Abstract
Plasmid pIP1206 was detected in Escherichia coli strain 1540 during the screening of clinical isolates of Enterobacteriaceae for high-level resistance to aminoglycosides. The sequence of this IncFI conjugative plasmid of ca. 100 kb was partially determined. pIP1206 carried the rmtB gene for a ribosome methyltransferase that was shown to modify the N7 position of nucleotide G1405, located in the A site of 16S rRNA. It also contained the qepA (quinolone efflux pump) gene that encodes a 14-transmembrane-segment putative efflux pump belonging to the major facilitator superfamily of proton-dependent transporters. Disruption of membrane proton potential by the efflux pump inhibitor carbonyl cyanide m-chlorophenylhydrazone in a transconjugant harboring the qepA gene resulted in elevation of norfloxacin accumulation. The transporter conferred resistance to the hydrophilic quinolones norfloxacin and ciprofloxacin.
Despite the development of new β-lactams and fluoroquinolones, aminoglycosides are still extensively used for the treatment of severe infections due to gram-negative bacteria.
Aminoglycosides act by causing translational errors and by inhibiting translocation (4). Their target sites include ribosomal domains in which the accuracy of the codon-anticodon is assessed (20), and in particular, they bind to a highly conserved motif of 16S rRNA (14, 28). Since their introduction into clinical use, five mechanisms of resistance to these drugs have been reported in bacterial human pathogens (12): (i) decreased intracellular accumulation of the antibiotic by alteration of outer membrane permeability, diminished inner membrane transport, or active efflux; (ii) enzymatic modification of the drug, which is the most common; (iii) modification of the target by mutation in ribosomal proteins or in 16S rRNA; (iv) trapping of the drug; and most recently, (v) posttranscriptional methylation of rRNA, using S-adenosyl-methionine as a cofactor (7). The last mechanism confers high-level resistance to all available aminoglycosides used for systemic therapy, except streptomycin. The first gene for this type of resistance, armA, was identified in France located on a self-transferable plasmid (7) and was shown to be part of the composite transposon Tn1548 (8) and to encode an enzyme that methylates the N7 position of nucleotide G1405 in 16S rRNA (10). Reports followed of four methyltransferases: RmtA (27) and RmtB (5), which share 82% identity; RmtC (24), with less than 29% identity with RmtA and RmtB; and, more recently, RmtD (6), sharing 40% and 42% identity with RmtA and RmtB, respectively, but less than 29% identity with RmtC. These methyltransferases are only 29% to 31% identical with ArmA.
Fluoroquinolones, by binding to complexes that form between DNA and type II topoisomerases, gyrase and topoisomerase IV, alter chromosome topology that is essential in replication, transcription, recombination, and DNA repair (9). Four mechanisms of resistance to quinolones have been described. The most common is mutational alteration in the so-called quinolone resistance-determining regions of the drug targets (9). The second is the reduction of fluoroquinolone accumulation by active export of the drugs via chromosomal (18) and, most recently, plasmid-borne (25) efflux pumps. Also recently, two additional plasmid-mediated low-level resistance mechanisms have been reported; the Qnr proteins, which protect type II topoisomerases from quinolones (13, 21), and AAC(6′)-Ib-cr, a variant aminoglycoside acetyltransferase that modifies ciprofloxacin (22).
During the screening of clinical isolates of enterobacteria for high-level resistance to aminoglycosides, Escherichia coli 1540 was found to harbor the IncFI self-transferable plasmid pIP1206 of ca. 100 kb, which was partially sequenced. The plasmid carried the rmtB gene (2), and the methylation position of RmtB on 16S rRNA was determined. pIP1206 was also found to confer resistance to hydrophilic quinolones due to the presence of the quinolone efflux pump (qepA) gene, which encodes a putative drug efflux pump belonging to the major facilitator superfamily (MFS).
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
E. coli strain 1540 was isolated in a Belgian hospital during screening of 15,386 nonreplicate clinical isolates of Enterobacteriaceae for aminoglycoside resistance (2). E. coli C600Rif and TOP10 (Invitrogen, Paisley, United Kingdom) strains were used as recipients for conjugation and transformation, respectively. E. coli AG100A, inactivated in the AcrAB drug efflux pump (16), was used to determine antibiotic susceptibility. Plasmids pCR2.1, pCR-Blunt, and pBAD/His (Invitrogen) were used for cloning of PCR products. The strains were grown in brain heart infusion broth or on agar at 37°C.
Susceptibility testing.
Antibiotic susceptibility was tested by disk diffusion on Mueller-Hinton agar, according to the Comité de l'Antibiogramme de la Société Française de Microbiologie standards (3). MICs of antimicrobial agents and dyes were determined on Mueller-Hinton agar by Etest (AB Biodisk, Combourg, France) or by agar dilution, respectively, with or without the efflux pump inhibitor 1-(1-naphthylmethyl)-piperazine (NMP) (Chess, Mannheim, Germany) or phenyl-arginine-β-naphthylamide (PAβN) (Sigma-Aldrich, Saint Quentin Fallavier, France) with 104 CFU per spot after 24 h of incubation. A fourfold or greater reduction in the MICs after addition of NMP or PAβN at 0.25× MIC was considered significant (17). MICs of NMP and PAβN were also performed by agar dilution.
Accumulation of norfloxacin.
Norfloxacin uptake was monitored by a fluorimetric assay slightly modified from that described previously (15). Cells were grown to an optical density at 600 nm of 0.8, pelleted carefully at 4°C, washed once with 50 mM sodium phosphate buffer (pH 7.0) at 4°C, and resuspended in the same buffer to an A600 of 15. The cells were equilibrated for 10 min at 37°C. Norfloxacin was then added to a final concentration of 10 μg/ml, and 0.5-ml samples were removed at different time intervals. Five minutes after the addition of norfloxacin, the efflux pump inhibitor carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added to the reaction mixture at a final concentration of 100 μM. The samples were immediately diluted in 1 ml of ice-cold sodium phosphate buffer and centrifuged for 5 min at 5,600 × g. The pellet was washed once with 1 ml of ice-cold buffer and resuspended in 1 ml of 0.1 M glycine hydrochloride (pH 3.0) for at least 15 h at room temperature. The samples were then centrifuged twice at 5,600 × g for 10 min. The fluorescence of the supernatant was measured with a Quanta Master C-60 spectrofluorimeter (Photon Technology International, Monmouth Junction, NJ) at excitation and emission wavelengths of 281 and 443 nm, respectively. The concentration of norfloxacin in the supernatant was calculated by comparison with a standard curve for norfloxacin (0.01 to 1 μg/ml) in 0.1 M glycine hydrochloride (pH 3.0). The results were expressed as nanograms of norfloxacin incorporated per milligram (dry weight) of bacteria.
DNA preparation and transformation.
Isolation of total and plasmid DNA was performed as described previously (23). Restriction with endonucleases was done according to the supplier's recommendations. Amplification of DNA was performed in a 4700 thermal cycler (Perkin-Elmer Cetus, Norwalk, CT) with Taq (Q-BIOgene, Inc., Carlsbad, CA) or Pfu (Stratagene, La Jolla, CA) DNA polymerase as recommended by the manufacturers. PCR elongation times and temperatures were adjusted to the expected size of the PCR product and to the nucleotide sequence of the primers, respectively. The amplification products were purified using the QIA-quick PCR purification kit (QIAGEN, Inc., Chatsworth, CA). Detection and identification of the rmtB gene in pIP1206 was done by PCR using previously described primers (26). Conjugation from E. coli 1450 to E. coli C600Rif was performed by the solid mating-out assay with selection on rifampin (250 μg/ml) and amikacin (250 μg/ml).
Overexpression of RmtB.
Two deoxyoligonucleotides, 5′-GGGGTACCCCATGAACATCAACGATGCCCT-3′ and 5′-GGAATTCCTTATCCATTCTTTTTTATCAAG-3′, containing KpnI and EcoRI restrictions sites (underlined), were used to amplify the rmtB gene from pIP1206 with Pfu polymerase. The PCR product was cloned in pCR-Blunt, resequenced, and subcloned under the control of an arabinose promoter in pBAD/His previously digested with the same enzymes, producing pAT790 (pBAD/HisΩrmtB).
Primer extension.
Primer extension was performed on total RNA extracted with the FastRNA ProBlue kit (Q-BIOgene) from an E. coli TOP10 strain carrying pAT790 (pBAD/HisΩ rmtB) or pBAD/His. Borohydride reduction and aniline treatment, which introduce a specific scission at N7-methylated guanine bases, were performed as described previously (28). For the sequencing ladder template, the rrnA gene (GenBank accession number NC_000913) encoding 16S rRNA was amplified from E. coli TOP10 genomic DNA and cloned in pCR2.1 (10).
DNA sequence determination and analysis.
Sequencing of inserts in the recombinant plasmids or of the PCR products was performed with a CEQ 2000 DNA Analysis System automatic sequencer (Beckman Coulter, Palo Alto, CA). The sequence of the pIP1206 plasmid was determined by the shotgun cloning method (MWG Biotech, Champlan, France) and analyzed with the GCG sequence analysis software package (version 10.1; Genetics Computer Group, Madison, WI). BLAST program searches were performed by using the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov). Transmembrane sequences were identified by using the TMHMM program at the ExPASy website (http://www.expasy.ch/).
Nucleotide sequence accession number.
The nucleotide sequence of the qepA gene and of the flanking regions has been deposited in the GenBank data library under accession number EF150886.
RESULTS AND DISCUSSION
Antibiotic resistance of E. coli 1540.
Clinical isolate E. coli 1540, selected for its high-level resistance to 4,6-disubstituted deoxystreptamines (MICs of amikacin, gentamicin, kanamycin, netilmicin, and tobramycin, >512 μg/ml), was found to also be resistant to ampicillin (MIC > 256 μg/ml), cefotaxime (MIC = 32 μg/ml), ciprofloxacin (MICs > 32 μg/ml), chloramphenicol (MIC > 256 μg/ml), tetracycline (MIC > 256 μg/ml), minocycline (MIC = 16 μg/ml), and trimethoprim-sulfamethoxazole (MIC > 320 μg/ml). Resistance to these drugs, except that to cefotaxime, was transferable en bloc by conjugation (2).
Analysis of the DNA content of transconjugant BM4650 indicated the presence of plasmid pIP1206 with a size of ca. 100 kb, as estimated by agarose gel electrophoresis after digestion with EcoRI or HindIII (data not shown). pIP1206 was partially sequenced, and its origin of replication was found to be typical of IncFI plasmids that have a broad range of hosts. The resistance genes blaTEM-1, dfrA17, sul1, tet(A), catI, ant3′′9, and rmtB carried by pIP1206 were closely related in structure to plasmid-borne determinants found commonly in enterobacteria.
Determination of the position methylated by RmtB.
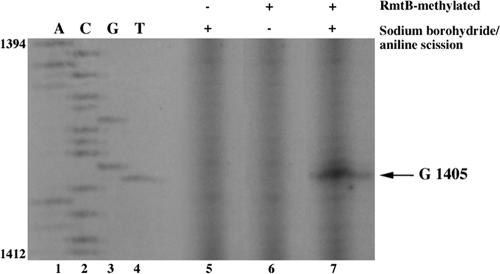
Sequence analysis of RmtB (5) and of the homologous ArmA (45% similarity) protein indicated approximately 43% similarity with 16S rRNA m7G methyltransferases from microorganisms that produce aminoglycosides (7). Methylation at N7 of guanine bases in RNA can be determined by treatment with borohydride and aniline, which creates a specific scission at N7-methylated guanine bases, followed by identification of the cleavage site by primer extension using this template (28).
Total RNA from E. coli TOP10 carrying pAT790 (pBAD/HisΩ rmtB) was analyzed for the presence of N7-methylated guanine. Primer extension of this template terminated at position T1406, indicating N7 methylation at G1405 (Fig. 1). The precise position of methylation at G1405 (E. coli numbering) within the 16S rRNA in the 30S subunit has been determined only for GmrA from Micromonospora purpurea (1) and for ArmA from Klebsiella pneumoniae (10). We thus confirmed identity in the methylation site of methyltransferases isolated from human bacterial pathogens or from aminoglycoside producers. Methylation at this position mediates aminoglycoside resistance by diminishing the affinity of the ribosome for gentamicin (10).
FIG. 1.
Autoradiograph of a primer extension reaction on a methylated 16S rRNA template. Lanes 1 to 4, dideoxy sequencing reactions of pAT787 (10) corresponding to the rrnA gene encoding 16S rRNA. Lanes 5 to 7, primer extension reactions of 16S rRNA templates extracted from wild-type or RmtB-methylated ribosomes, untreated or treated with sodium borohydride and aniline, as indicated. G1405 methylation by RmtB is indicated by primer extension termination at T1406.
Characterization of fluoroquinolone resistance.
The MICs of fluoroquinolones for transconjugant BM4650 (E. coli C600Rif/pIP1206) were determined by Etest (Table 1 and data not shown). A 10-fold or greater increase in the level of resistance to the hydrophilic quinolones, norfloxacin and ciprofloxacin, was observed against BM4650 relative to its plasmid-free counterpart. However, the increase in the MICs of norfloxacin and ciprofloxacin did not lead to a change in clinical categorization. By contrast, the MICs of relatively hydrophilic (pefloxacin, sparfloxacin, gatifloxacin, levofloxacin, and moxifloxacin) and hydrophobic (nalidixic acid) molecules were not significantly altered. In the presence of the NMP or PAβN efflux pump inhibitors, a 10-fold decrease in the level of resistance to norfloxacin and ciprofloxacin was observed (Table 1 and data not shown). These data suggest that efflux was responsible for resistance.
TABLE 1.
MICs of antimicrobial agents and dyes for the strains
| E. coli strain/plasmid | Efflux pump inhibitorb | MIC (μg/ml)a
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| NOR | CIP | PFX | GAT | LVX | MXF | NAL | EBR | ||
| C600Rif | 0 | 0.064 | 0.023 | 0.094 | 0.023 | 0.032 | 0.064 | 2 | 128 |
| +NMP | 0.047 | 0.016 | 0.064 | 0.016 | 0.016 | 0.032 | 1.5 | 64 | |
| +PAβN | 0.032 | 0.012 | 0.047 | 0.016 | 0.012 | 0.032 | 1.5 | 128 | |
| C600Rif/pIP1206 | 0 | 1.5 | 0.25 | 0.094 | 0.032 | 0.032 | 0.094 | 2 | 128 |
| +NMP | 0.064 | 0.016 | 0.064 | 0.016 | 0.016 | 0.032 | 1.5 | 64 | |
| +PAβN | 0.047 | 0.008 | 0.032 | 0.012 | 0.012 | 0.006 | 1.5 | 64 | |
| TOP10 | 0 | 0.032 | 0.003 | 0.023 | 0.003 | 0.006 | 0.004 | 1.5 | 128 |
| +NMP | 0.016 | 0.002 | 0.016 | 0.002 | 0.002 | 0.002 | 1 | 64 | |
| +PAβN | 0.016 | 0.002 | 0.016 | 0.002 | 0.002 | 0.002 | 1 | 64 | |
| TOP10/pIP1206 | 0 | 0.38 | 0.023 | 0.023 | 0.003 | 0.006 | 0.006 | 1.5 | 128 |
| +NMP | 0.016 | 0.003 | 0.016 | 0.002 | 0.002 | 0.002 | 1 | 64 | |
| +PAβN | 0.016 | 0.002 | 0.016 | 0.002 | 0.002 | 0.002 | 1 | 64 | |
| TOP10/pAT851 | 0 | 0.19 | 0.016 | 0.023 | 0.003 | 0.008 | 0.004 | 1.5 | 128 |
| +NMP | 0.016 | 0.002 | 0.016 | 0.002 | 0.002 | 0.002 | 1 | 64 | |
| +PAβN | 0.016 | 0.002 | 0.016 | 0.002 | 0.002 | 0.002 | 1 | 128 | |
CIP, ciprofloxacin; EBR, ethidium bromide; GAT, gatifloxacin; LVX, levofloxacin; MXF, moxifloxacin; NAL, nalidixic acid; NOR, norfloxacin; PFX, pefloxacin. MICs of fluoroquinolones and EBR were determined by E-test and agar dilution, respectively.
NMP used at 40 μg/ml; PAβN used at 50 μg/ml.
Sequence analysis of the fluoroquinolone resistance gene and of the deduced protein.
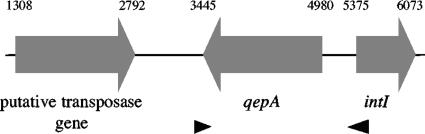
Comparison of the nucleotide sequences obtained after shotgun cloning of pIP1206 DNA with those in the GenBank data library revealed no identity with genes for efflux pumps conferring fluoroquinolone resistance. BLASTX, which allows searching for regions of local similarity between translated query sequences versus protein sequences in the database, resulted in the identification of three open reading frames (ORFs), ORF1 to -3, in a 6,150-bp contig. The deduced sequence of ORF1 was found to be almost identical to that of a transposase-like protein in Salmonella enterica serovar Typhimurium (99% identity over 480 amino acids; GenPept accession number AAK02053). The sequence predicted from ORF2 was homologous to that of proteins belonging to the MFS from Polaromonas sp. (58% identity over 245 amino acids; GenPept accession number YP_548216) and from Nitrosomonas eutropha (55% identity over 230 amino acids; GenPept accession number YP_747938). The translated sequence of ORF3 was identical to that of a class 1 integron integrase in various microorganisms (100% identity over 227 amino acids; GenPept accession number AAK02045) (Fig. 2).
FIG. 2.
Schematic representation of the flanking regions of the qepA gene. The gray arrows indicate the direction of transcription. The arrowheads represent oligodeoxynucleotides MFS8 and MFS9, used to generate pAT851 (pCR2.1ΩqepA). The coordinates correspond to GenBank accession number EF150886.
Within ORF2, a putative GGAAG ribosome binding site (positions 4993 to 4989) was found 8 nucleotides upstream from an ATG initiation codon (position 4980) leading to a 1,536-bp putative coding sequence. The putative fluoroquinolone efflux gene, named qepA for quinolone efflux pump, specified a protein with a calculated mass of 53,080 Da.
The 511-amino-acid deduced QepA protein showed homology with various 14-transmembrane-segment (14-TMS) putative proton-dependent MFS transporters (Table 2 and Fig. 3) (19). These include SgcB from Streptomyces globisporus C-1027 (GenPept accession number AAL06672), Lic from Leptospira interrogans L1-130 (GenPept accession number YP_000642), Nfa from Nocardia farcinica IFM 10152 (GenPept accession number YP_118761), Neut from N. eutropha C91 (GenPept accession number YP_747938), and Bpro from Polaromonas sp. strain JS666 (GenPept accession number YP_548216). The levels of identity of QepA with these proteins varied from 45 to 56%, and the levels of similarity were between 61 and 70% (Table 2). TMS prediction with the TMHMM program indicated that the 14 TMSs of QepA were at positions similar to those in other 14-TMS proton-dependent efflux pumps (19), with the N and C termini located in the cytoplasm. The translocase consensus sequence (motif A), located in the loop between TMS 2 and TMS 3, and the drug extrusion consensus sequence (motif C) at the end of TMS 5 were conserved in all six sequences. Other motifs, in particular, D1, E, F, and H, which are exclusive to the 14-TMS cluster, confirmed that QepA is a new member of this family (Fig. 3) (19).
TABLE 2.
Amino acid identities and similarities between 14-TMS MFS efflux proteins
| Efflux protein (species)a | % Amino acid identity or similarityb
|
|||||
|---|---|---|---|---|---|---|
| QepA | Bpro | Neut | Nfa | Lic | SgcB | |
| QepA (E. coli) | 56.3 | 51.3 | 50.1 | 48.3 | 44.9 | |
| Bpro (Polaromonas sp.) | 70.3 | 54.0 | 52.7 | 60.3 | 43.1 | |
| Neut (N. eutropha) | 65.4 | 67.2 | 44.2 | 47.5 | 39.5 | |
| Nfa (N. farcinica) | 64.2 | 69.1 | 63.1 | 48.8 | 41.9 | |
| Lic (L. interrogans) | 65.6 | 75.6 | 65.9 | 68.3 | 43.5 | |
| SgcB (S. globisporus) | 60.9 | 60.5 | 57.6 | 59.1 | 59.8 | |
The MFS efflux proteins were QepA from E. coli 1450 (this study and GenBank accession number EF150886), Bpro from Polaromonas sp. JS666 (GenPept accession number YP_548216), Neut from N. eutropha C91 (GenPept accession number YP_747938), Nfa from N. farcinica IFM 10152 (GenPept accession number YP_118761), Lic from L. interrogans L1-130 (GenPept accession number YP_000642), SgcB from S. globisporus C-1027 (GenPept accession number AAL06672).
Percent amino acid identity values are presented in the upper half (above the diagonal space), and percent similarity values are given in the lower half (below the diagonal space).
FIG. 3.
Alignment of the QepA protein from E. coli 1450 with the sequences of 14-TMS MFS proteins. SgcB, from S. globisporus C-1027 (GenPept accession number AAL06672); Lic, from L. interrogans L1-130 (GenPept accession number YP_000642); Nfa, from N. farcinica IFM 10152 (GenPept accession number YP_118761); Neut, from N. eutropha C91 (GenPept accession number YP_747938); Bpro, from Polaromonas sp. strain JS666 (GenPept accession number YP_548216); and Qep, from E. coli 1450 (this study and GenBank accession number EF150886). Identical amino acids are indicated by a star, weakly conserved groups by dots, and strongly conserved groups by double dots. The horizontal lines above the alignment indicate the locations of the 14 TMSs (1 to 14). Highly conserved motifs (motifs A, B, C, D1, H, E, and F) are displayed below the alignment: x is any amino acid, a capital letter indicates that the amino acid occurs in >70% of examined sequences, and a lowercase letter indicates that the amino acid occurs in <40% of examined sequences (19).
Resistance conferred by the qepA gene.
A 2,095-bp fragment corresponding to the structural gene for the QepA efflux pump and its flanking regions was amplified by PCR with primers MFS8 (positions 3405 to 3422), 5′-AGCAGCGCGCTGAATCCA-3′, and MFS9 (positions 5499 to 5482), 5′-CGAACCCAGTGGACATAA-3′, and cloned in pCR2.1 to generate pAT851 (pCR2.1ΩqepA). The recombinant plasmid conferred on E. coli TOP10 and AG100A a fivefold or greater increase in resistance to the hydrophilic fluoroquinolones norfloxacin and ciprofloxacin only. In the presence of NMP or PAβN efflux pump inhibitors, the MICs of norfloxacin and ciprofloxacin were reduced at least sixfold (Table 1). The MICs of other antimicrobial agents (chloramphenicol, erythromycin, tetracycline, minocycline, tigecycline, and rifampin) and of dyes (acridine orange, acriflavine, and ethidium bromide) known to be substrates for efflux pumps remained unchanged against the strain harboring pAT851 (pCR2.1ΩqepA) (Table 1 and data not shown). Taken together, these results suggested that QepA acts through drug-specific efflux.
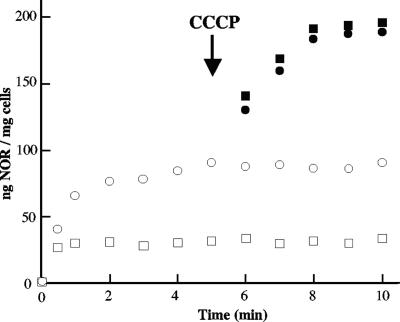
To confirm that the difference in norfloxacin susceptibility between E. coli strains carrying pCR2.1 or pAT851 (pCR2.1Ω qepA) was due to an efflux mechanism, the accumulation of norfloxacin was measured and found to be approximately three times lower in E. coli carrying pAT851 than in the control strain (Fig. 4). Dissipation of the membrane proton motive force by addition of the protonophore CCCP increased norfloxacin accumulation in both strains, resulting in similar concentrations.
FIG. 4.
Accumulation of norfloxacin (NOR) by E. coli cells carrying pCR2.1 (circles) or pAT851 (pCR2.1ΩqepA) (squares), without (open symbols) or with (closed symbols) CCCP. Addition of CCCP (100 μM) is indicated by an arrow.
The 72% overall guanine-plus-cytosine (%GC) content of the qepA gene significantly differed from that of the flanking DNA (51% upstream and 60% downstream, respectively). This observation suggests a recent acquisition by plasmid pIP1206 of a gene from a microorganism with a high %GC content. However, because of a lack of sequences with a high degree of identity to qepA in the data banks, the actual progenitor of the resistance determinant remains unknown. The %GCs of genes for efflux pumps in antibiotic-producing streptomycetes, which have to protect themselves against suicide by the products of their secondary metabolism, range from 64% to 72% (11). This is in contrast with the base composition of the genomes of members of the family Enterobacteriaceae, which is ca. 50%. These observations favor an origin in actinomycetes for qepA, which is rather surprising in view of the fact that quinolones are synthetic antibiotics. It is therefore likely that we did not test the preferred substrate for the QepA pump.
In the conjugative plasmid pIP1206, qepA is flanked by genes known to be associated with the movement of genetic elements and is also physically linked to the rmtB and blaTEM-1 resistance determinants. This genetic organization could further enhance the dissemination of qepA among human pathogens, in particular by coselection by aminoglycosides and β-lactams. In fact, a plasmid-mediated 14-TMS fluoroquinolone efflux pump has been reported recently in an aminoglycoside-resistant E. coli strain in Japan (25).
The qepA gene, together with the qnr family and aac(6′)-Ib-cr, is the third recently detected plasmid-borne determinant of resistance to the fluoroquinolones. These genes confer only low-level resistance, but their presence could potentially facilitate evolution of the bacterial host toward higher levels of resistance by mutational alterations in the target type II topoisomerases.
Acknowledgments
We thank Pierre Bogaerts for the gift of E. coli 1540, Thierry Lambert for drawing our attention to the fluoroquinolone resistance of E. coli BM4650, and Bruno Baron for norfloxacin accumulation experiments.
Footnotes
Published ahead of print on 30 April 2007.
REFERENCES
- 1.Beauclerck, A. A., and E. Cundliffe. 1987. Sites of action of two ribosomal RNA methylases responsible for resistance to aminoglycosides. J. Mol. Biol. 193661-671. [DOI] [PubMed] [Google Scholar]
- 2.Bogaerts, P., M. Galimand, C. Bauraing, A. Deplano, R. Vanhoof, R. De Mendonca, H. Rodriguez-Villalobos, M. Struelens, and Y. Glupczynski. 2007. Emergence of ArmA and RmtB aminoglycoside resistance 16S rRNA methylases in Belgium. J. Antimicrob. Chemother. 59459-464. [DOI] [PubMed] [Google Scholar]
- 3.Comité de l'Antibiogramme de la Société Française de Microbiologie. 2006. Communiqué 2006. http://www.sfm.asso.fr.
- 4.Davies, J., and B. D. Davis. 1968. Misreading of ribonucleic acid code words induced by aminoglycoside antibiotics. The effect of drug concentration. J. Biol. Chem. 2433312-3316. [PubMed] [Google Scholar]
- 5.Doi, Y., K. Yokoyama, K. Yamane, J. Wachino, N. Shibata, T. Yagi, K. Shibayama, H. Kato, and Y. Arakawa. 2004. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob. Agents Chemother. 48491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doi, Y., D. de Oliveira Garcia, J. Adams, and D. L. Paterson. 2007. Coproduction of novel 16S rRNA methylase RmtD and metallo-β-lactamase SPM-1 in a panresistant Pseudomonas aeruginosa isolate from Brazil. Antimicrob. Agents Chemother. 51852-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galimand, M., P. Courvalin, and T. Lambert. 2003. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 472565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galimand, M., S. Sabtcheva, P. Courvalin, and T. Lambert. 2005. Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548. Antimicrob. Agents Chemother. 492949-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawkey, P. M. 2003. Mechanisms of quinolone action and microbial response. J. Antimicrob. Chemother. 51(Suppl. 1)29-35. [DOI] [PubMed] [Google Scholar]
- 10.Liou, G. F., S. Yoshizawa, P. Courvalin, and M. Galimand. 2006. Aminoglycoside resistance by ArmA-mediated ribosomal 16 S methylation in human bacterial pathogens. J. Mol. Biol. 359358-364. [DOI] [PubMed] [Google Scholar]
- 11.Liu, W., and B. Shen. 2000. Genes for production of the enediyne antitumor antibiotic C-1027 in Streptomyces globisporus are clustered with the cagA gene that encodes the C-1027 apoprotein. Antimicrob. Agents Chemother. 44382-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magnet, S., and J. S. Blanchard. 2005. Molecular insights into aminoglycoside action and resistance. Chem. Rev. 105477-497. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Martinez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351797-799. [DOI] [PubMed] [Google Scholar]
- 14.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327389-394. [DOI] [PubMed] [Google Scholar]
- 15.Mortimer, P. G. S., and L. J. V. Piddock. 1991. A comparison of methods used for measuring the accumulation of quinolones by Enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus. J. Antimicrob. Chemother. 28639-653. [DOI] [PubMed] [Google Scholar]
- 16.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panneck, S., P. G. Higgins, P. Steinke, D. Jonas, M. Akova, J. A. Bohnert, H. Seifert, and W. V. Kern. 2006. Multidrug efflux inhibition in Acinetobacter baumannii: comparison between 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-β-naphthylamide. J. Antimicrob. Chemother. 57970-974. [DOI] [PubMed] [Google Scholar]
- 18.Poole, K. 2004. Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 1012-26. [DOI] [PubMed] [Google Scholar]
- 19.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purohit, P., and S. Stern. 1994. Interactions of a small RNA with antibiotic and RNA ligands of the 30S subunit. Nature 370659-662. [DOI] [PubMed] [Google Scholar]
- 21.Robicsek, A., G. A. Jacoby, and D. C. Hooper. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6629-640. [DOI] [PubMed] [Google Scholar]
- 22.Robicsek, A., J. Strahilevitz, G. A. Jacoby, M. Macielag, D. Abbanat, C. H. Park, K. Bush, and D. C. Hooper. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 1283-88. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., and D. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 24.Wachino, J., K. Yamane, K. Shibayama, H. Kurokawa, N. Shibata, S. Suzuki, Y. Doi, K. Kimura, Y. Ike, and Y. Arakawa. 2006. Novel plasmid-mediated 16S rRNA methylase, RmtC, found in a Proteus mirabilis isolate demonstrating extraordinary high-level resistance against various aminoglycosides. Antimicrob. Agents Chemother. 50178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamane, K., J. Wachino, K. Kimura, S. Suzuki, N. Shibata, and Y. Arakawa. 2006. Novel plasmid-mediated fluoroquinolone efflux pump, Qep, identified in Escherichia coli, abstr. C1-586. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 26.Yan, J. J., J. J. Wu, W. C. Ko, S. H. Tsai, C. L. Chuang, H. M. Wu, Y. J. Lu, and J. D. Li. 2004. Plasmid-mediated 16S rRNA methylases conferring high-level aminoglycoside resistance in Escherichia coli and Klebsiella pneumoniae isolates from two Taiwanese hospitals. J. Antimicrob. Chemother. 541007-1012. [DOI] [PubMed] [Google Scholar]
- 27.Yokoyama, K., Y. Doi, K. Yamane, H. Kurokawa, N. Shibata, K. Shibayama, T. Yagi, H. Kato, and Y. Arakawa. 2003. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet 3621888-1893. [DOI] [PubMed] [Google Scholar]
- 28.Yoshizawa, S., D. Fourmy, and J. D. Puglisi. 1998. Structural origins of gentamicin antibiotic action. EMBO J. 176437-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]