Abstract
Mycolic acids are a key component of the mycobacterial cell wall, providing structure and forming a major permeability barrier. In Mycobacterium tuberculosis mycolic acids are synthesized by type I and type II fatty acid synthases. One of the enzymes of the type II system is encoded by fabG1. We demonstrate here that this gene can be deleted from the M. tuberculosis chromosome only when another functional copy is provided elsewhere, showing that under normal culture conditions fabG1 is essential. FabG1 activity can be replaced by the corresponding enzyme from the closely related species Mycobacterium smegmatis but not by the enzyme from Escherichia coli. M. tuberculosis carrying FabG from M. smegmatis showed no phenotypic changes, and both the mycolic acids and cell wall permeability were unchanged. Thus, M. tuberculosis and M. smegmatis enzymes are interchangeable and do not control the lengths and types of mycolic acids synthesized.
The mycobacterial cell wall is a complex and lipid-rich structure composed of an outer layer of mycolic acids covalently linked to arabinogalactan, which in turn is linked to peptidoglycan. This structure is surrounded by the outermost layer, the “capsule,” in pathogenic species (8). Much work has focused on understanding the role and biosynthesis of the components of the cell wall, particularly the mycolic acids. Current models propose that the mycolic acids form the inner leaflet of an asymmetric bilayer outside the peptidoglycan-arabinogalactan layer, with free lipids forming the outer leaflet (5, 8). Numerous other molecules, including the important immune modulator lipoarabinomannan, are also found in this outer layer. This extremely hydrophobic layer forms an important permeability barrier and is one of the reasons for the intrinsic resistance of mycobacteria to many antibacterial agents.
Mycolic acids are α-alkyl, β-hydroxy fatty acids with two alkyl chains, a “short” chain (C22 to C26) and a long “meromycolate” chain (C>50). Mycobacterium tuberculosis synthesizes three distinct types of mycolic acids, alpha-, methoxy-, and keto-mycolic acids with C24 to C26 and C54 to C63 side chains. Synthesis of mycolic acids involves two types of fatty acid synthase (FAS). FAS I is a multienzyme, single-polypeptide complex which is responsible for de novo synthesis of C22 to C26 fatty acids. FAS II is composed of a number of individual enzymes, including enzymes encoded by inhA and fabG, which elongate the FAS I products to produce the long-chain fatty acids (for a review, see reference 29).
Mycolic acids are of particular interest, since their biosynthesis is the target of antibiotics, such as isoniazid and ethionamide. A number of other compounds, including triclosan and thiolactomycin, affect mycolic acid biosynthesis by inhibiting enzymes of the FAS II pathway. Global gene expression profiling after drug treatment has demonstrated that isoniazid, thiolactomycin, and triclosan have different effects on whole cells and therefore are likely to have different modes of action (2). Triclosan and isoniazid both inhibit InhA (21), whereas thiolactomycin inhibits KasA and KasB (12, 15, 26, 27). There has been much debate concerning the primary target of isoniazid, which is a prodrug that is activated by the mycobacterial KatG catalase (13, 33, 34). However, recent work has demonstrated that a single point mutation in inhA which reduces the binding of the isoniazid-NAD adduct is sufficient to confer isoniazid resistance (31). Interestingly, isoniazid also inhibits FabG1 in vitro (11).
We were interested in the M. tuberculosis inhA operon, which consists of three genes, fabG1 (mabA), inhA, and hemZ, and is cotranscribed as a polycistronic message (1). We have previously demonstrated that hemZ is required for heme biosynthesis and that hemZ mutants are hemin auxotrophs (22). Although intuitively we would expect both inhA and fabG1 to be essential genes, no definitive genetic proof of this has been obtained. The fact that the FAS II system is essential has been demonstrated with Mycobacterium smegmatis, where inactivation of InhA or KasA leads to cell lysis (4, 31). However, the results of saturating transposon mutagenesis in M. tuberculosis suggested that kasA and kasB are essential for growth, while both inhA and fabG1 are not (25). In addition, mycolic acid-defective mutants of M. smegmatis and Mycobacterium bovis BCG have been isolated (3, 18), suggesting that mycolic acids may not be essential for cell viability. In order to determine whether the FAS II complex is essential, we investigated whether one of the key genes, fabG1, which encodes a 3-ketoacyl reductase (1), is essential. We also examined the specificity of homologs of fabG in M. smegmatis and Escherichia coli to determine whether they could complement the function of the M. tuberculosis enzyme.
MATERIALS AND METHODS
Gene replacement in M. tuberculosis.
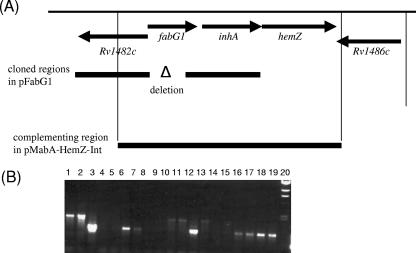
A fabG1 deletion delivery vector was constructed by amplifying the flanking regions of fabG1 using primers fabG1-1 (AAG CTT GTA CTC GGC GAC GAC C) and fabG1-2 (GTT TCC TCC GGT AAC CAG) and primers fabG1-3 (TAT ATC TCC GGT GCG GTC A) and fabG1-4 (AAC AGG TAC CCG CGT CCA ACC GTT CG). Primers fabG1-1 and fabG1-4 were engineered to carry HindIII and KpnI sites (underlined). The products were cloned into the HindIII-KpnI sites of p2NIL (23). The PacI cassette from pGOAL19 (23) carrying hyg, lacZ, and sacB was inserted to obtain the final vector, pFabG1 (data available on request). A two-step recombination method was used to isolate a single-crossover strain (Mess 4) first and double crossovers subsequently (23). Double crossovers were screened by PCR using primers fabG1 for (5′ CGG CCG CGG CGA GAC GAT AG 3′) and fabG1 rev (5′ GGT CGC CGG CAG CCA GTC AGA 3′). The complementing vector pMabA-HemZ-Int (22), carrying the complete fabG1 operon and gentamicin resistance in an L5 integrating vector, was used to generate a merodiploid strain (Mess 10) (Table 1). Double crossovers were isolated from the merodiploid strain screened by PCR as described above and were confirmed by Southern hybridization.
TABLE 1.
M. tuberculosis strains
| Name | Description | Genotype |
|---|---|---|
| H37Rv | Wild-type laboratory strain | fabG1 |
| Mess 4 | Single crossover | fabG1 fabG1Δ kan hyg lacZ sacB |
| Mess 10 | Merodiploid | fabG1 fabG1Δ kan hyg lacZ sacB [fabG1 inhA hemZ gm L5 int] |
| Mess 19 | Delinquent strain | fabG1Δ [fabG1 inhA hemZ gm L5 int] |
| Mess 25 | Switched strain (pDUCK1) | fabG1Δ [fabG1 inhA hemZ kan L5 int] |
| Mess 26 | Switched strain (pDUCK2) | fabG1Δ [fabG1 inhA hemZ hyg L5 int] |
| Mess 29 | Switched strain (pDUCK8) | fabG1Δ [fabG1 hyg L5 int] |
| Mess 27 | Switched strain (M. smegmatis fabG1) | fabG1Δ [fabGMsmgm L5 int] |
| Mess 30 | Cointegrated strain | fabG1Δ [fabG1 inhA hemZ hyg L5 int][fabGEcE233K,A154Tgm L5 int] |
| Mess 31 | Cointegrated strain | fabG1Δ [fabG1 inhA hemZ hyg L5 int][fabGEcA154Tgm L5 int] |
| Mess 32 | Cointegrated strain | fabG1Δ [fabG1 inhA hemZ hyg L5 int][fabGEcE233Kgm L5 int] |
Switching experiments with “delinquent” strain.
Integrating vectors carrying the whole fabG1 operon with different resistance markers were constructed (data available on request). pDUCK1 was constructed by cloning the KpnI-HindIII fragment from pMabA-HemZ-Int into pMV306 (kan) (28), and pDUCK2 was constructed by replacing the gm-int fragment in pMabA-HemZ-Int with the hyg-int HindIII cassette from pUC-Hyg-Int (19). pDUCK8 carrying only fabG1 was constructed by amplifying the M. tuberculosis gene with primers Messy8 (5′ GGT ACC ATG GAA ATC GAC TGG TCA GG 3′) and Messy9 (5′ AAG TCC ATC GAC GAG TCG GTG ATG AT 3′), subcloning the product into pGEM-T Easy, and adding the hyg-int cassette from pUC-Hyg-Int (19) as a HindIII fragment. M. smegmatis fabG with its native promoter was amplified from genomic DNA using primers MAP3 (5′ GGT ACC CCG CGC TTC TGA TCA ACC 3′) and MAP4 (5′ AAG TCC TGA CTC CTT GTG AGC GAG AA 3′), cloned into pGEM-T Easy (Promega), excised as an EcoRI fragment, and cloned into the L5 integrating vector pINT3 (22) to obtain pDUCK9. E. coli fabG alleles were amplified from plasmids pCL33, pCL79, and pCL80 (16) using primers Ec-FabG-for (5′ CCT TAA TTA AGC GCT CGA GCT TTA AAA GAG 3′) and Ec-FabG-rev (5′ CCT TAA TTA ACA ACT AAA TCC CGG CAG GTC T 3′). Products were cloned as PacI fragments (sites in primers underlined) into pAPA3 (a derivative of pINT3 carrying the mycobacterial Ag85a promoter and a single PacI site for cloning inserts) to generate plasmids pDUCK15, pDUCK16, and pDUCK18, respectively.
The switching experiment to replace the integrated pMabA-HemZ-Int vector with various versions of alternative integrating vectors was carried out as previously described (24). Mess 19 (Table 1) was electroporated, and transformants were isolated with the appropriate antibiotic selection (for the incoming vector). Transformants were patch tested for antibiotic resistance as required.
Phenotypic analyses.
Colony morphology was assessed on 7H10-OADC agar plates (19 g liter−1 Middlebrook 7H10, 10% [vol/vol] oleic acid-albumin-dextrose-catalase [OADC] supplement [Becton Dickinson]) without antibiotics. To determine resistance to isoniazid, M. tuberculosis strains were plated onto 7H10-OADC agar plates containing various concentrations of isoniazid (0, 0.01, 0.05, 0.1, 0.5, 1, and 5 μg ml−1), and growth was scored after 7 days. Growth was also monitored in 5 ml of 7H9-OADC-Tw liquid medium (4.7 g liter−1 Middlebrook 7H9, 10% [vol/vol] OADC supplement, 0.05% [wt/vol] Tween 80) containing various concentrations of isoniazid (0, 0.01, 0.05, 0.1, 0.5, 1, and 5 μg ml−1); the cultures were stirred at 250 rpm with an 8-mm flea, and growth was monitored for 21 days. Detergent sensitivity was determined in 7H9-OADC-Tw liquid medium or on 7H10-OADC agar plates. Growth curves were obtained by using 12-mm-diameter borosilicate tubes containing 4 to 5 ml medium that was stirred at 250 rpm with an 8-mm flea.
Mycolic acid analyses.
Cultures were grown to the late log phase in 100 ml of 7H9-OADC medium (containing no Tween 80) and autoclaved. Pellets were recovered and extracted with organic solvents (14). The bacterial residues obtained after lipid extraction were saponified with a mixture of 40% KOH and methoxyethanol (1:7, vol/vol) at 110°C for 3 h in a screw-cap tube (9). After acidification, fatty acids were extracted with diethyl ether and methylated with an ethereal solution of diazomethane. The mycolate patterns of the strains were determined by analytical thin-layer chromatography (TLC) on Silica Gel 60 (Macherey-Nagel) using either eluent A (petroleum ether-diethyl ether, 9:1 [vol/vol]; five runs) or eluent B (dichloromethane). Lipid spots were revealed by spraying the plates with molybdophosphoric acid (10% in ethanol), followed by charring. The crude mycolate fraction was obtained by precipitating an ethereal solution of fatty acid methyl esters with methanol at 4°C, followed by centrifugation at 4,000 × g for 20 min (10). The different classes of mycolates were separated by preparative TLC using eluent A. The lengths of the various types of purified mycolates were determined by matrix-assisted laser desorption ionization—time of flight (MALDI-TOF) mass spectrometry as previously described (17). The spectra were acquired in reflectron mode with an Applied Biosystems 4700 Analyser mass spectrometer (Applied Biosystems, Framingham, MA) equipped with an Nd:YAG laser (wavelength, 355 nm; pulse, <500 ps; repetition rate, 200 Hz). A total of 2,500 shots were accumulated in positive ion mode, and mass spectrometry data were acquired using the instrument default calibration. Mycolate samples were dissolved in chloroform at a concentration of 1 mM and were directly spotted onto the target plate as 0.5-μl droplets, followed by addition of 0.5 μl of matrix solution. Samples were allowed to crystallize at room temperature. The matrix used was 2,5-dihydroxybenzoic acid (10 mg/ml) in CHCl3-CH3OH (1:1, vol/vol).
RESULTS AND DISCUSSION
Essentiality of M. tuberculosis fabG1.
There has been much work on understanding the biosynthesis of mycolic acids because of their key role in the cell wall of mycobacteria and their attractiveness as drug targets. The target of isoniazid, a key antitubercular agent, is mycolic acid biosynthesis (21). The FAS II system is required for the elongation of mycolic acids and thus is predicted to be an essential cellular component. The majority of the individual enzymes which make up the FAS II elongation system have been identified (6, 7), and it has been confirmed that both InhA and KasA are required for cellular survival and integrity of M. smegmatis (4, 31). However, there have been contradictory findings which have raised the possibility that certain enzymes in the FAS II cycle may not be essential. These findings include the prediction based on transposon mutagenesis that inhA and fabG1 are not required for growth in M. tuberculosis (25) and the isolation of mycobacterial mutants defective in mycolic acid synthesis (3, 18). In addition, genes encoding several other potential homologs of FabG (FabG2 to FabG5), which may have overlapping functions, have been identified in the M. tuberculosis genome (7).
In order to determine whether fabG1 is essential in vitro, we used a two-step method to try to generate a deletion strain (23). A suicide delivery vector containing an unmarked in-frame deletion allele of fabG1 was constructed and used to electroporate M. tuberculosis (Fig. 1). Since the deletion allele was unmarked and in frame, polar effects on the expression of inhA and hemZ were not expected. Single-crossover strains were isolated in which the entire vector was incorporated by homologous recombination. One strain (Mess 4) (Table 1) was verified by Southern blotting and was used to isolate double-crossover strains. We screened 40 such strains by PCR to determine if they had the wild-type or deletion allele of fabG1. All of them had the wild-type allele, indicating that we could not isolate a mutant. A merodiploid strain was constructed by transforming the single-crossover strain Mess 4 with the L5 integrating plasmid pMabA-HemZ-Int carrying the M. tuberculosis complete operon (fabG1, inhA, and hemZ) with its native promoter. In this merodiploid background, we were able to isolate both wild-type and deletion double-crossover strains; 10 of 27 strains had the deletion allele (Fig. 2). The fact that a chromosomal deletion could be obtained only in the presence of another functional copy is formal genetic proof that fabG1 is indeed an essential gene under these culture conditions. One “delinquent” strain, Mess 19 (fabG1Δ [fabG1 inhA hemZ gm L5 int]) (Table 1), was analyzed by Southern hybridization and used for further study.
FIG. 1.
Chromosomal arrangement of fabG1 and isolation of mutant strains. (A) Chromosomal arrangement of fabG1 in M. tuberculosis. Regions amplified for the delivery vector and complementing vector are indicated by bars. (B) PCR screening of double crossovers isolated from merodiploid strain Mess 10. Primers fabG1 for and fabG1 rev were used to amplify fabG1 alleles; the expected sizes were 1.5 kb for the wild type and 0.9 kb for the deletion. Lane 1, 100-bp marker; lane 2, lambda HindIII marker; lanes 3 to 18, double crossovers generated from Mess 10; lane 19, Mess 10 (merodiploid); lane 20, Mess 4 (single crossover).
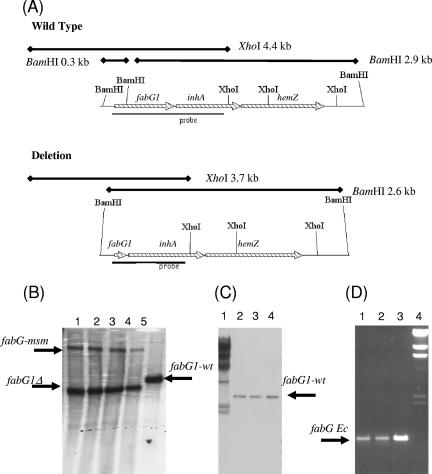
FIG. 2.
Genomic analyses of recombinant strains. (A) Restriction maps of chromosomal wild-type and deletion alleles of fabG1. The expected sizes of the fragments are indicated on the maps along with the positions of the probes used. (B) Switching with M. smegmatis fabG. Genomic DNA was digested with XhoI. Lanes 1 to 4, switched strains carrying M. smegmatis fabG from which M. tuberculosis fabG1 was deleted; lane 5, wild type. fabG-msm, M. smegmatis fabG; fabG1-wt, wild-type fabG1. (C) Switching with E. coli fabG. Genomic DNA was digested with BamHI. Lane 1, lambda HindIII marker; lanes 2 to 4, cointegrated strains Mess 30, Mess 31, and Mess 32 carrying M. tuberculosis wild-type and E. coli fabG genes. (D) PCR amplification of E. coli fabG from cointegrated strains. Lane 1, Mess 30; lane 2, Mess 31; lane 3, Mess 32; lane 4, lambda HindIII marker. fabG Ec, E. coli fabG. The expected product size (0.9 kb) and the product identity were confirmed by sequencing.
Functional complementation by M. smegmatis fabG.
Different bacterial species produce structural variants of the mycolic acids. For example, M. smegmatis produces ethylenic α-, α′-, and epoxy-mycolic acids that are different lengths, whereas M. tuberculosis synthesizes cyclopropanated α-, methoxy-, and keto-mycolic acids. In addition, the oxygenated mycolates are longer in M. tuberculosis than in M. smegmatis (32). It is not known how the FAS II enzymes control chain length, although it is probable that there is some structural limitation to the length of a fatty acid that can be accommodated in the active site of the FAS II enzymes (20). In addition, protein-protein interactions between FabG and other members of the FAS II complex have been demonstrated (30). We wanted to determine whether the FabG enzyme plays a role in determining chain length or mycolic acid type. We first determined whether the M. smegmatis fabG gene could complement the fabG1 defect in M. tuberculosis.
We previously demonstrated that high-efficiency replacement of L5-based integrating vectors occurs in M. tuberculosis (22, 24). We used this “switching” technique to test fabG homologues from other species in order to determine whether they are functional in M. tuberculosis. The basic premise of the method was that we could efficiently replace the resident integrated vector, which carried the only functional copy of fabG1, by transformation and selection for a second version of the integrating vector carrying alternative alleles. If the incoming allele was functional, then we would obtain viable cells; if it was not, then no switched strains would be obtained.
First, we confirmed that the switching method worked efficiently. Three plasmids carrying the complete M. tuberculosis fabG1-hemZ operon but different resistance markers (gm, kan, and hyg) were used. Transformation of the delinquent strain Mess 19 (fabG1Δ [fabG1 inhA hemZ gm L5 int]) with either the pDUCK1 (fabG1 inhA hemZ kan L5 int) or pDUCK2 (fabG1 inhA hemZ hyg L5 int) vector resulted in high-efficiency vector replacement (Table 2). To confirm that vector replacement rather than cointegration had occurred, 24 samples of each transformant were patch tested for gentamicin resistance, and all were gentamicin sensitive, confirming that the original plasmid was lost. One transformant obtained with each switching plasmid was selected for further study. The genotypes of strains Mess 25 (fabG1Δ [fabG1 inhA hemZ kan L5 int]) and Mess 26 (fabG1Δ [fabG1 inhA hemZ hyg L5 int]) were confirmed by Southern hybridization (not shown). We also confirmed that complementation could be performed using the fabG1 gene alone. Transformation of Mess 25 (containing the M. tuberculosis fabG1 [fabG1Mtb] operon) with pDUCK8 (containing only fabG1Mtb) occurred at a high efficiency (Table 2), confirming that only fabG1 was required and that there were no polar effects on inhA or hemZ.
TABLE 2.
Functional complementation of M. tuberculosis with fabG homologs
| Strain | Plasmid | Allele | Antibiotic selection | Transformation efficiencya |
|---|---|---|---|---|
| Mess 19 | pUC-Hyg-Int | None | Hygromycin | 10 |
| Mess 19 | pDUCK1 | M. tuberculosis operon | Kanamycin | 1 × 105 |
| Mess 19 | pDUCK2 | M. tuberculosis operon | Hygromycin | 2 × 106 |
| Mess 25 | pDUCK8 | M. tuberculosis fabG1 | Hygromcyin | 1.7 × 105 |
| Mess 26 | pDUCK9 | M. smegmatis fabG | Gentamicin | 1 × 105 |
| Mess 26 | pDUCK15 | E. coli (Ts) | Gentamicinb | 1 |
| Mess 26 | pDUCK16 | E. coli | Gentamicinb | 1 |
| Mess 26 | pDUCK18 | E. coli (Ts) | Gentamicinb | 2 |
Transformation efficiencies for the plasmids are expressed as the number of transformants per μg of plasmid DNA.
Transformants were isolated at 30°C.
Next, we examined functional complementation by M. smegmatis FabG. M. smegmatis fabG (fabGMsm) was cloned with its own promoter into an integrating vector to obtain plasmid pDUCK9 (fabGMsm gm L5 int) and transformed into strain Mess 26 (fabG1Δ [fabG1 inhA hemZ hyg L5 int]). Gentamicin-resistant transformants were obtained at a high frequency (Table 2) and tested to determine their sensitivities to hygromycin (24 of 24 transformants were Gmr Hygs). PCR and Southern analysis were used to confirm the expected genotypes of transformants carrying the deletion allele of the M. tuberculosis fabG1 gene and a functional copy of M. smegmatis fabG (Fig. 2). The results confirmed that the M. smegmatis gene was able to complement the function of the M. tuberculosis gene.
Lack of functional complementation by E. coli fabG.
Fatty acid biosynthesis is a critical metabolic process in all bacteria. Although E. coli does not produce mycolic acids, other fatty acids are synthesized using a type II FAS system. Hence, homologs of the mycobacterial FAS II enzymes, including FabG, are present in E. coli (Fig. 3). We tested the ability of E. coli fabG (fabGEc) to complement M. tuberculosis fabG1. Vectors carrying fabGEc expressed from a mycobacterial promoter were constructed. We tested three E. coli alleles, one encoding a protein with normal activity (FabGA154T) and two encoding temperature-sensitive derivatives (FabGE233K and FabGE233K,A154T), the latter of which was nonpermissive at 42°C in E. coli (16). We reasoned that if the E. coli allele was functional, we would be able to construct a temperature-sensitive strain of M. tuberculosis. Electroporation of Mess 26 (fabG1Δ [fabG1 inhA hemZ hyg L5 int]) with plasmids pDUCK15, pDUCK16, and pDUCK18 resulted in very low frequencies of transformation when organisms were selected on gentamicin at 30°C (Table 2). This suggested that vector replacement had not occurred. PCR amplification using primers specific for either the M. tuberculosis or E. coli gene (Fig. 3D), followed by sequencing, revealed that in each case the strain had a copy of each gene, indicating that the incoming plasmid had cointegrated into the L5 site (which occurred at a very low frequency). Southern analysis confirmed the presence of a functional fabG1Mtb gene (Fig. 3C). The transformation procedure was repeated, and the same results were obtained. These results indicated that E. coli FabG cannot complement a defect in the fabG1 gene in M. tuberculosis. Since the wild-type gene was nonfunctional, we were not able to isolate temperature-sensitive mutants.
FIG. 3.
Multiple-sequence alignment of fabG alleles. fabG genes from M. tuberculosis H37Rv (Mtb), M. smegmatis (Msm), and E. coli (Eco) were aligned using Clustal (www.ebi.ac.uk).
Phenotypic consequences of FabG replacement.
Although replacement of the native M. tuberculosis fabG1 gene by the M. smegmatis fabG homolog resulted in viable cells, it was still possible that changes in mycolic acid biosynthesis and/or cell wall structure occurred. We investigated a number of phenotypes which might have indicated that there were cell wall changes and the mycolic acid profiles of the recombinant strains.
No differences in colony morphology or sensitivity to the detergent sodium dodecyl sulfate or isoniazid (in liquid media or on solid media) between strains carrying fabGMtb and strains carrying fabGMsm were observed (data not shown). In addition, no differences in the growth rate in liquid medium were observed (Fig. 4). The specificity of FabG in relation to the type, length, and quantity of the mycolic acids synthesized was investigated. Strains carrying either fabG1Mtb or fabGMsm or both fabG1Mtb and fabGEc were grown in medium lacking Tween 80, and total mycolic acid profiles were generated (Fig. 5). TLC analysis of the mycolates from the different strains showed that the same mycolates were produced (Fig. 5A). The three types of mycolates were purified from the various strains, and their lengths were determined by MALDI-TOF mass spectrometry (17). Superimposable mass spectra were obtained for the different α-mycolates (Fig. 5B), methoxy-mycolates (Fig. 5C), and keto-mycolates (Fig. 5D). The pseudomolecular mass (M+Na)+ peaks observed in the spectra corresponded to the peaks determined previously for the mycolates synthesized by the wild-type strain of M. tuberculosis (strain H37Rv) (10). No additional lipid spots or structural changes in the transformants were detected by TLC and MALDI-TOF mass spectrometry (data not shown).
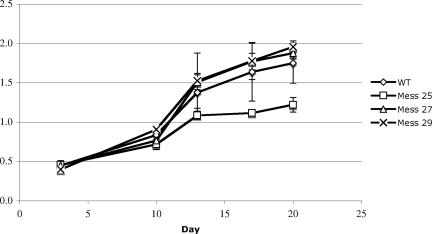
FIG. 4.
Growth of recombinant M. tuberculosis strains: growth of M. tuberculosis wild type (WT), Mess 25 ([fabG1 inhA hemZ]Mtb), Mess 27 (fabGMsm), and Mess 29 (fabGMtb) in liquid culture. The data are averages ± standard deviations of three cultures. Growth curves were constructed three times, and similar results were obtained.
FIG. 5.
Mycolic acid analyses: TLC of mycolates (A) and MALDI-TOF mass spectra of purified α-mycolates (B), methoxy-mycolates (C), and keto-mycolates from different strains of M. tuberculosis expressing FabG1 from either M. tuberculosis (Mtb), M. smegmatis (Msm), or E. coli (Eco). TLC was performed with eluent A (petroleum ether-diethyl ether, 9:1 [vol/vol]; five runs). Lipid spots were revealed by spraying the plates with molybdophosphoric acid (10% in ethanol), followed by charring.
These results indicated that the M. smegmatis FabG protein is functionally equivalent to the M. tuberculosis FabG1 protein and can utilize the endogenous substrates normally. Thus, FabG does not define the length of the fatty acid chains. In addition, there was no interference with FabG1 function in the cells which also contained the E. coli enzyme.
Unusually, the strain carrying the complete M. tuberculosis operon in the integrated vector (Mess 25) grew slower than the strain carrying fabG1 alone (Mess 29) grew (Fig. 4). The reason for the difference is not immediately obvious, although the difference could indicate that the relative amounts of fabG1 and inhA are important; the strain carrying the complete operon had two functional copies of inhA and so would be expected to produce more of this enzyme than the strain carrying fabG1 (containing only one functional copy of the gene). Since the enzymes of the FAS II pathway do interact to form a complex (30), this is a possibility.
Conclusions.
We confirmed that fabG1 is an essential gene using robust genetic methods. This conclusion contrasts with the prediction based on transposon mutagenesis results. However, the transposon method predicts only essentiality and does result in a percentage of false negatives. For example, if a transposon insertion occurred near the 3′ end of a gene, it might still result in partially or fully functional proteins. Our data confirm that fabG1 is essential and that other fabG homologs are not able to complement its function under normal culture conditions. However, at this stage we cannot say whether this is due to a lack of expression of the genes or to a lack of the required enzymatic activity in the FabG proteins. The equivalence of the M. smegmatis homolog was demonstrated by the fact that no changes in viability or mycolic acid structure were observed when the only functional copy in the cell was fabGMsm. In contrast, E. coli fabG was not functional; it is unlikely that this was related to a lack of expression, since we provided a mycobacterial promoter for expression. It is more likely that the structure of the E. coli enzyme is not able to accommodate the long-chain fatty acids or that protein-protein interactions that are necessary for the FAS II system to function are disrupted (20, 30).
Acknowledgments
G.R. was funded by the GlaxoSmithKline Action TB Initiative. The work on mycolic acids was partially funded by grant QLK2-CT-2000-01761 from the European Community.
We thank J. Cronan for providing plasmids pCL33, pCL79, and pCL80.
Published ahead of print on 2 March 2007.
REFERENCES
- 1.Banerjee, A., M. Sugantino, J. C. Sacchettini, and W. R. Jacobs. 1998. The mabA gene from the inhA operon of Mycobacterium tuberculosis encodes a 3-ketoacyl reductase that fails to confer isoniazid resistance. Microbiology 144:2697-2704. [DOI] [PubMed] [Google Scholar]
- 2.Betts, J. C., A. McLaren, M. G. Lennon, F. M. Kelly, P. T. Lukey, S. J. Blakemore, and K. Duncan. 2003. Signature gene expression profiles discriminate between isoniazid-, thiolactomycin-, and triclosan-treated Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47:2903-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhakta, S., G. S. Besra, A. M. Upton, T. Parish, C. Sholto-Douglas-Vernon, K. J. C. Gibson, S. Knutton, S. Gordon, R. P. daSilva, M. C. Anderton, and E. Sim. 2004. Arylamine N-acetyltransferase is required for synthesis of mycolic acids and complex lipids in Mycobacterium bovis BCG and represents a novel drug target. J. Exp. Med. 199:1191-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhatt, A., L. Kremer, A. Z. Dai, J. C. Sacchettini, and W. R. Jacobs. 2005. Conditional depletion of KasA, a key enzyme of mycolic acid biosynthesis, leads to mycobacterial cell lysis. J. Bacteriol. 187:7596-7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennan, P. J. 2003. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis 83:91-97. [DOI] [PubMed] [Google Scholar]
- 6.Camus, J. C., M. J. Pryor, C. Medigue, and S. T. Cole. 2002. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology 148:2967-2973. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Daffe, M., and P. Draper. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39:131-203. [DOI] [PubMed] [Google Scholar]
- 9.Daffe, M., M.-A. Lanéelle, C. Asselineau, V. Lévy-Frébault, and H. L. David. 1983. Intérêt taxonomique des acides gras des Mycobactéries: proposition d'une méthode d'analyse. Ann. Microbiol. Inst. Pasteur 134:241-256. [PubMed] [Google Scholar]
- 10.Daffé, M., M.-A. Lanéelle, and P. L. Valero-Guillen. 1988. Tetraenoic and pentaenoic mycolic acids from Mycobacterium thamnopheos. Structure, taxonomic and biosynthetic implications. Eur. J. Biochem. 177:339-344. [DOI] [PubMed] [Google Scholar]
- 11.Ducasse-Cabanot, S., M. Cohen-Gonsaud, H. Marrakchi, M. Nguyen, D. Zerbib, J. Bernadou, M. Daffe, G. Labesse, and A. Quemard. 2004. In vitro inhibition of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein reductase MabA by isoniazid. Antimicrob. Agents Chemother. 48:242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi, T., O. Yamamoto, H. Sasaki, H. okazaki, and A. Kawaguchi. 1984. Inhibition of fatty acid synthesis by the antibiotic thiolactomycin. J. Antibiot. 37:1456-1461. [DOI] [PubMed] [Google Scholar]
- 13.Heym, B., P. M. Alzari, N. Honore, and S. T. Cole. 1995. Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol. Microbiol. 15:235-245. [DOI] [PubMed] [Google Scholar]
- 14.Jackson, M., C. Raynaud, M. A. Laneelle, C. Guilhot, C. LaurentWinter, D. Ensergueix, B. Gicquel, and M. Daffe. 1999. Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Mol. Microbiol. 31:1573-1587. [DOI] [PubMed] [Google Scholar]
- 15.Kremer, L., J. D. Douglas, A. Baulard, C. Morehouse, M. R. Guy, D. Alland, L. G. Dover, J. H. Lakey, W. R. Jacobs, P. J. Brennan, D. E. Minnikin, and G. S. Besra. 2000. Thiolactomycin and related analogues as novel anti-mycobacterial agents targeting KasA and KasB condensing enzymes in Mycobacterium tuberculosis. J. Biol. Chem. 275:16857-16864. [DOI] [PubMed] [Google Scholar]
- 16.Lai, C.-Y., and J. E. Cronan. 2004. Isolation and characterization of β-ketoacyl-acyl carrier protein reductase (fabG) mutants of Escherichia coli and Salmonella enterica serovar Typhimurium. J. Bacteriol. 186:1869-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laval, F., M. A. Laneelle, C. Deon, B. Monsarrat, and M. Daffe. 2001. Accurate molecular mass determination of mycolic acids by MALDI-TOF mass spectrometry. Anal. Chem. 73:4537-4544. [DOI] [PubMed] [Google Scholar]
- 18.Liu, J., and H. Nikaido. 1999. A mutant of Mycobacterium smegmatis defective in the biosynthesis of mycolic acids accumulates meromycolates. Proc. Natl. Acad. Sci. USA 96:4011-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahenthiralingam, E., B. I. Marklund, L. A. Brooks, D. A. Smith, G. J. Bancroft, and R. W. Stokes. 1998. Site-directed mutagenesis of the 19-kilodalton lipoprotein antigen reveals no essential role for the protein in the growth and virulence of Mycobacterium intracellulare. Infect. Immun. 66:3626-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marrakchi, H., S. Ducasse, G. Labesse, H. Montrozier, E. Margeat, L. Emorine, X. Charpentier, M. Daffe, and A. Quemard. 2002. MabA (FabG1), a Mycobacterium tuberculosis protein involved in the long-chain fatty acid elongation system FAS-II. Microbiology 148:951-960. [DOI] [PubMed] [Google Scholar]
- 21.Parikh, S. L., G. Xiao, and P. J. Tonge. 2000. Inhibition of InhA, the enoyl reductase from Mycobacterium tuberculosis, by triclosan and isoniazid. Biochemistry 39:7645-7650. [DOI] [PubMed] [Google Scholar]
- 22.Parish, T., M. Schaeffer, G. Roberts, and K. Duncan. 2005. HemZ is essential for the growth of Mycobacterium tuberculosis. Tuberculosis 85:197-204. [DOI] [PubMed] [Google Scholar]
- 23.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969-1975. [DOI] [PubMed] [Google Scholar]
- 24.Pashley, C. A., and T. Parish. 2003. Efficient switching of mycobacteriophage L5-based integrating plasmids in Mycobacterium tuberculosis. FEMS Microbiol. Lett. 229:211-215. [DOI] [PubMed] [Google Scholar]
- 25.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 26.Schaeffer, M. L., G. Agnihotri, C. Volker, H. Kallender, P. J. Brennan, and J. T. Lonsdale. 2001. Purification and biochemical characterization of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthases KasA and KasB. J. Biol. Chem. 276:47029-47037. [DOI] [PubMed] [Google Scholar]
- 27.Slayden, R. A., R. E. Lee, J. W. Armour, A. M. Cooper, I. M. Orme, P. J. Brennan, and G. S. Besra. 1996. Antimycobacterial action of thiolactomycin: an inhibitor of fatty acid and mycolic acid synthesis. Antimicrob. Agents Chemother. 40:2813-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stover, C. K., V. F. Delacruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, S. B. Snapper, R. G. Barletta, W. R. Jacobs, and B. R. Bloom. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 29.Takayama, K., C. Wang, and G. S. Besra. 2005. Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 18:81-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veyron-Churlet, R., O. Guerrini, L. Mourey, M. Daffe, and D. Zerbib. 2004. Protein-protein interactions within the fatty acid synthase-II system of Mycobacterium tuberculosis are essential for mycobacterial viability. Mol. Microbiol. 54:1161-1172. [DOI] [PubMed] [Google Scholar]
- 31.Vilcheze, C., H. R. Morbidoni, T. R. Weisbrod, H. Iwamoto, M. Kuo, J. C. Sacchettini, and W. R. Jacobs. 2000. Inactivation of the inhA-encoded fatty acid synthase II (FASII) enoyl-acyl carrier protein reductase induces accumulation of the FASI end products and cell lysis of Mycobacterium smegmatis. J. Bacteriol. 182:4059-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong, M. Y. H., P. A. Steck, and G. R. Gray. 1979. The major mycolic acids of Mycobacterium smegmatis. J. Biol. Chem. 5734-5740. [PubMed]
- 33.Zhang, Y., T. Garbe, and D. Young. 1993. Transformation with katG restores isoniazid-sensitivity in Mycobacterium tuberculosis isolates resistant to a range of drug concentrations. Mol. Microbiol. 8:521-524. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Y., and D. Young. 1994. Strain variation in the katG region of Mycobacterium tuberculosis. Mol. Microbiol. 14:301-308. [DOI] [PubMed] [Google Scholar]