. J. Bonten, and M. K. Hayden. 1996. Rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. J. Clin. Microbiol. 342598-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKinney, T. K., V. K. Sharma, W. A. Craig, and G. L. Archer. 2001. Transcription of the gene mediating methicillin resistance in Staphylococcus aureus (mecA) is corepressed but not coinduced by cognate mecA and beta-lactamase regulators. J. Bacteriol. 1836862-6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically, approved standard M7-A4, 4th ed. National Committee for Clinical Laboratory Standards, Villanova, PA.
- 25.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 404289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 462155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinho, M. G., H. de Lencastre, and A. Tomasz. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. USA 9810886-10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinho, M. G., S. R. Filipe, H. de Lencastre, and A. Tomasz. 2001. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP 2) by the drug resistance protein PBP 2A in Staphylococcus aureus. J. Bacteriol. 1836525-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinho, M. G., A. M. Ludovice, S. Wu, and H. de Lencastre. 1997. Massive reduction in methicillin resistance by transposon inactivation of the normal PBP2 in a methicillin-resistant strain of Staphylococcus aureus. Microb. Drug Resist. 3409-413. [DOI] [PubMed] [Google Scholar]
- 30.Rosato, A., J. Pierre, D. Billot-Klein, A. Buu-Hoi, and L. Gutmann. 1995. Inducible and constitutive expression of resistance to glycopeptides and vancomycin dependence in glycopeptide-resistant Enterococcus avium. Antimicrob. Agents Chemother. 39830-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosato, A. E., W. A. Craig, and G. L. Archer. 2003. Quantitation of mecA transcription in oxacillin-resistant Staphylococcus aureus clinical isolates. J. Bacteriol. 1853446-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosato, A. E., B. N. Kreiswirth, W. A. Craig, W. Eisner, M. W. Climo, and G. L. Archer. 2003. mecA-blaZ corepressors in clinical Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 471460-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma, V. K., C. J. Hackbarth, T. M. Dickinson, and G. L. Archer. 1998. Interaction of native and mutant MecI repressors with sequences that regulate mecA, the gene encoding penicillin binding protein 2a in methicillin-resistant staphylococci. J. Bacteriol. 1802160-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sifaoui, F., and L. Gutmann. 1997. Vancomycin dependence in a vanA-producing Enterococcus avium strain with a nonsense mutation in the natural d-Ala-d-Ala ligase gene. Antimicrob. Agents Chemother. 411409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomasz, A., S. Nachman, and H. Leaf. 1991. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob. Agents Chemother. 35124-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Bambeke, F., M. Chauvel, P. E. Reynolds, H. S. Fraimow, and P. Courvalin. 1999. Vancomycin-dependent Enterococcus faecalis clinical isolates and revertant mutants. Antimicrob. Agents Chemother. 4341-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wisplinghoff, H., A. E. Rosato, M. C. Enright, M. Noto, W. Craig, and G. L. Archer. 2003. Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob. Agents Chemother. 473574-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Worthington, T., J. White, P. Lambert, S. Adlakha, and T. Elliott. 1999. Beta-lactam-dependent coagulase-negative staphylococcus associated with urinary-tract infection. Lancet 3541097. [DOI] [PubMed] [Google Scholar]
- 40.Yin, S., R. S. Daum, and S. Boyle-Vavra. 2006. VraSR two-component regulatory system and its role in induction of pbp2 and vraSR expression by cell wall antimicrobials in Staphylococcus aureus. Antimicrob. Agents Chemother. 50336-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, H. Z., C. J. Hackbarth, K. M. Chansky, and H. F. Chambers. 2001. A proteolytic transmembrane signaling pathway and resistance to beta-lactams in staphylococci. Science 2911962-1965. [DOI] [PubMed] [Google Scholar]