Abstract
Microcins are gene-encoded peptide antibiotics produced by enterobacteria that act on strains of gram-negative bacteria. In this work, we concentrated on higher-molecular-mass microcins, i.e., those possessing 60 or more amino acids. They can be subdivided into unmodified and posttranslationally modified peptides. In both cases, they exhibit conserved C-terminal sequences that appear to be characteristic of each subgroup. In the hypothesis that these sequences could correspond to domains, gene fusions between the activity genes for the unmodified microcin colicin V and the modified microcin H47 were constructed. These two microcins differ in their mode of synthesis, uptake, target, and specific immunity. Through this experimental approach, chimeric peptides with exchanged C-terminal sequences were encoded. Cells carrying the fusions in different genetic contexts were then assayed for antibiotic production. Many of them produced antibiotic activities with recombinant properties: the toxicity of one microcin and the mode of uptake of the other. The results led to the identification of a modular structure of colicin V and microcin H47, with the recognition of two domains in their peptide chains: a toxic N-terminal domain and an uptake C-terminal domain. This modular design would be shared by other microcins from each subgroup.
Some enterobacterial strains produce peptide antibiotics called microcins that inhibit the growth of gram-negative bacteria phylogenetically related to the producing strains. Microcins are synthesized as gene-encoded peptides, and many of them undergo subsequent posttranslational modifications to convert into mature molecules. Two main subgroups can be recognized: one comprising modified small peptides with molecular masses below 3.5 kDa (microcins B17, C7, and J25) (11, 22, 26) and one comprising higher-molecular-mass microcins (5 to 9 kDa) (7, 9, 17, 23). The latter subgroup is the subject of the present communication.
Higher-molecular-mass microcins have 60 to 90 amino acid residues. They are ribosomally synthesized as larger precursors with a double-glycine-type motif at their N terminus. This motif is responsible for directing the molecule through an ATP-binding cassette (ABC) secretion apparatus that delivers the antibiotic into the external milieu and concomitantly processes the precursor's N-terminal extension (13). In turn, these microcins can be subdivided into unmodified and modified peptides. Unmodified microcins are peptides that need only be secreted in order to act on sensitive cells. Their genetic clusters, located in plasmids, are very simple. They comprise four genes: the activity gene, encoding the microcin precursor; the immunity gene, which codes for a peptide that specifically protects the producing cell against its cognate microcin; and two genes encoding proteins that integrate the ABC secretion apparatus. Colicin V (ColV) and microcin L (MccL) are examples of unmodified microcins (10, 18). On the other hand, modified microcins of the higher-molecular-mass class are peptides that undergo posttranslational modifications before being secreted. Their precursors follow a similar maturation process by which the peptides would gain a salmochelin attached to their C terminus (17, 23). Salmochelins are mono- or diglucosylated versions of the catechol-type siderophore enterobactin that are produced by Salmonella enterica and by several extraintestinal pathogenic enterobacteria to perform iron-scavenging functions (8, 12). Strains producing these modified microcins synthesize enterobactin, convert it into salmochelins, and employ salmochelins in microcin synthesis (2, 12, 17). Owing to this catechol group, these modified microcins enter the target cells through the pathway employed by the siderophores of the catechol type (17, 24). The genetic clusters for modified microcins are located in the bacterial chromosome and exhibit a complex structure. In addition to the activity, immunity, and secretion genes, they contain four genes devoted to microcin modification, as well as regulatory genes. This group of activities is integrated by microcins H47 (MccH47), I47 (MccI47), E492 (MccE492), and M (MccM) (17). They are referred to below as “catechol microcins.”
Comparison of the amino acid sequences of the precursor peptides of ColV and MccL reveals strong similarity at their C-terminal portions (Fig. 1A). Likewise, the sequences of the precursors of the four catechol microcins can be aligned in a serine-rich region at their C terminus (Fig. 1B). It was thus suspected that such conserved sequences might correspond to a motif involved in a common feature of unmodified microcins on the one hand and of catechol microcins on the other. Considering that the posttranslational modification of catechol microcins would occur at their C terminus, it was thought that their C-terminal sequences could be the target of the modification enzymes. Since the added catechol group is employed as a key to enter target cells through the catechol receptors (Cir, Fiu, and FepA in Escherichia coli K-12), these serine-rich sequences could be at the basis of the mode of uptake of catechol microcins. By extrapolation, a similar assumption was adopted for the conserved C-terminal sequences of ColV and MccL, which, besides being highly identical, both have a disulfide bond between two cysteines (Fig. 1A) (18). As to the uptake pathways followed by these microcins, it is known that ColV employs the receptor Cir (6), while no data are available for MccL. In the context of our hypothesis, the C-terminal sequences would direct these microcins through the Cir receptor to enter E. coli K-12 cells. The possibility that higher-molecular-mass microcins could possess an uptake domain also led us to consider that their toxicity, i.e., the ability to disrupt their targets, could be determined by the remaining peptide sequences.
FIG. 1.
Alignments of the amino acid sequences of microcin precursors. The alignment was performed with Multalin, version 5.4.1 (5). (A) Precursors of ColV and MccL, with a disulfide bond at the C-terminal region; (B) precursors of catechol microcins. Identical or conserved residues are boxed. Lines indicate the secretion domain (double-glycine signal peptide) and the putative uptake domain, with two possible extensions in panel B. Arrows point to the site of processing during secretion.
In this work, evidence of the modular structures of ColV and MccH47 is provided by the construction of fusions between their activity genes. Chimeric peptides with antibiotic activity were produced, and a correlation between their C-terminal portions and their mode of uptake was observed. In addition, these C-terminal sequences proved to be unrelated to toxicity, thus revealing the existence of a toxic domain in the N-terminal sequences of ColV and MccH47.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. coli K-12 strains used in this study are listed in Table 1. Luria-Bertani rich medium and M63 minimal medium supplemented with glucose were used (16). Antibiotics were added to media at the following final concentrations: ampicillin, 100 μg/ml; chloramphenicol, 60 μg/ml. Constructions carrying gene fusions were introduced into RYC1000 cells. This was also the context of the control strains producing MccH47 and ColV. The indicator strains were MC4100 and derivatives bearing different mutations or plasmids. The cir mutant strain FGB202 was constructed by mating pop3000 with FGB1870 (cir fepA::Tn10), and selecting fepA+ cir recombinants on minimal plates supplemented with 200 μM 2,2′-dipyridyl (to create iron deprivation conditions), and with an extract of ColV (to keep the cir mutation). Plasmid pUY271, carrying the cvi and cvaC genes from the ColV system, was obtained by performing EcoRV deletion on pUY270 (1). Plasmid pUY272 carries a DNA segment containing the cvi gene, which was amplified using primers V-L (Table 2) and V-I (5′-CACCACCAGAAACAGAA-3′). This segment was cloned into the HincII site of the pUC13 vector, with cvi expressed opposite to the lac promoter.
TABLE 1.
E. coli K-12 strains
| Strain | Relevant genotype or phenotype | Source |
|---|---|---|
| RYC1000 | araD gyrA lacU169 rbs relA rpsL thiA | Laboratory collection |
| MC4100 | araD lacU169 relA rpsL thiA | Laboratory collection |
| FGB103 | MC4100 Δ(ompT-fepC)267 (Ent−) | 17 |
| FGB1876 | MC4100 rpsL+cir fepA::Tn10 fiu::Mud1X | 17 |
| FGB1870 | MC4100 rpsL+cir fepA::Tn10 | 17 |
| FGB1728 | MC4100 rpsL+cir fiu::Mud1X | 17 |
| FGB1594 | MC4100 rpsL+fiu::Mud1X | 17 |
| FGB102 | MC4100 fepA::Tn10 | 2 |
| FGB202 | MC4100 rpsL+cir | This work |
| FGB099 | MC4100 atpB::Tn599 | 20 |
| pop3000 | HfrH | Laboratory collection |
TABLE 2.
Primers used for building constructions carrying the gene fusions
| Name | Sequence 5′ to 3′ |
|---|---|
| For constructions with cvaC-mchB fusionsa | |
| V-L | CCTCCTACCCTTCACTC |
| V-R103 | CAATTGCTAAACAAACATCACTAA |
| V-R90 | CAATTGCCAATCTTCCCGCAGCAT |
| V-R71 | CAATTGTTCCCCCTAAACC |
| H-L19 | CAATTGGCTCGACCGTG |
| H-L11 | CAATTGGTAGTGCCAGTTCTT |
| H-R | TAAATCATATCTTCATCAGT |
| For constructions with mchB-cvaC fusionsb | |
| H-L | ATGGACAATATGACACTT |
| H-R75 | CTCGAGCTACCGCCACCAGCAGA |
| H-R66 | CTCGAGCCACTTCCCACGGTC |
| V-L32 | CTCGAGCAAGCAAAAACCCGAA |
| V-R | GAGGAATTACAAGCGTATGAGG |
The underlined sequence is the MfeI site.
The underlined sequence is the XhoI site.
Construction of gene fusions.
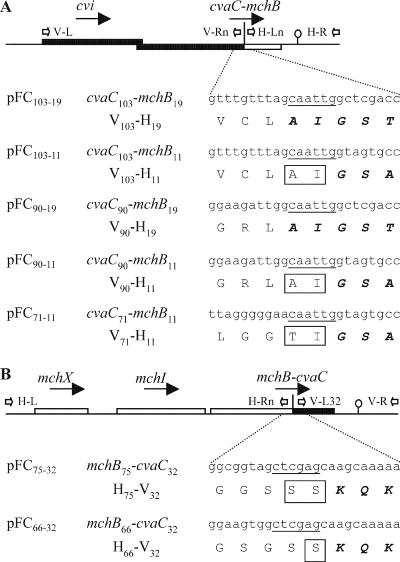
Fusions between the activity genes for ColV (cvaC) and MccH47 (mchB) were constructed. The building blocks were PCR-amplified DNA segments containing the required portions from the ColV and MccH47 genetic clusters (Fig. 2). The primers used are listed in Table 2. For the cvaC-mchB fusions, (i) three cvaC portions were synthesized with primer V-L on the left and, on the right, V-R103 for cvaC103, V-R90 for cvaC90, and V-R71 for cvaC71, and (ii) two mchB portions were amplified with H-R on the right and, on the left, H-L19 for mchB19 and H-L11 for mchB11. For the mchB-cvaC fusions, (i) two mchB portions were synthesized with primer H-L on the left and, on the right, H-R75 for mchB75 and H-R66 for mchB66, and (ii) a cvaC portion was amplified with V-L32 on the left and V-R on the right. For the subsequent joining of the DNA segments in the construction of fusions, primers corresponding to the junction region carried a restriction site: MfeI for cvaC-mchB fusions and XhoI for mchB-cvaC fusions (Table 2; Fig. 2). The amplified DNA segments were digested, ligated in different combinations, and cloned into a polylinker-containing intermediate vector. In these constructions, the expected gene fusions were confirmed by DNA sequencing. Finally, the fusion-bearing DNA segments were subcloned into the pBR322 vector: those carrying cvaC-mchB fusions were subcloned between the AvaI and HindIII sites, and those with mchB-cvaC fusions were subcloned between the SalI and EcoRV sites. The cvaC-mchB fusions were expressed in the AvaI-to-HindIII direction and the mchB-cvaC fusions in the SalI-to-EcoRV direction. The plasmids thus originated were named as shown in Fig. 2.
FIG. 2.
Gene fusions encoding chimeric peptides. Shown are genetic constructions containing the cvaC-mchB fusions, coding for V-H peptides (A), and the mchB-cvaC fusions, coding for H-V peptides (B). The general genetic structure of the constructions is diagramed at the top of each panel. Solid bars, genes from the ColV system; open bars, genes from the MccH47 system; vertical lines, MfeI site in panel A and XhoI site in panel B; stem-loops, transcription terminators; open arrows, primers used for the constructions (where V-Rn is V-R103, V-R90, or V-R71, H-Ln is H-L19 or H-L11, and H-Rn is H-R75 or H-R66). Below each general structure, the DNA sequences of the junction regions are given, with the MfeI or XhoI sites underlined, and with the encoded amino acids. Residues of the N-terminal portions are represented by roman capital letters; residues of the C-terminal portions are boldfaced and italicized; residues added at the junction site are boxed. The names of the plasmids carrying the constructions are given on the left, followed by the names of the gene fusions and their chimeric protein products.
Microcin production and sensitivity assays.
Microcin production was assayed by patch test on minimal M63 glucose plates as previously described (21). The requirements for ColV production were met by the presence of the mchEF secretion genes from the MccH47 system carried by plasmid pMVD41 (17), and the requirements for MccH47 production were met by plasmid pEX100::Tn576, which carries the entire MccH47 genetic system with an insertion mutation in the mchB activity gene (17). The control strains used in the assays were RYC1000(pUY271, pMVD41), which produces ColV, and RYC1000(pEX100), which produces MccH47 (17). To analyze the maturation and secretion of the chimeric peptides in the context of the MccH47 requirements, the fusions were introduced into strains carrying a collection of pEX100-derived plasmids with mutations in the MccH47 genes mchA (pEX100::Tnlac7.2), mchC (pEX100::Tnlac7.7), and mchD (pEX100::Tnlac3.5), all three devoted to MccH47 maturation, and mchE (pEX100::Tnlac7.1), required for MccH47 secretion. None of these plasmids is able to determine MccH47 production (17). Intracellular antibiotic activity was detected by cell lysis as previously described (2).
Several isogenic indicator strains were employed: (i) MC4100, sensitive to ColV and MccH47; (ii) FGB099, an atpB mutant resistant to MccH47; (iii) MC4100(pUY272), immune to ColV; and (iv) MC4100(pUY69), immune to MccH47 (19). Of these, MC4100 was used as the general indicator strain to detect antibiotic production and the three latter strains were used as a set to elucidate the toxic specificities of the chimeric activities, i.e., whether they were of the ColV or of the MccH47 type. To analyze the receptors employed by these activities for their uptake, strains mutated for the different catechol receptors (Cir, Fiu, FepA) were assayed for their sensitivities to the chimeric activities.
RESULTS
Gene fusions encoding chimeric peptides.
The work was performed with the unmodified microcin ColV and the catechol microcin H47. These microcins differ in their mode of synthesis, in their uptake, in their targets. and in their cognate immunity peptides. However, they share the same mode of secretion, since their ABC export systems are quasi-identical. ColV is synthesized as a 103-residue peptide precursor and is then secreted while its N-terminal extension is processed, and the 88-residue mature ColV enters E. coli K-12 cells through the Cir receptor. ColV finally acts by dissipating the membrane potential, and its action is counteracted by the immunity peptide encoded by the cvi gene (6, 10, 25). Genetic analyses indicated that MccH47 is synthesized as a 75-residue peptide precursor that is posttranslationally modified by the acquisition of a salmochelin group and is then secreted and processed, and that mature MccH47, a 60-residue modified peptide, enters cells of E. coli K-12 through any of its three catechol receptors, Cir, Fiu, and FepA. To exert its final action, MccH47 requires the presence of the proton channel of ATP synthase, which was proposed as its target. Specific immunity to MccH47 is conferred by a peptide encoded by the mchI gene (2, 19, 20, 21, 24).
Fusions between the activity genes cvaC and mchB, for ColV and MccH47, respectively, were constructed in order to produce chimeric peptides (Fig. 2). These peptides consisted of an “N-terminal portion,” corresponding to all or part of one of the precursor peptides, followed by a “C-terminal portion,” corresponding to a C-terminal segment from the other peptide. Thus, ColV-H47 (V-H) and H47-ColV (H-V) chimeras were encoded. All the genetic constructions carried, besides the gene fusion, sequences naturally upstream of the gene encoding the N-terminal portion, so that they included the cognate immunity gene. This inclusion would guarantee cell viability in the case of chimeras with toxic activity due to their N-terminal portion. When this portion was encoded by mchB, the constructions also contained the regulatory gene mchX, which is located upstream of the MccH47 immunity and activity genes. It has been proposed that the three genes mchXIB form an operon (19). In addition, all the fusions were followed by sequences naturally downstream of the gene encoding the C-terminal portion, which included a putative transcription terminator.
Taking into account the similarities exhibited by the microcin precursors at their C termini (Fig. 1), the following C-terminal portions were selected: H11 and H19, containing the last 11 and 19 residues of MccH47, respectively, and V32, containing the last 32 residues of ColV. As to the N-terminal portions, two MccH47 segments and three ColV segments were chosen. The MccH47 N-terminal portions were H75, encompassing the entire sequence of the precursor, and H66, lacking most of the C-terminal serine-rich region. The ColV N-terminal portions were V103, corresponding to the entire sequence of the precursor; V90, which lacked the C-terminal segment carrying the two cysteines involved in the disulfide bond; and V71, lacking the last 32 residues. For the gene fusions and their products, the number of codons or amino acids of each portion was indicated, as shown in Fig. 2.
Plasmids carrying the gene fusions were introduced into different E. coli K-12 genetic contexts to analyze whether the chimeric peptides could be converted into secreted molecules with antibiotic activity. The strains constructed were assayed by patch tests to detect growth inhibition halos on the lawn of an indicator strain as described in Materials and Methods. Strains only carrying plasmids with the gene fusions did not generate antibiosis halos (Table 3). Considering that ColV and MccH47 precursors have different requirements for being converted into antibiotics, gene fusions were introduced into cells with the genetic contexts enabling the production of these microcins. To test ColV requirements, cells contained the secretion determinants for the ABC export apparatus, while conditions for MccH47 production were provided by cells carrying the corresponding modification and secretion genes.
TABLE 3.
Antibiotic production of strains encoding the chimeric peptides
| Chimera | Antibiotic productiona in the following genetic context relative to microcin genes:
|
||
|---|---|---|---|
| None | ColV | MccH47 | |
| V103-H19 | − | + | + |
| V103-H11 | − | + | + |
| V90-H19 | − | − | ++ |
| V90-H11 | − | − | ++ |
| V71-H11 | − | − | ++ |
| H75-V32 | − | ++ | ++ |
| H66-V32 | − | ++ | ++ |
Assessed by the production of growth inhibition halos in a patch test. −, no halo; +, minimal halos; ++, halos of about 10 mm in diameter.
Antibiotic activity of chimeric peptides produced in the context for ColV production.
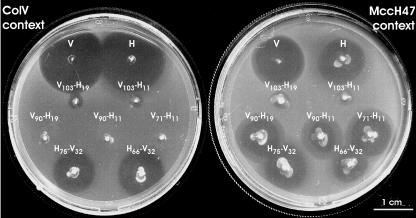
For practical purposes in the experimental design, the secretion genes employed to fulfill the requirements for ColV production were those from the MccH47 genetic system. These genes, mchE and mchF, are almost identical to the ColV secretion determinants, and their products efficiently secreted ColV when put in the presence of plasmid pUY271, which carries the wild-type activity and immunity genes for ColV (Fig. 3). Then gene fusions were introduced into cells carrying plasmid pMVD41, which contains the mchEF genes, and the resulting strains were assayed for antibiotic production (Fig. 3; Table 3). Those with constructions encoding V-H chimeric peptides gave rise to barely detectable inhibition halos (V103-H19 and V103-H11) or generated no halo (V90-H19, V90-H11, and V71-H11). In contrast, cells producing H-V chimeras (H75-V32 and H66-V32) gave rise to conspicuous inhibition zones. In view of the scarcity or absence of antibiotic production by strains coding for the V-H chimeras, intracellular antibiotic activity was searched for as described in Materials and Methods. In these assays no activity was detected (data not shown). Thus, a secretion defect of the chimeric molecules could be ruled out.
FIG. 3.
Antibiotic production of strains carrying gene fusions. Patch tests were performed on the indicator strain MC4100. V, the colicin V-producing control strain RYC1000(pUY271, pMVD41); H, the MccH47-producing control strain RYC1000(pEX100). For stabs of strains carrying gene fusions, RYC1000 derivatives encoding the indicated chimeric peptides in the context for ColV production (pMVD41) and in the context for MccH47 production (pEX100::Tn576) were used.
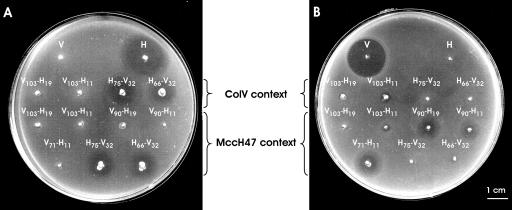
The specificity of the resulting antibiotic activities were assayed on three strains: one immune to ColV, one immune to MccH47, and one resistant to MccH47 (atp mutant) (Fig. 4 and data not shown). Strains coding for V-H peptides did not inhibit the growth of the ColV-immune cells, while halos could be detected on both the MccH47-immune and the MccH47-resistant cells. The opposite result was observed with strains encoding H-V peptides. Therefore, the specific toxicity of each chimeric peptide was conferred by its N-terminal portion, and interestingly, the elimination of the nine C-terminal residues from the MccH47 peptide in H66-V32 did not affect toxicity. It should also be noted that the addition of the 32 C-terminal residues of ColV to the MccH47 N-terminal portions determined the production of antibiotic activity in the sole presence of the secretion genes.
FIG. 4.
Toxic specificity of the chimeric antibiotic activities according to the immunity criterion. Patch tests were performed on the indicator strains MC4100(pUY272), immune to ColV (A), and MC4100(pUY69), immune to MccH47 (B). The stabbed strains are indicated as explained in the legend to Fig. 3, and their genetic contexts are shown.
Antibiotic activity of chimeric peptides produced in the context for MccH47 production.
The requirements for MccH47 production were fulfilled by the presence of plasmid pEX100::Tn576, which carries the MccH47 modification and secretion genes. Although this plasmid also includes the genetic determinants for production of MccI47, this microcin is not produced under the conditions employed in this work (17). In this context, all strains carrying constructions encoding V-H chimeric peptides gave rise to inhibition halos (Fig. 3; Table 3). Chimeras with the V103 N-terminal portion still produced minimal halos, but those with smaller N-terminal portions (V90 and V71) now produced significant inhibition zones. The length of the C-terminal portion (H19 and H11) had little influence on the size of the halos. Cells producing H-V chimeras behaved as they did in the context for the ColV requirements (Fig. 3; Table 3). In all cases, the specificity of the activity was conferred by the N-terminal portion, as deduced from the analysis on the set of immune and resistant strains (Fig. 4 and data not shown). Here again, extracts from strains producing low levels of activity, i.e., those carrying the fusions for V103-H19 and V103-H11, were assayed. No intracellular antibiotic activity was detected (data not shown).
Three of the V-H chimeras showed the most informative results: V90-H19, V90-H11, and V71-H11, which produced no antibiotic activity in the sole presence of the secretion genes, acquired the ability to inhibit the growth of the indicator strain when put in the context for MccH47 production. Their N-terminal portion could be reduced to 90 and even to 71 residues without affecting the toxicity of the chimeras. As to their C-terminal portions, both extensions H19 and H11 were proficient to lead the chimeric peptides through the MccH47 modification pathway.
To corroborate the modification and secretion requirements that were necessary for V90-H19, V90-H11, and V71-H11 to acquire antibiotic activity, the corresponding fusions were introduced into four strains, each carrying a plasmid with the entire MccH47 system but with a mutation in a single gene for microcin maturation (mchA, mchC, or mchD) or microcin secretion (mchE). Thus, the requirement for each of these genes was assessed separately. These strains did not give rise to antibiosis halos, confirming that, to gain antibiotic activity, the chimeric peptides needed the presence of the modification and secretion genes. Considering that these chimeric peptides would be modified like the MccH47 precursor, the synthesis of salmochelins and of their precursor, enterobactin, would also be needed for the modification process. Therefore, the constructs were introduced into a mutant deficient in enterobactin synthesis (FGB103) containing pEX100::Tn576. As expected, no antibiotic production was observed (data not shown).
Similarly, fusions encoding H75-V32 and H66-V32 were introduced into the same genetic contexts, deficient in MccH47 modification or secretion. In this case, mutants in which the MccH47 maturation genes or the enterobactin synthesis pathway was affected still generated antibiosis halos, while the mchE secretion mutant did not. These results revealed that only the secretion functions were required (data not shown).
Receptors employed by the chimeric antibiotic activities.
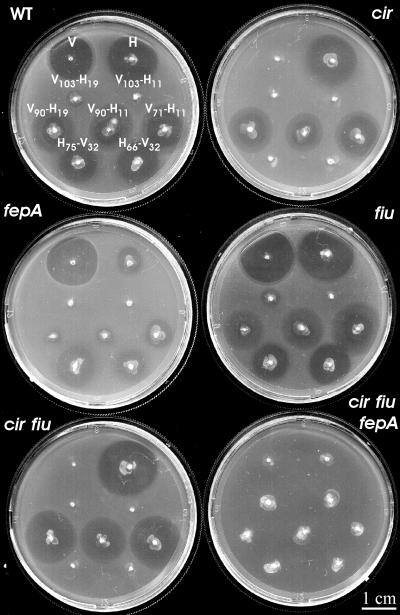
The receptors involved in the uptake of each chimeric activity were analyzed by patch test. A collection of isogenic indicator strains comprising single, double, and triple mutants for the catechol receptors Cir, Fiu, and FepA was employed. Regardless of the genetic context assayed, analysis of the activities derived from V103-H19 and V103-H11 did not lead to conclusive results due to the very small size of the halos, which did not always appear, even on the wild-type indicator strain. For the remaining chimeric activities, a clear pattern of receptor requirements was observed. In sum, chimeric activities derived from the V-H peptides employed any of the three catechol receptors, while those derived from the H-V peptides employed only the Cir receptor (Fig. 5 and data not shown).
FIG. 5.
Sensitivity of catechol receptor mutants to chimeric antibiotic activities. Patch tests were performed on the lawn of strains with mutations in the indicated genes for catechol receptors. In the plate corresponding to the wild-type (WT) indicator strain (MC4100), the stabbed strains are indicated as explained in the legend to Fig. 3. The same order is followed in the remaining plates. The V-H activities were produced in the context for MccH47 production (pEX100::Tn576) and the H-V activities in the context for ColV production (pMVD41).
In principle, it was important to discern whether the activities could possess recombinant properties, i.e., the toxicity of one microcin and the mode of uptake of the other microcin. Therefore, it was of particular interest to elucidate the uptake pathway followed by the chimeric activities produced in the context for the microcin providing the C-terminal portion (Fig. 5). In the context for MccH47 production, the activities derived from V90-H19, V90-H11, and V71-H11 inhibited the growth of all the indicator strains except for the triple mutant, indicating that they employed any of the three catechol receptors. Therefore, these antibiotic activities, although possessing ColV toxicity, behaved like MccH47 in regard to their uptake. In the context for ColV production, cells producing H75-V32 and H66-V32 generated antibiosis halos on every indicator strain provided it bore the wild-type cir allele. Thus, these activities also exhibited recombinant properties: MccH47 toxicity and the ability to follow the ColV uptake pathway.
Finally, the activities derived from H75-V32 and H66-V32 in the context for MccH47 production employed Cir as their sole receptor. In this case, the presence of the MccH47 modification genes did not determine that the chimeras would follow the MccH47 uptake pathway. Therefore, these activities also exhibited recombinant properties: MccH47 toxicity and ColV mode of uptake (data not shown).
DISCUSSION
In this work, microcins ColV and H47 are shown to possess a modular structure in their peptide chains. Although they are clearly different, they share a common general design: a C-terminal sequence confers uptake properties on the molecules, and the remaining N-terminal portion is responsible for the specific antibiotic toxicity.
Fusions between the activity genes for ColV and MccH47 encoded chimeric peptides which, as the natural microcin precursors, could be the starting point for the production of molecules with antibiotic activity. In several cases, this was indeed what occurred when the fusions were introduced in the contexts required to convert the precursors of ColV or of MccH47 into the corresponding microcins. Most of the resulting antibiotic activities exhibited recombinant properties, with the toxic specificity of one microcin and the mode of uptake of the other. Hence, these were chimeric microcins, whose properties revealed the modular structure of ColV and MccH47. As precedents to these findings, colicins and pediocin-like bacteriocins should be mentioned. Colicins are large proteinaceous antibiotics produced by enterobacteria which possess a well-known modular structure, and pediocin-like bacteriocins are peptide antibiotics produced by gram-positive bacteria with a C-terminal domain responsible for their antimicrobial spectrum (4, 15).
The toxic domain of ColV was identified through analysis of the antibiotic activities derived from the ColV-H47 peptides. These activities had the specific toxicity of ColV, as proved by the immunity criterion and by the fact that MccH47-type toxicity could be ruled out. The antibiotic activity of the chimeras did not disappear when their ColV N-terminal portion was reduced from 103 to 90 or 71 residues; on the contrary, it clearly increased. Considering that the 15 N-terminal residues of the ColV precursor are processed during export, the ColV toxic domain must then be contained between residues 16 and 71 of the peptide precursor, i.e., in a segment of 56 amino acids (Fig. 6). Therefore, the 32-residue C-terminal region of ColV is unrelated to toxicity. This was precisely the segment that determined the uptake of H47-ColV chimeric microcins by the single Cir receptor. It is then deduced that ColV has an uptake domain contained in its 32 C-terminal residues (Fig. 6).
FIG. 6.
Modular structure of ColV and MccH47 peptides. Regions containing the toxic domain are indicated by arrows, and those containing the uptake domain are boxed.
The H47-ColV chimeric microcins had the specific toxicity of MccH47, and this property was not affected when the MccH47 N-terminal portion was reduced to 66 residues. Here again, an MccH47 toxic domain, contained between residues 16 and 66 of the peptide precursor, is identified (Fig. 6). Therefore, although MccH47 is a modified microcin, its toxic properties reside exclusively in a segment of its peptide portion, a finding that agrees with previous descriptions of the toxicity of the MccH47 peptide precursor (21). With respect to MccH47 uptake, ColV-H47 chimeras showed that this function was related to the C-terminal portion of this microcin. V90-H19, V90-H11, and V71-H11 acquired antibiotic activity only in the context for MccH47 production; they were modified and secreted as catechol microcins. Concordantly, the resulting chimeric microcins employed the MccH47 uptake pathway. The 11 C-terminal residues from MccH47 proved to be sufficient to accomplish this function, and therefore, this segment must contain the MccH47 uptake domain (Fig. 6). It is then presumed that during MccH47 maturation this domain is modified by the acquisition of a catechol group, and as such, it would be the signal responsible for the interaction with the catechol receptors.
As to the activities derived from the chimeric peptides containing both ColV and MccH47 uptake domains, further aspects could be discussed. The activity corresponding to the H75-V32 peptide was always imported through the Cir receptor, even when produced in the context for MccH47 production. In this context, the chimeric peptide would be in the presence of the MccH47 modification proteins and thus could eventually contain both uptake signals. However, the exclusive use of the Cir receptor indicates the complete predominance of the ColV motif, a fact that could be due to interference exerted by the ColV C-terminal sequence in catechol modification or in the accessibility of the MccH47 signal to the receptors. In the case of the chimeras with a V103 N-terminal portion, the antibiotic activities generated halos of minimal size. Taking into account the fact that no intracellular antibiotic activity was detected in cells carrying the corresponding gene fusions, this result could be explained by the small amounts of active molecules or by difficulties in their uptake. The latter possibility seems more likely: mutual interference between the uptake signals could be the reason for such inefficiency and would also explain the marked increase in the size of the halos produced by the activities derived from chimeras with the V90 and V71 N-terminal portions, which are molecules with a single uptake domain.
In view of the results presented above, it could be suspected that other microcins might also possess a modular structure with nonoverlapping domains for toxicity and uptake. For instance, MccL, which shares the C-terminal sequence with ColV, could be presumed to employ the Cir receptor for its uptake and to contain an N-terminal toxic domain. Likewise, the catechol microcins E492, I47, and M, which bear a C-terminal serine-rich sequence very similar to that of MccH47 and which employ the three E. coli K-12 catechol receptors for their uptake, would also be structured with separate domains for toxicity and uptake. Along these lines, it has recently been reported that the 11 C-terminal residues of the MccE492 precursor could be removed without affecting toxicity. However, the truncated molecule was unable to enter target cells, indicating that the C terminus of the precursor would be involved in MccE492 outer membrane recognition (3).
Although the higher-molecular-mass microcins considered in this work may be modified or unmodified peptides, may employ different uptake pathways, and may affect different targets, they now appear to follow a similar design based on a modular structure. This property makes us wonder about the degree of relatedness existing among them. Moreover, the fact that hybrid microcins could be produced in the laboratory raises the question whether natural events could have generated diverse antibiotic molecules of this type following a combinatorial design. Some clues appear to point in this direction. The amino acid sequences of ColV and the catechol microcin M can be aligned in a 51-residue segment that presents 41% identity (Fig. 7). This segment is located precisely in the 56-residue portion where we found that the ColV toxic domain was contained. In MccM, the aligned sequence lies in the N-terminal portion and does not reach the proposed C-terminal serine-rich uptake domain. Another example arises from the comparison of the sequences of MccE492, a catechol microcin, and Mcc24 (Swiss-Prot accession no. Q46971), an unmodified 74-residue microcin whose C-terminal sequence is different from those of ColV and MccH47. In this case, 55% identity was found in a 67-residue overlap that is located in the putative toxic domain of MccE492 and extends over most of the sequence of Mcc24 (Fig. 7). These observations were further supported by the identities exhibited, in most of their sequence, by the immunity peptides for ColV and MccM (24% in a 67-residue overlap) and for MccE492 and Mcc24 (43% in an 88-residue overlap). All these data suggest that natural events of genetic shuffling could have occurred which gave rise to new microcins with combined domains.
FIG. 7.
Similarities between the amino acid sequences of mature microcins. Identical or conserved residues following the best local alignment with the LALIGN program are highlighted (14). Arrows indicate regions containing the toxic domain, and regions containing the uptake domain are boxed: for ColV, as presented in the text; for microcins M and E492, as deduced from the determinations for MccH47. Regions are not indicated in the Mcc24 sequence.
Acknowledgments
This work was supported by Comisión Sectorial de Investigación Científica and by Programa de Desarrollo de las Ciencias Básicas, Uruguay.
We are indebted to Eliana Rodríguez for critical reading of the manuscript. We are also grateful to María Parente for excellent technical assistance.
Footnotes
Published ahead of print on 23 April 2007.
REFERENCES
- 1.Azpiroz, M. F., E. Rodríguez, and M. Laviña. 2001. The structure, function, and origin of the microcin H47 ATP-binding cassette exporter indicate its relatedness to that of colicin V. Antimicrob. Agents Chemother. 45969-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azpiroz, M. F., and M. Laviña. 2004. Involvement of enterobactin synthesis pathway in production of microcin H47. Antimicrob. Agents Chemother. 481235-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieler, S., F. Silva, C. Soto, and D. Belin. 2006. Bactericidal activity of both secreted and nonsecreted microcin E492 requires the mannose permease. J. Bacteriol. 1887049-7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, V., S. I. Patzer, and K. Hantke. 2002. Ton-dependent colicins and microcins: modular design and evolution. Biochimie 84365-380. [DOI] [PubMed] [Google Scholar]
- 5.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1610881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies, J. K., and P. Reeves. 1975. Genetics of resistance to colicins in Escherichia coli K-12: cross-resistance among colicins of group B. J. Bacteriol. 12396-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fath, M. J., L. H. Zhang, J. Rush, and R. Kolter. 1994. Purification and characterization of colicin V from Escherichia coli culture supernatants. Biochemistry 336911-6917. [DOI] [PubMed] [Google Scholar]
- 8.Fischbach, M. A., L. Hening, D. R. Liu, and C. T. Walsh. 2005. In vitro characterization of IroB, a pathogen-associated C-glycosyltransferase. Proc. Natl. Acad. Sci. USA 102571-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaillard-Gendron, S., D. Vignon, G. Cottenceau, M. Graber, N. Zorn, A. Van Dorsselaer, and A.-M. Pons. 2000. Isolation, purification and partial amino acid sequence of a highly hydrophobic new microcin named microcin L produced by Escherichia coli. FEMS Microbiol. Lett. 193:95-98. [DOI] [PubMed] [Google Scholar]
- 10.Gilson, L., H. K. Mahanty, and R. Kolter. 1990. Genetic analysis of an MDR-like export system: the secretion of colicin V. EMBO J. 93875-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guijarro, J. I., J. E. González-Pastor, F. Baleux, J. L. San Millán, M. A. Castilla, M. Rico, F. Moreno, and M. Delepierre. 1995. Chemical structure and translation inhibition studies of the antibiotic microcin C7. J. Biol. Chem. 27023520-23532. [DOI] [PubMed] [Google Scholar]
- 12.Hantke, K., G. Nicholson, W. Rabsch, and G. Winkelmann. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. USA 1003677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Havarstein, L. S., H. Holo, and I. F. Nes. 1994. The leader peptide of colicin V shares consensus sequences with leader peptides that are common among peptide bacteriocins produced by Gram-positive bacteria. Microbiology 1402383-2389. [DOI] [PubMed] [Google Scholar]
- 14.Huang, X., and W. Miller. 1991. A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 12373-381. [Google Scholar]
- 15.Johnsen, L., G. Fimland, and J. Nissen-Meyer. 2005. The C-terminal domain of pediocin-like antimicrobial peptides (class IIa bacteriocins) is involved in specific recognition of the C-terminal part of cognate immunity proteins and in determining the antimicrobial spectrum. J. Biol. Chem. 2809243-9250. [DOI] [PubMed] [Google Scholar]
- 16.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 17.Poey, M. E., M. F. Azpiroz, and M. Laviña. 2006. Chromosome-encoded microcins: a comparative analysis. Antimicrob. Agents Chemother. 501411-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pons, A.-M., F. Delalande, M. Duarte, S. Benoit, I. Lanneluc, S. Sablé, A. Van Dorsselaer, and G. Cottenceau. 2004. Genetic analysis and complete primary structure of microcin L. Antimicrob. Agents Chemother. 48505-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodríguez, E., and M. Laviña. 1998. Genetic analysis of microcin H47 immunity. Can. J. Microbiol. 44:692-697. [DOI] [PubMed] [Google Scholar]
- 20.Rodríguez, E., and M. Laviña. 2003. The proton channel is the minimal structure of ATP synthase necessary and sufficient for microcin H47 antibiotic action. Antimicrob. Agents Chemother. 47181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodríguez, E., C. Gaggero, and M. Laviña. 1999. The structural gene for microcin H47 encodes a peptide precursor with antibiotic activity. Antimicrob. Agents Chemother. 432176-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosengren, K. J., R. J. Clark, N. L. Daly, U. Göransson, A. Jones, and D. J. Craik. 2003. Microcin J25 has a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone. J. Am. Chem. Soc. 12512464-12474. [DOI] [PubMed] [Google Scholar]
- 23.Thomas, X., D. Destoumieux-Garzon, J. Peduzzi, C. Afonso, A. Blond, N. Birlirakis, C. Goulard, L. Dubost, R. Thai, J. C. Tabet, and S. Rebuffat. 2004. Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J. Biol. Chem. 27928233-28242. [DOI] [PubMed] [Google Scholar]
- 24.Trujillo, M., E. Rodríguez, and M. Laviña. 2001. ATP synthase is necessary for microcin H47 antibiotic action. Antimicrob. Agents Chemother. 453128-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, C. C., and J. Konisky. 1984. Colicin V-treated Escherichia coli does not generate membrane potential. J. Bacteriol. 158757-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yorgey, P., J. Lee, J. Kördel, E. Vivas, P. Warner, D. Jebaratnam, and R. Kolter. 1994. Posttranslational modifications in microcin B17 define an additional class of DNA gyrase inhibitor. Proc. Natl. Acad. Sci. USA 914519-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]