Abstract
Bacterial conjugation is the process by which a single strand of a conjugative plasmid is transferred from donor to recipient. For F plasmid, TraI, a relaxase or nickase, binds a single plasmid DNA strand at its specific origin of transfer (oriT) binding site, sbi, and cleaves at a site called nic. In vitro studies suggest TraI is recruited to sbi by its accessory proteins, TraY and integration host factor (IHF). TraY and IHF bind conserved oriT sites sbyA and ihfA, respectively, and bend DNA. The resulting conformational changes may propagate to nic, generating the single-stranded region that TraI can bind. Previous deletion studies performed by others showed transfer efficiency of a plasmid containing F oriT decreased progressively as increasingly longer segments, ultimately containing both sbyA and ihfA, were deleted. Here we describe our efforts to more precisely define the role of sbyA and ihfA by examining the effects of multiple base substitutions at sbyA and ihfA on binding and plasmid mobilization. While we observed significant decreases in in vitro DNA-binding affinities, we saw little effect on plasmid mobilization even when sbyA and ihfA variants were combined. In contrast, when half or full helical turns were inserted between the relaxosome protein-binding sites, mobilization was dramatically reduced, in some cases below the detectable limit of the assay. These results are consistent with TraY and IHF recognizing sbyA and ihfA with limited sequence specificity and with relaxosome proteins requiring proper spacing and orientation with respect to each other.
Bacterial conjugation is an example of horizontal gene transfer by which a conjugative plasmid in single-stranded form is transmitted from a donor to a recipient (10, 12). Despite the diversity of conjugative plasmids isolated and examined, several general features of the transfer process are conserved among those plasmids that have been studied (45). For instance, a complex of several proteins, often called a relaxosome, forms at the plasmid origin of transfer (oriT) prior to transfer (3, 8, 12, 14, 27, 29, 33). The relaxosome serves at least two functions. One function is to facilitate the activity of the relaxase, a protein that cleaves one plasmid strand in preparation for transfer of that cut strand. The second function is to somehow participate in a sensing mechanism, detecting that the donor and recipient cells have formed a stable mating pair and that DNA transfer should initiate. The identity of the signal that indicates formation of the stable mating pair and the precise molecular effects caused by this signal remain central questions in the field of conjugation.
To better understand the activity and regulation of relaxosomes, we have been examining transfer of F factor or F plasmid. For F plasmid, a much-studied model of conjugative transfer, the relaxosome includes plasmid-encoded TraY and TraI as well as host-encoded integration host factor (IHF) (17). TraY and IHF are double-stranded DNA-binding accessory proteins that bend DNA (25, 31, 32), possibly creating a single-stranded DNA (ssDNA) conformation suitable for TraI binding. TraI, a sequence-specific ssDNA nuclease or relaxase, cleaves a single plasmid strand, which is subsequently transferred to the recipient. In the recipient, the complementary strand is synthesized and the plasmid is circularized (10, 20, 41).
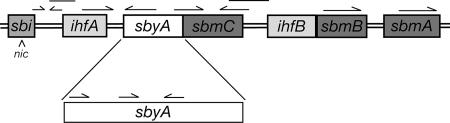
The F relaxosome forms within a region of F oriT (Fig. 1). The F oriT is complex, containing several protein-binding sites (sbi, ihfA, ihfB, sbyA, and sbmC-sbmA) and structural elements (intrinsic bends, direct repeats [DR], inverted repeats [IR], and A-tracts). As shown in Fig. 1, the position of TraI cleavage, nic, is located on the bottom DNA strand, and all protein-binding sites and the majority of the structural elements are located 5′ to nic (to the right as depicted) (7, 12, 19, 26, 38, 39). Sites ihfA and sbyA are positioned closest to nic, and these three protein-binding sites (sbi, ihfA, and sbyA) along with their associated proteins are implicated in the nicking reaction (13, 27). Three TraM-binding sites (sbmC, sbmB, and sbmA) and the lower-affinity IHF site, ihfB, are located beyond sbyA (to the right as shown in Fig. 1). The sbmC site, which is bound with lowest affinity and partially overlaps sbyA (7, 12), is not necessary for nicking but is required for transfer (13). Sites sbmB and sbmA are involved in the regulation of TraM expression (11, 30, 37). Site ihfB is positioned between sbmC and sbmB (12, 39) and is not implicated in nicking (13, 27).
FIG. 1.
Map of F oriT. Protein-binding sites are boxed and labeled. Binding sites bound by the same protein are shaded similarly. nic is indicated by the arrowhead on the bottom strand. IRs and DRs are depicted by arrows above the protein-binding sites, and intrinsic bends are shown as horizontal lines. An expanded view of sbyA is provided to show the IR and DR TraY subsites.
The existence of the ihfA site in F oriT was first established by footprinting and electrophoretic mobility shift assay (EMSA) (39), but the precise positioning is unclear because of the relatively low sequence specificity of IHF. The IHF consensus binding site includes the sequence 5′-WWWCAANNNNTTR-3′ (where W = A/T and R = A/G) and a 5′ A/T-rich region (4, 16, 21). The most likely IHF-binding sequence within ihfA is 5′-AAATAGAGTGTTA-3′ (bp 182 to 194, numbered according to Frost et al. [12]). Participation of this sequence in IHF binding potentially can be confirmed through binding studies with variant sites. Others have shown using other IHF-binding sites that substitutions in the WWWCAA (40) and TTR (4, 40, 43) regions cause the most significant reductions in binding affinity and IHF binding site occupation.
Our earlier footprinting results suggested that the sbyA site consists of three subsites each bound by a TraY monomer (22). Two subsites are arranged as an imperfect IR (Fig. 1) that contains the GA(T/G)A high-affinity subsites to which TraY binds cooperatively. The third subsite, which appears to be arranged as a DR with its adjacent site, is either bound at higher TraY concentrations or bound cooperatively with the TraY molecules at the IR, depending on the DNA construct and experimental method used (22). Our previous work demonstrated that substitutions within the IR yielded the greatest reductions in in vitro binding (22). The addition or subtraction of two base pairs between the sbyA subsites also altered binding affinity (22).
Here we describe our initial examination of the F relaxosome. In this series of experiments, we undertook to determine the importance of the TraY and IHF oriT binding sites and their spacing to plasmid transfer. First, using EMSA we measured TraY and IHF binding affinities for oligonucleotides with multiple substitutions in the high-affinity sbyA IR and throughout ihfA. We confirmed the location and sequence of the IHF-binding site, ihfA, and identified variant sbyA and ihfA sites bound with significantly reduced affinity. We then introduced the same substitutions into plasmids and assayed them for mobilization efficiency. We also inserted half and full helical turns between the relaxosome protein-binding sites and assayed for mobilization efficiency. We found that while base substitutions can significantly reduce in vitro binding, these substitutions have little effect on mobilization. In contrast, the insertions between TraI-, IHF-, and TraY-binding sites dramatically reduce mobilization efficiency.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli strains ER2738 [F′ proA+B+ lacIq Δ(lacZ)M15 zzf::Tn10(Tetr)/fhuA2 glnV Δ(lac-proAB) thi-1 Δ(hsdS-mcrB)5], ER2267 [F′ proA+B+ lacIq Δ(lacZ)M15 zzf::mini-Tn10 (Kanr)/Δ(argF-lacZ)U169 glnV44 e14−(McrA−) rfbD1? recA1 relA1? endA1 spoT1? thi-1 Δ(mcrC-mrr)114::IS10], and TB1 {F− ara Δ(lac-proAB) [φ80dlac Δ(lacZ)M15] rpsL (Strr) thi hsdR}, and plasmids pNEB193 and pACYC177 were obtained from New England BioLabs. E. coli strain XL-1 Blue was purchased from Stratagene. Plasmids pACYC177-ForiT (42) and pNEB193-ForiT (36) were constructed as described elsewhere. The question marks included in the ER2267 genotype indicate genes that ER2267 should contain based on the ancestral strain but that have not been verified by direct testing of ER2267.
Site-directed mutagenesis.
Base substitutions in the F oriT region of pNEB193-ForiT and pACYC177-ForiT and insertions in the F oriT region of pACYC177-ForiT were introduced using a QuikChange kit (Stratagene). After confirming mutations using DNA sequencing, each plasmid variant was transformed into ER2267 and/or ER2738.
Plasmid mobilization assay.
Mobilization assays were performed as described previously (42). Variants of pACYC177-ForiT were tested using F′ strain ER2738 and the recA− F′ strain ER2267 as donors. Variants were also created in pNEB193-ForiT and tested using ER2738 as a donor strain. Overnight cultures of ER2738 with wild-type (WT) or variant pNEB193-ForiT (using pNEB193 as a negative control) or WT or variant pACYC177-ForiT (using pACYC177 as a negative control) were grown in Luria-Burtani (LB) broth containing tetracycline (25 μg/ml) and ampicillin (100 μg/ml) (LB+Tet+Amp). Overnight cultures of ER2267 with WT or variant pACYC177-ForiT constructs, using pACYC177 as a negative control, were grown in LB containing kanamycin (30 μg/ml) and ampicillin (100 μg/ml) (LB+Kan+Amp). To follow plasmid mobilization, serial dilutions of each assay were plated onto LB+Amp to select for donors and LB+Amp+Strep to select for transconjugates. To follow F′ transfer, dilutions were plated onto LB+Tet (ER2738) or LB+Kan (ER2267) to select for donors and on LB+Tet+Strep (ER2738) or LB+Kan+Strep (ER2267) to select for transconjugates. Mobilization efficiencies were calculated as the number of transconjugates per donor. These measurements are presented as an average of 3 to 35 experiments with the standard errors.
Expression and purification of TraY.
TraY was expressed as described previously (34) with minor modifications. Strain BL21(DE3)/pET-21a(+)-traY was grown in 0.5 liters of LB+Amp at 37°C to an optical density at 600 nm of 0.5. Protein expression was induced by adding isopropyl-1-thio-β-d-galactopyranoside to a final concentration of 1 mM. Cells were grown for an additional 3 h at 37°C. Cells were centrifuged at 10,000 × g for 20 min, and pellets were frozen at −80°C. TraY was purified as described previously (23) with some changes. Cell pellets were thawed at 4°C and resuspended in 50 ml of ice-cold buffer A (50 mM Tris [pH 7.5], 1 mM EDTA, and 5 mM β-mercaptoethanol [β-ME]) with 100 mM NaCl. Phenylmethylsulfonyl flouride was added to a final concentration of 200 μM. Cells were lysed by sonication, and membranes were pelleted by centrifugation at 10,000 rpm for 10 min at 4°C. Supernatant was loaded onto a 5-ml HiTrap Q column (GE Healthcare) equilibrated with buffer A plus 100 mM NaCl. A gradient from 100 mM to 2 M NaCl was applied to the column at 5 ml/min over a 100-ml volume. Flowthrough was loaded on a 5-ml HiTrap heparin column (GE Healthcare) equilibrated with buffer B (20 mM NaPO4 [pH 7.4], 1 mM EDTA, and 5 mM β-ME) with 100 mM NaCl. A salt gradient was run from 100 mM to 1 M NaCl. Protein eluting at 500 to 670 nM NaCl was loaded onto a 5-ml HiTrap Blue column (GE Healthcare) equilibrated with buffer B plus 500 mM NaCl, and a 500 mM to 3.8 M salt gradient was applied. Fractions eluting between 1.7 M and 2.8 M NaCl were combined and dialyzed overnight into buffer B with 100 mM NaCl. The dialysate from the Blue column was loaded onto a 5-ml HiTrap SP Sepharose column (GE Healthcare) equilibrated with buffer B plus 100 mM NaCl, and a gradient was run from 100 mM to 1 M NaCl. The single TraY peak that eluted from 450 mM to 630 mM NaCl was mixed 1:1 with 4 M NaCl and concentrated over a 1-ml butyl-Sepharose 4 Fast Flow column (GE Healthcare) equilibrated with buffer B plus 3.8 M NaCl. TraY fractions were pooled and dialyzed in buffer B with 100 mM NaCl. Dialysate was further concentrated with a centrifugal filter device (Millipore). The protein concentration was calculated from the A280 using the extinction coefficient of 11,460 M−1 cm−1. Purity of TraY was determined to be >95% as seen by Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Expression and purification of IHF.
IHF was expressed and purified as previously described (9) with minor modifications. HN880 pPLhip.himA-5 cells were grown in LB with 50 μg/ml ampicillin at 30°C to an optical density at 600 nm of 0.5. Overexpression was induced by shifting the culture temperature to 42°C for 3 h. Cells were pelleted by centrifugation at 3,700 rpm for 30 min at 4°C, and the pellet was frozen at −80°C. The pellet was resuspended in buffer C (25 mM Tris-HCl pH 7.4, 1 mM EDTA, 20 mM NaCl, 3 mM β-ME) at 0.2 ml/g pellet and 5 mM MgCl2 and 1 mM phenylmethylsulfonyl fluoride (final concentrations). Cells were lysed by sonication, incubated on ice for 30 min, and then pelleted by centrifugation at 43,000 × g for 35 min. A two-step ammonium sulfate precipitation was performed. First, 0.33 g ammonium sulfate/ml supernatant was added to the supernatant followed by centrifugation at 43,000 × g for 20 min at 4°C. Second, 0.56 g ammonium sulfate/ml supernatant was added to the supernatant from the first step followed by another centrifugation step. The pellet was resuspended in buffer C (30 ml/500 ml of original culture) and dialyzed against 4 liters of buffer C. The dialysate was filtered through a 0.45-μm filter and was applied to a 5-ml HiTrap heparin column equilibrated with buffer C. A salt gradient was run from 20 mM to 2 M NaCl. IHF fractions were pooled and dialyzed in buffer C with 20 mM NaCl. Dialysate was further concentrated with a centrifugal filter device (Millipore). The protein concentration was calculated from the A280 using the extinction coefficient 5,960 M−1 cm−1. Purity of IHF was determined to be >95% as seen by Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Labeling oligonucleotides.
Single stranded WT and mutant sbyA or ihfA oligonucleotides were 5′ end-labeled with [γ-32P]ATP (GE Healthcare) using T4 polynucleotide kinase (New England BioLabs) according to the manufacturer's instructions. The complementary strand was then mixed 3:1 with the labeled strand, heated to 95°C for 5 min, and slow cooled for 3 h. Unincorporated nucleotides were separated from labeled oligonucleotide using the Sephadex G-50 Fine quick spin columns (Roche).
Electrophoretic mobility shift assays.
EMSAs were performed as described elsewhere (23) with minor modifications. In EMSA buffer, β-ME replaced dithiothreitol and 10 μg/ml replaced 1 μg/ml sonicated calf thymus DNA. Reaction mixtures were incubated for 60 instead of 90 min. The concentration range of γ-32P-end-labeled WT or mutant sbyA oligonucleotide was 42.5 to 755 pM. The same procedure was followed using WT IHF protein and 51.0 to 95.7 pM of WT or mutant ihfA DNA sequence.
Data analysis.
Intensities of radioactive bands for each EMSA were determined using ImageQuant software v.1.2 (Molecular Dynamics) and were used without background correction. At each protein concentration, the fraction bound (θ) was calculated for each bound species. We refer to these values as a partial fraction bound, θx. For example, the EMSA of WT TraY binding to the WT 39-bp sbyA oligonucleotide results in two predominate bound species (see Fig. 2, left upper panel, below). Therefore, a partial fraction bound was calculated for bound species 1, θ1, and bound species 2, θ2. Our interpretation of the data for the 39-bp oligonucleotides (see Results, below) is that bound species 1 represents two molecules of TraY bound, occupying the sbyA IR subsites GA(T/G)A, and bound species 2 consists of three bound TraY molecules occupying both the sbyA IR and an additional, possibly nonspecific site 3′ to the GATA site. This latter site is referred to as the alternative site. (The 39-bp oligonucleotides do not contain an intact TTTC subsite, and TraY therefore will not bind to the sbyA DR.) TraY binding to the 50-bp oligonucleotides also yields two bound species, but we interpret θ1 as representing cooperative binding of three TraY proteins to both the IR and DR, while θ2 represents binding to the IR, DR, and the alternative binding site. Binding of the 44-bp oligonucleotide results in three bound species: θ1 represents two TraY molecules binding cooperatively to the IR, θ2 represents occupation of the IR and either the alternative binding site or the DR, and θ3 represents occupation of the IR, the alternative binding site, and the DR. From these studies we cannot distinguish between TraY occupation of the DR and the alternative binding site; therefore, we cannot assign either binding event to the second or third bound species. (The 44-bp oligonucleotide does not extend far beyond the end of the TTTC subsite, and this potentially incomplete DR may exclude some contacts essential for the highest-affinity binding.) For all oligonucleotides, each binding event was graphed separately. For example, when using 39-bp oligonucleotides the IR is occupied in both bound species; therefore, θ1 and θ2 are added to give the total IR fraction bound at each protein concentration. The third bound TraY molecule is present only in the second bound species and, therefore, θ2 equals the total alternative binding site fraction bound. Curves reflecting IR binding were fit with a two-site cooperative binding model: fraction IR occupied = 1/{1+(KD/[TraY]2)}. Curves reflecting cooperative IR/DR binding (50-bp oligonucleotides) were fit with a three-site cooperative binding model: fraction IR occupied = 1/{1+(KD/[TraY]3)}. Curves depicting alternative site binding, or DR binding in the 44-bp oligonucleotides, were fit with a single-site independent binding model: fraction alternative site occupied or fraction DR occupied = 1/{1+(KD/[TraY])}.
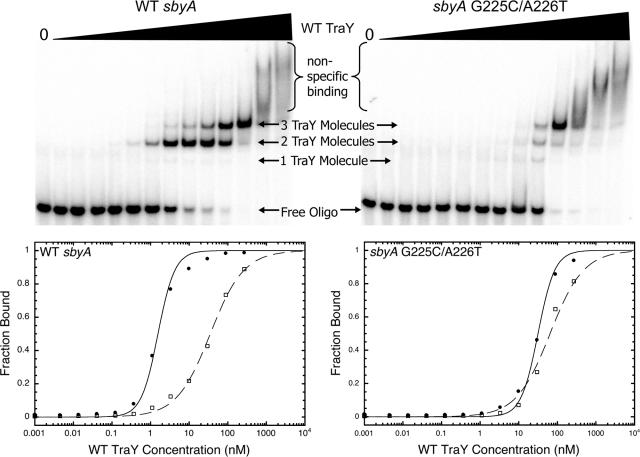
FIG. 2.
Representative EMSAs and graphs of TraY binding to WT and variant 39-bp sbyA binding sites. The upper panels depict TraY binding to the WT 39-bp sbyA binding site (upper left) or the G225C/A226T variant 39-bp sbyA site (upper right). The concentration of TraY increases from left to right (0 to 2.4 μM). The fastest migrating band illustrates free labeled DNA, whereas the slower migrating bands represent TraY bound to oligonucleotide. Smearing at the highest protein concentrations indicates nonspecific binding. Data from each EMSA were plotted as TraY concentration versus fraction bound. Each graph is displayed beneath its corresponding EMSA. TraY binding to WT and variant 39-bp sbyA oligonucleotides is plotted as two distinct binding events representing two TraY molecules binding the IR (filled circles) and a third molecule of TraY binding the alternative binding site (open squares). The IR data are fit with a two-site cooperative model, while the third bound TraY molecule is fit with a single independent binding site model.
For all IHF EMSAs, multiple bound species were present and were assumed to each represent a single IHF molecule binding independently. When comparing EMSAs using WT and variant ihfA oligonucleotides, only the first bound species was affected by ihfA substitutions. We therefore assume that the first bound species represents IHF occupying its specific site and other bound species represent nonspecific binding. Therefore, the total fraction bound of the specific IHF sites occupied in all WT and variant IHF oligonucleotides was the sum of each partial fraction bound, θtot = (θ1 + θ2 + θ3). IHF curves were fit with a one-site independent binding model: fraction specific IHF site occupied = 1/{1+(KD/[IHF])}.
To better compare binding affinities, all KD values were converted to half-maximal DNA binding concentrations (half-max, or [protein]1/2). The half-max is the protein concentration required to occupy half of a given binding site. For data fit with the single-site binding model, the KD equals the half-max. For data fit with the two-site cooperative model, the square root of the KD value equals the half-max, and for data fit with the three-site cooperative model, the cube root of the KD value equals the half-max. A ratio of the average half-max for each variant construct compared to the average half-max of its corresponding WT construct was calculated and reported as the relative half-max (relative [protein]1/2).
RESULTS
Binding affinities of F oriT sbyA and ihfA binding site variants.
The F oriT TraY site, sbyA, is located between the ihfA and sbmC binding sites (bp 205 to 240, numbered according to the scheme of Frost et al. [12]). Site sbyA has a high A/T content and contains an imperfect IR at bp 216 to 219 (TCTC) and bp 225 to 228 (GATA) that, when mutated, affects TraY binding (22). Hydroxyl radical footprinting shows that a third sbyA subsite (possibly TTTC, bp 206 to 209) is protected, but binding to this subsite appears less affected by substitutions than the others (22). To extend our previous work and in an effort to identify sbyA variants bound by TraY with significantly reduced affinity, we tested combinations of single base substitutions known to reduce binding. Three categories of variants were constructed, including those with multiple substitutions in each half of the IR and those containing substitutions in both halves of the repeat.
The binding affinities of TraY for the WT and variant sbyA sites were determined using EMSA. In Fig. 2, the upper left panel shows a representative EMSA for TraY binding to the WT 39-bp sbyA oligonucleotide. The fastest migrating bands represent free oligonucleotide. As TraY concentration increases, the oligonucleotide becomes bound, ultimately resulting in three slower migrating species. Of these three bound species, one is marginally populated and likely represents one molecule of TraY binding, while the other two are the predominate bound species. We interpret the faster migrating of these two bound species (first bound species) as representing two cooperatively bound TraY molecules at the IR. The slower migrating band (second bound species), which appears at higher TraY concentrations, represents an additional bound TraY molecule at a location we refer to as the “alternative binding site” (see below). At the two highest TraY concentrations, the intensity of the two bound species begins to diminish, replaced by smearing, indicating nonspecific binding of additional TraY molecules.
The upper right panel of Fig. 2 shows a representative EMSA of TraY binding to a variant 39-bp sbyA site. The variant sbyA IR is bound with lowered affinity, as reflected in the greater TraY concentrations that were required to populate the first bound species of the 39-bp sbyA site variant, relative to the WT sequence. The second bound species of the 39-bp sbyA variant, however, appears at a TraY concentration similar to the concentration at which the second species appears for the WT sbyA site, indicating its affinity was not altered. Additionally, the appearance of a single bound molecule of TraY is more pronounced in 39-bp sbyA variants, presumably resulting from reduced binding to the subsite(s).
The oligonucleotides used for the binding experiments were also employed to engineer the same substitutions into plasmids utilized in the in vivo mobilization assays. The lengths of oligonucleotides used in the EMSAs varied depending on the area of substitution. The TCTC subsite variants were incorporated into 50-bp oligonucleotides, the GATA subsite substitutions consisted of 39-bp oligonucleotides, and the 44-bp oligonucleotides included variants with substitutions in both regions of interest. The differences in the oligonucleotides may have effects on protein-binding affinity; therefore, the binding affinity of each variant oligonucleotide is compared to the binding affinity of the WT sequence of the same length and reported as the relative half-max, relative [TraY]1/2.
Curves depicting TraY binding to a WT or variant 50-bp sbyA site were divided into two binding events: occupation of the IR/DR subsites ([TraY]1/2 = 0.6 nM) and binding to a less specific alternative binding site ([TraY]1/2 = 47 nM) (see Fig. S1, lower panels, in the supplemental material). (The alternative binding site is located to the right of the sbyA sites in Table 1.) The 39-bp sbyA oligonucleotides do not contain the third sbyA subsite, TTTC, included in the DR. The two binding events observed for the 39-bp oligonucleotides are therefore occupation of the IR ([TraY]1/2 = 2 nM) and binding to the alternative site ([TraY]1/2 = 50 nM) (Fig. 2). Three binding events were observed with the 44-bp oligonucleotides (see Fig. S2 in the supplemental material). We interpret the first of these as binding to the IR ([TraY]1/2 = 1.6 nM). We cannot unambiguously identify the second ([TraY]1/2 = 18 nM) and third ([TraY]1/2 = 120 nM) binding events. One represents binding to the TTTC subsite of the DR with an affinity reduced relative to the 50-bp oligonucleotides due to the absence of some base pairs involved in recognition of this subsite (the oligonucleotide extends only 1 bp beyond the TTTC sequence). The other represents binding to the alternative site. Affinities for both of these binding events are within threefold of the alternative sites of the 39-bp and 50-bp sites. We therefore refer to these two bands as the second and third bound species of the 44-bp oligonucleotide. The binding curves representing association with the IR (44-bp and 39-bp oligonucleotides) or the IR/DR (50-bp oligonucleotides) were fit with a cooperative model, whereas the binding curves representing binding to the alternative binding site or the DR were fit with an independent site binding model. The difference between the concentration of TraY needed for binding to the IR or IR/DR site and for binding to the alternative binding site of WT sbyA is greater than 1 order of magnitude. For the variant sbyA sites, the IR transition was noticeably shifted to higher TraY concentrations while the alternative binding site transition was modestly altered, resulting in an overlap of the two.
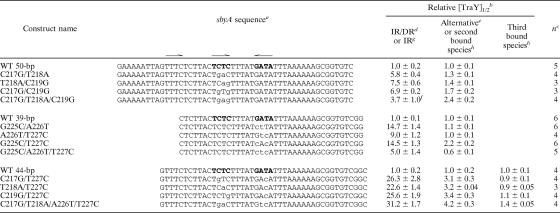
TABLE 1.
Relative binding of WT TraY to WT and mutant sbyA F oriT binding sites
aThe IR of the sbyA WT sequence is shown in bold. Specific substitutions are in lowercase letters. IR and DR subsites are indicated by arrows above the sequence.
bRelative half-maximal binding. Values were averaged, and averages and standard errors were divided by the average for WT oligo.
cNumber of measurements.
dAll three sbyA subsites are bound in a single cooperative binding event, occupying both the IR and the DR.
eAdditional bound species binding 3′ to GATA (to the right as depicted).
fComparison between IR binding of the C217G/T218A/C219G variant with IR/DR binding of the WT 50-bp oligonucleotide. See text for further details.
gOccupation of the IR represents the first bound species for the 39-bp and 44-bp oligonucleotides.
hThe DR and alternative binding sites of the 44-bp oligonucleotides could not be accurately assigned to a particular bound species.
Individual C217G, T218A, and C219G substitutions within the TCTC subsite (bp 216 to 219) reduce TraY-binding affinity (22). Combining substitutions in variant C217G/T218A reduced binding, causing a sixfold increase in the half-max binding concentration (Table 1). The C219G substitution combined with either the C217G or T218A substitution further decreased binding, raising the half-max binding concentration to ∼7-fold of WT. When all three of the above substitutions were combined, the binding affinity showed an improvement relative to the double substitutions, displaying a moderate fourfold decrease from WT. This result may be due to the creation of an alternative imperfect IR within the sbyA site. The C217G/T218A/C219G variant combined with T220 generates a GAGT subsite that can combine with a TCTC subsite (bp 208 to 211), creating an alternative imperfect IR. This new subsite pair, GAG(T/A), shifts the cooperative binding sites within sbyA while maintaining the preferred 5-bp spacing between subsites. This analysis of the binding is supported by the appearance of a third shifted band ([TraY]1/2 = 9 nM) not seen in EMSA for other 50-bp oligonucleotides, indicating an altered binding pattern for the C217G/T218A/C219G variant. We list values in Table 1 for this variant assuming that the first shifted band represents binding to an IR and the third shifted band represents binding to the alternative binding site, although we are not certain of this interpretation.
Substitutions within the GATA subsite (bp 225 to 228) also showed reduced binding with a 5- to 15-fold increase in relative [TraY]1/2 for the IR (Table 1). When the G225C substitution was combined with either A226T or T227C, the half-max binding concentration increased 15-fold, while the A226T/T227C variant displayed a 9-fold difference. The G225C/A226T/T227C substitutions show only a fivefold decrease in binding affinity compared to WT. It is possible that like the C217G/T218A/C219G variant, the G225C/A226T/T227C substitutions have created an alternative pair of high-affinity binding sites for TraY, although we have not identified one.
Each of the variants incorporating a single base pair substitution in each half of the imperfect IR (C217G/T227C, T218A/T227C, and C219G/T227C) yielded 23- to 26-fold reductions in binding affinity (Table 1). The combination of two 2-bp substitutions, C217G/T218A/A226T/T227C, resulted in a 31-fold decrease in binding compared to WT. In contrast, the relative [TraY]1/2 for the DR and alternative binding sites of all sbyA variants remained similar to WT, with a maximal fourfold change within the 44-bp sbyA variants.
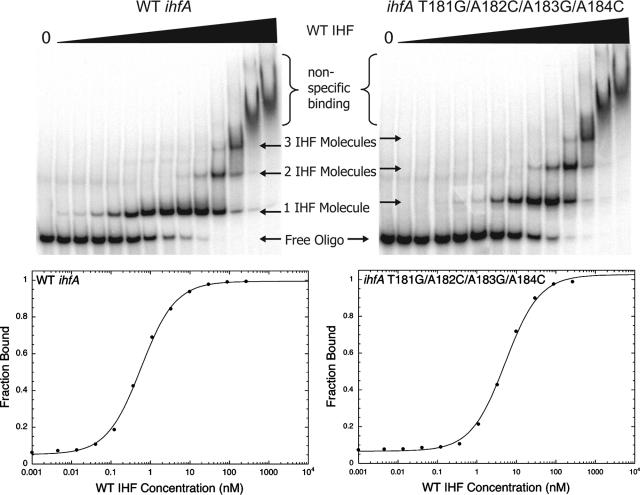
In addition to investigating the binding of TraY to variant sbyA sites, binding of the F relaxosome accessory protein IHF was examined. The F oriT IHF site, ihfA, is positioned between the sbi and sbyA binding sites (bp 168 to 194) (12, 39). This region contains the sequence AAATAGAGTGTTA (bp 182 to 194), which resembles the IHF consensus sequence WWWCAANNNNTTR. To confirm this sequence as the IHF-binding site and to identify ihfA variants bound by IHF with reduced affinity, we generated ihfA variants with substitutions in this region and in an A/T-rich region 5′ to this sequence. The upper panels of Fig. 3 show representative EMSAs of IHF binding to either WT ihfA (left panel) or a variant ihfA (right panel) oligonucleotide. Both show multiple bound species with presumably the first bound species representing specific binding to ihfA and the other slower migrating bound species illustrating nonspecific binding. Binding to the variant oligonucleotide requires a higher concentration of IHF to occupy the first bound species, while the concentrations at which the second and third bound species become occupied were unaltered. This indicates that the first bound species represents IHF binding to its conserved ihfA site, whereas the other bound species are binding nonspecifically.
FIG. 3.
Representative EMSAs and graphs of IHF binding to WT and variant ihfA binding sites. The upper panels depict IHF binding to the WT 46-bp ihfA binding site (upper left) or the T181G/A182C/A183G/A184C variant 46-bp ihfA site (upper right). The concentration of IHF increases from left to right (0 to 2.4 μM). The fastest migrating band illustrates free labeled DNA, whereas the slower migrating bands represent IHF bound to oligonucleotide. Smearing at the highest protein concentrations indicates nonspecific binding. Data from each EMSA were plotted as IHF concentration versus fraction bound. Each graph of the first IHF-binding event is displayed beneath its corresponding EMSA. IHF binding to WT and variant 46-bp ihfA sites was fit with a single-site independent binding model.
The ihfA WT and variant oligonucleotides differ in length (40, 46, 50, and 52 bp) and, to assist interpretation, we converted the KD values into relative [IHF]1/2 values. The graphical representations for representative WT and variant IHF EMSAs are shown in Fig. 3, lower panels. Both were fit best with a one-site independent binding model. IHF binding to variant ihfA sites resulted in a 4- to 18-fold decrease in binding affinity compared to WT (Table 2). The location of the substitutions that yielded the greatest reductions in binding affinities strongly suggest that bp 182 to 194 is the specific IHF binding site within ihfA.
TABLE 2.
Relative binding of WT IHF to WT and mutant ihfA F oriT binding sites
| Construct name | ihfA sequencea | Relative [IHF]1/2b | nc |
|---|---|---|---|
| WT 50-bp | CACGCAAAAACAAGTTTTTGCTGATTTTTCTTTATAAATAGAGTGTTATG | 1.0 ± 0.2 | 3 |
| T172G/T174C/T175G | CACGCAAAAACAAGTTTTTGCTGATgTcgCTTTATAAATAGAGTGTTATG | 4.7 ± 0.4 | 4 |
| WT 40-bp | CAAGTTTTTGCTGATTTTTCTTTATAAATAGAGTGTTATG | 1.0 ± 0.1 | 3 |
| T178C/T179A/A180G | CAAGTTTTTGCTGATTTTTCTcagTAAATAGAGTGTTATG | 3.8 ± 0.7 | 3 |
| WT 46-bp | GTTTTTGCTGATTTTTCTTTATAAATAGAGTGTTATGAAAAATTAG | 1.0 ± 0.04 | 3 |
| T181G/A182C/A183G/A184C | GTTTTTGCTGATTTTTCTTTAgcgcTAGAGTGTTATGAAAAATTAG | 17.8 ± 3.9 | 3 |
| WT 52-bp | GCTGATTTTTCTTTATAAATAGAGTGTTATGAAAAATTAGTTTCTCTTACTC | 1.0 ± 0.1 | 3 |
| T192G/T193G/A194C | GCTGATTTTTCTTTATAAATAGAGTGggcTGAAAAATTAGTTTCTCTTACTC | 14.2 ± 2.4 | 3 |
The region most similar to the IHF consensus sequence is shown in bold, and the 5′ A/T-rich region is underlined. Substitutions are in lowercase letters.
Relative half-maximal binding reported with standard errors.
Number of measurements.
Mobilization efficiencies of F oriT sbyA and ihfA binding site variants.
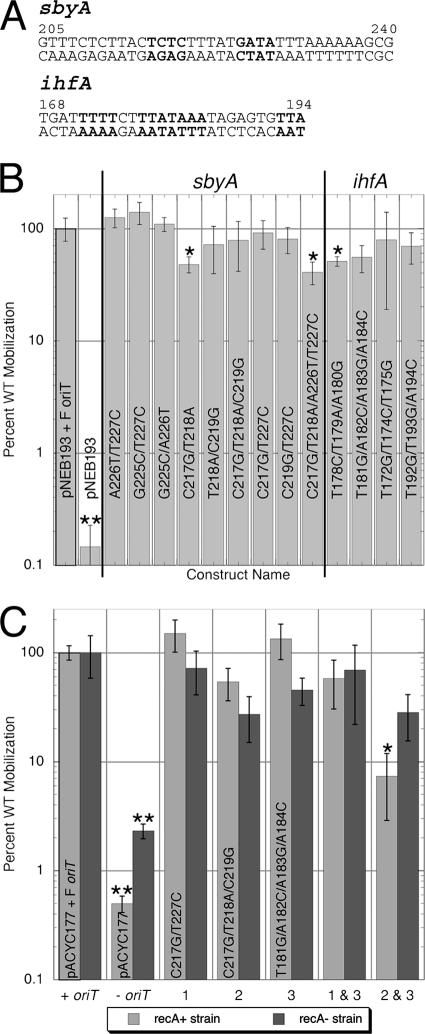
To determine the effects of the sbyA and ihfA binding site substitutions in vivo, the variants were constructed in an F oriT cloned into pNEB193 or pACYC177. Mobilization of WT and variant plasmids from F′ strains ER2738 and ER2267 to TB1 cells was measured. All substitutions made in the high-copy-number plasmid pNEB193-F oriT resulted in mobilization efficiencies of at least 40% of WT (Fig. 4B). Most of the constructs tested yielded mobilization efficiencies that were not statistically different from the WT values. The reductions in mobilization for C217G/T218A, C217G/T218A/A226T/T227C, and T178C/T179A/A180G were statistically different from WT using a Student's t test with a P value of <0.06 but only reflected minor decreases of 40% of WT mobilization. To test whether the high copy number of pNEB193 was somehow affecting transfer, several variants were recreated in the lower-copy-number plasmid pACYC177-ForiT. These variants demonstrated mobilization efficiencies equivalent to WT (Fig. 4C).
FIG. 4.
Mobilization efficiencies of WT and variant sbyA and ihfA constructs. (A) F oriT sbyA and ihfA sequences. Bolded bases indicate regions of substitution. (B) Percent WT mobilization of pNEB193 with cloned wild-type, variant, or no oriT is shown with error bars indicating standard errors of the measurements. Asterisks indicate constructs showing mobilization efficiencies that differed from WT (P < 0.06) as determined by a two-sided Student's t test. Two asterisks highlight mobilization assays resulting in no transconjugates. The value displayed gives the average percent WT mobilization if one transconjugate were present; therefore, the percent WT mobilization of pNEB193 is <0.25%. (C) Percent WT mobilizations of pACYC177 with cloned wild-type, variant, or no oriT are shown with error bars indicating standard errors of the measurements. Light gray bars indicate mobilization assays done in a recA+ strain (ER2738). The dark gray bars indicate experiments done in a recA-deficient strain (ER2267). Bar sets: 1, pACYC177-ForiT C217G/T227C; 2, pACYC177-ForiT C217G/T218A/C219G; 3, pACYC177-ForiT T181G/A182C/A183G/A184C. A single asterisk indicates those constructs showing mobilization efficiencies that differed from WT with a P value of <0.1. The negative control, pACYC177, resulted in no transconjugates whether in the recA+ or the recA-deficient strain, yielding a relative mobilization efficiency of <0.4% in ER2738 and <2.3% in ER2267 (double asterisks).
We were concerned that the mobilization results might be affected by recombination between the F′ and the pACYC177 plasmids, returning the cloned, variant oriT to the wild-type sequence and improving its apparent mobilization efficiency. To examine this possibility, we repeated the mobilization assays using the F′ recA-deficient strain ER2267 as the donor. The results using ER2267 cells resembled those using the F′ recA+ strain, ER2378, with a maximal decrease in mobilization to 27% of WT. These results suggest that recombination activities of the donor cells have little effect on the mobilization efficiencies of the sbyA and ihfA variants.
Plasmids with oriT sequences containing combinations of sbyA and ihfA substitutions were also tested. The combination variants assayed in the ER2267 (recA deficient) cell line reduced mobilization to 28% and 69% of WT, similar to individual sbyA and ihfA substitutions (Fig. 4C). These same variants examined in the ER2738 (recA+) cell line reduced mobilization to 7% and 58%, respectively, of WT. The C217G/T218A/C219G/T181G/A182C/A183G/A184C substitution in ER2738 is statistically different from WT with 99% confidence. However, the mobilization efficiencies of each construct in the recA+ background are not statistically different from the mobilization efficiencies of the same constructs in the recA-deficient background.
Mobilization efficiencies of F oriT spacer variants.
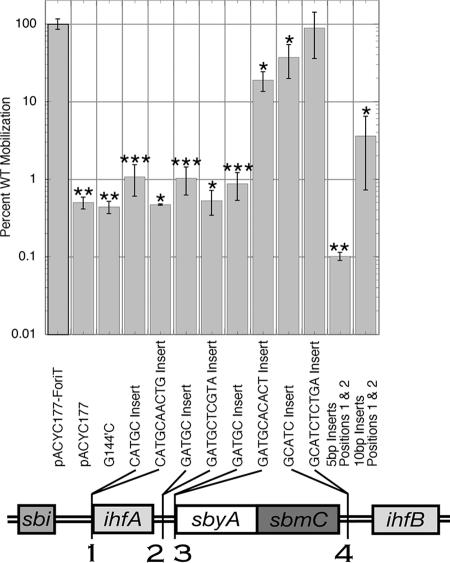
Although most substitutions we tested in F oriT sbyA and ihfA measurably reduced in vitro binding, only one showed a significant, albeit modest, decrease in transfer. These results suggest several possibilities: (i) the reductions in binding affinities were not great enough to prevent in vivo occupation of the binding sites, (ii) the specific sequences of the sbyA and ihfA binding sites may not be the only factors required for TraY and IHF binding, or (iii) binding to sbyA and ihfA is not essential for transfer. To further investigate the role of the F oriT relaxosome protein-binding sites in transfer, their spacing and orientation with respect to each other were examined. We reasoned that if TraY and IHF were required for transfer, altering the spacing and/or phasing of their sites might alter transfer efficiency. Insertions of 5 or 10 bp were made at four oriT sites: (i) one between the sbi and ihfA binding sites, (ii) two between the ihfA and sbyA binding sites (one interrupts an A-tract in this region at bp 198 and the other, at bp 204, does not), and (iii) one between the sbyA and ihfB binding sites.
Single 5-bp insertions between sbi and ihfA or ihfA and sbyA decreased mobilization efficiency to ∼1% of WT (Fig. 5). The 10-bp insertion between sbi and ihfA along with the 10-bp insertion between ihfA and sbyA positioned at bp 198 showed 0.5% of WT mobilization. These results indicate the importance of the distance between oriT protein-binding sites and their orientation with respect to one another. If the reduction in mobilization were solely the result of altering the phasing of a relaxosome protein-binding site with respect to its neighboring protein-binding sites, then the 10-bp insertions would have restored mobilization efficiency to WT levels. The 10-bp insert between ihfA and sbyA at bp 204 decreased mobilization to 19% of WT, which is a significant reduction but not nearly as great as the reduction shown by the 5-bp insertion at this site or the 10-bp insertion at bp 198. The 5-bp insertion between sbyA and ihfB reduced mobilization to 37% of WT, while the 10-bp insertion between sbyA and ihfB yielded a mobilization efficiency similar to WT. In general, as the distance between nic and the insertions increases, the negative effect on mobilization decreases. The length of the spacing between the sbyA and ihfB binding sites and their distance from nic may provide sufficient flexibility to this region of DNA and, therefore, 5 or 10 additional base pairs do not drastically change the DNA topography.
FIG. 5.
Mobilization efficiencies of spacer variants. Five- or 10-bp insertions were made in pACYC177-ForiT. The specific sequence inserted is listed below each bar. The location of the insertions is illustrated in the schematic of F oriT located below the graph. All constructs yielding efficiencies statistically different from WT (P ≤ 0.02) are indicated by one asterisk. Two asterisks highlight constructs for which no transconjugates were detected. The values given represent the average percent WT mobilization if one transconjugate was present. Three asterisks show constructs where the majority, but not all, of the mobilization assays resulted in no transconjugates.
The combination of the 5-bp insertion between sbi and ihfA and the 5-bp insertion between ihfA and sbyA positioned at bp 198 resulted in no transconjugates, with a mobilization efficiency at least 1 order of magnitude below the single 5-bp insertions (Fig. 5). The 5-bp insert between sbi and ihfA alone causes both ihfA and sbyA to be out of phase with sbi. Combined with the 5-bp insert between ihfA and sbyA, ihfA remains out of phase with sbi, but sbyA is restored to its WT position with respect to sbi. Together, the 10-bp insert between sbi and ihfA plus the 10-bp insert between ihfA and sbyA at bp 198 resulted in a mobilization efficiency of less than 4% of WT, demonstrating a seven- to eightfold increase in mobilization efficiency compared to the individual 10-bp insertions. However, the mobilization efficiency of plasmids with the combination of 10-bp inserts at bp 167 and 198 is not statistically different from that of the plasmids containing the individual 10-bp inserts. Both the spacing and orientation between relaxosome protein-binding sites must be conserved for optimal mobilization.
DISCUSSION
Complexes of proteins assemble at the oriT of conjugative plasmids to generate the plasmid nick that is essential for transferring one strand of the plasmid to the recipient. Here we have examined the relaxosome complex in F plasmid, attempting to identify some of the DNA sequence requirements for relaxosome assembly and plasmid transfer. In an effort to determine the importance of the F oriT TraY- and IHF-binding sites, we examined TraY and IHF affinities for variants of their respective sbyA and ihfA sites. While we identified sbyA and ihfA substitutions that caused significant decreases in in vitro binding compared to WT, mobilization efficiencies were only modestly reduced if at all. In contrast, we observed dramatic decreases in mobilization efficiency when we increased spacing between oriT binding sites and altered orientation of the relaxosome binding proteins with respect to each other.
Based on our previous work (22) and the data presented here, we conclude that one molecule of TraY binds to each of three subsites in the F oriT sbyA binding site. Two of the three subsites, TCTC (bp 216 to 219) and GATA (bp 225 to 228), separated by a 5-bp spacer, form an imperfect IR to which two molecules of TraY bind cooperatively. A third TraY molecule binds cooperatively to a third sbyA subsite (possibly TTTC, bp 206 to 209) oriented as a DR with the TCTC subsite. An additional TraY molecule can bind to a sequence outside of the IR and DR. This result was unexpected, given that binding in this region was not observed in earlier footprinting studies (22). We have no evidence that this binding is functionally relevant.
The IHF binding data presented here are consistent with a single molecule of IHF binding specifically to ihfA. The reduced affinity of the ihfA variants tested supports our contention that the ihfA sequence AAATAGAGTGTTA (bp 182 to 194), which resembles the IHF consensus sequence, WWWCAANNNNTTR, is the region within ihfA specifically bound by IHF.
Our analysis of sbyA variants underscores the relatively limited sequence specificity of TraY recognition for sbyA. Even combinations of base substitutions have relatively small effects on in vitro binding by TraY. We also see evidence of the relative complexity of the sbyA site. For example, the C217G/T218A/C219G triple variant shows improved binding compared to the C217G/C219G and T218A/C219G double substitutions, probably due to the creation of an alternative IR binding sequence within sbyA. IHF, like TraY, exhibits a relatively low level of sequence specificity. Binding specificity of IHF relies upon “indirect readout” of a DNA conformation in addition to direct readout of bases (31). TraY may also rely upon indirect readout to bind to its site, helping explain why it can recognize its site even though it is located in the midst of similar sequences.
When sbyA and ihfA substitutions were incorporated into plasmids and the plasmids tested for mobilization efficiency, most showed no statistically significant difference in efficiency from plasmids containing the wild-type oriT. The single exception was the variant C217G/T218A/C219G/T181G/A182C/A183G/A184C, and this variant showed a significantly reduced transfer efficiency only when strain ER2738, but not ER2267, was used as a donor strain. This apparent lack of correlation between the binding and mobilization results may be due to a failure to reduce binding affinities sufficiently to eliminate occupation of the binding sites in vivo. sbyA is bound with a severalfold-higher affinity than sbyB (22, 28), a TraY-binding site near the traY promoter, yet both sites appear to play functional roles in vivo (1, 15, 35). IHF also plays multiple roles in the cell and has many binding sites in both the chromosome and plasmid DNA. Apparent dissociation constants of IHF for binding sites in λ phage range from 2 to 128 nM (40, 44). Presumably, to prevent in vivo binding to variant sbyA and ihfA sites, the affinities for the variant sites would have to be well below the affinities for their naturally occurring sites. We are not convinced that the affinity for the engineered sites was sufficiently reduced.
Another possibility for the lack of effect of the substitutions on transfer is that IHF and TraY are not essential to transfer. This seems unlikely, because although our changes to the specific binding sites for sbyA and ihfA did not greatly affect plasmid mobilization, inserting spacers between the oriT protein-binding sites disrupts transfer. Insertions of a half or full helical turn positioned between sbi and ihfA or ihfA and sbyA were poorly tolerated, reducing mobilization efficiencies to 0.5 to 1% of WT. These results indicate that the spacing of the protein-binding sites and perhaps the phasing of the sites cannot be altered without adversely affecting transfer.
We are not certain why insertions between sbi and ihfA and between ihfA and sbyA cause reduced transfer efficiency, but there are a number of possibilities. First, if TraY and IHF bind cooperatively to their sites within the relaxosome, with cooperativity arising either from direct physical interaction between the proteins or from an induced DNA conformation, altering the distance between the ihfA and sbyA could reduce cooperativity. The reduced cooperativity would cause reduced binding of TraY and IHF, leading to reduced TraI activity and therefore reduced transfer. Second, if the effects of TraY and IHF are due to a DNA conformational change propagated to sbi, increasing the distance between bound TraY and/or IHF and sbi could blunt the effect on sbi conformation. Third, IHF and/or TraY may directly interact with TraI to stabilize TraI binding or to affect TraI cleavage activity. Altering the distance between binding sites or altering the phasing of the bound proteins relative to each other would prevent direct protein-protein interactions. Fourth, altered spacing might affect events following ssDNA cleavage by TraI. TraI from F and related plasmids possesses a helicase activity in addition to its relaxase activity, and both activities are required for efficient transfer. Work from the Zechner lab on R1 TraI demonstrates that the helicase activity requires a significant ssDNA region 5′ to the duplex to load onto DNA and initiate DNA strand separation (5, 6). If loading the TraI helicase at the relaxosome is a necessary step in initiating DNA transfer, then DNA insertions in the region might prevent helicase loading and dramatically reduce transfer efficiency.
As the distance between the inserted sequences and nic increase, the effect of the insertions on mobilization decreases. Plasmids with 5- and 10-bp insertions between the sbyA and ihfB sites have mobilization efficiencies similar to those with a wild-type oriT. This is not surprising. Occupation of sbi, ihfA, and sbyA is required for the nicking reaction (13, 17) while, using deletion analysis, ihfB was found to have no effect on in vitro (27) or in vivo (13) nicking. This result apparently distinguishes F from R100, a plasmid closely related to F. In R100, IHF binding to ihfB was shown to be important for transfer in vivo (1) and was suggested to form a complex with the TraM molecule occupying sbmC to aid in DNA transfer (2). Assuming that the same IHF-TraM complex is formed in F, the requirements for the spacing and orientation of the sbmC and ihfB binding sites, relative to nic, are not as stringent as for R100.
In F oriT, 10-bp insertions at either bp 198 or bp 204 are positioned between the ihfA and sbyA sites, yet there is a 36-fold difference in their mobilization efficiencies. The 5-bp inserts at these positions have a mobilization efficiency of ∼1% of WT, similar to the 10-bp insert at position 198. The 10-bp sequences we inserted at 198 and 204 are similar in sequence and G/C content, but the 10-bp insertion at position 198 interrupts an A-tract. The source of the differences in the mobilization efficiencies of plasmids containing these insertions is not clear. The insertions at different sites could have different effects on the transmission of a conformational change to nic. Alternatively, there may be an additional factor involved in the initiation of transfer. The possibility of direct protein-protein interactions between TraI and TraY has been previously proposed (18, 24, 27). Conceivably, the 10-bp insertions between ihfA and sbyA could decrease DNA flexibility of the surrounding region, making it difficult to closely position sbi and sbyA in order to promote protein-protein interactions. The flexibility of the DNA region between ihfA and sbyA may be affected differently by the two 10-bp insertions, explaining the variations in their mobilization efficiencies. Further experimentation to directly test for F relaxosome protein interactions will be necessary.
Insertions between F oriT relaxosome protein-binding sites reduce mobilization, even though substitutions within the specific binding sites are tolerated. Both the distance between binding sites and their orientation with respect to one another must be maintained for optimal mobilization. These results support a model including both indirect and possibly direct interactions between relaxosome proteins during the initiation of transfer. The changes in DNA topology at sbi caused by TraY and IHF bending do not fully explain the mechanism required for the recruitment of TraI to sbi. However, the conformational changes of F oriT DNA during the initiation of transfer also position TraY closer to sbi, highlighting the possibility of direct TraY-TraI interactions to encourage TraI binding (27).
Supplementary Material
Acknowledgments
We thank Christine DeGennaro for cloning F oriT into pACYC177, Jennifer C. Stern for cloning F oriT into pNEB193, and Robert Schleif for use of equipment. We thank Michael E. Rodgers for helpful discussions and suggestions.
This work was supported by National Institutes of Health grant GM61017.
Footnotes
Published ahead of print on 9 March 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abo, T., and E. Ohtsubo. 1995. Characterization of the functional sites in the oriT region involved in DNA transfer promoted by sex factor plasmid R100. J. Bacteriol. 177:4350-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abo, T., and E. Ohtsubo. 1993. Repression of the traM gene of plasmid R100 by its own product and integration host factor at one of the two promoters. J. Bacteriol. 175:4466-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clewell, D. B., and D. R. Helinski. 1969. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an open circular DNA form. Proc. Natl. Acad. Sci. USA 62:1159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craig, N. L., and H. A. Nash. 1984. E. coli integration host factor binds to specific sites in DNA. Cell 39:707-716. [DOI] [PubMed] [Google Scholar]
- 5.Csitkovits, V. C., D. Dermic, and E. L. Zechner. 2004. Concomitant reconstitution of TraI-catalyzed DNA transesterase and DNA helicase activity in vitro. J. Biol. Chem. 279:45477-45484. [DOI] [PubMed] [Google Scholar]
- 6.Csitkovits, V. C., and E. L. Zechner. 2003. Extent of single-stranded DNA required for efficient TraI helicase activity in vitro. J. Biol. Chem. 278:48696-48703. [DOI] [PubMed] [Google Scholar]
- 7.Di Laurenzio, L., L. S. Frost, and W. Paranchych. 1992. The TraM protein of the conjugative plasmid F binds to the origin of transfer of the F and ColE1 plasmids. Mol. Microbiol. 6:2951-2959. [DOI] [PubMed] [Google Scholar]
- 8.Everett, R., and N. Willetts. 1980. Characterisation of an in vivo system for nicking at the origin of conjugal DNA transfer of the sex factor F. J. Mol. Biol. 136:129-150. [DOI] [PubMed] [Google Scholar]
- 9.Filutowicz, M., H. Grimek, and K. Appelt. 1994. Purification of the Escherichia coli integration host factor (IHF) in one chromatographic step. Gene 147:149-150. [DOI] [PubMed] [Google Scholar]
- 10.Firth, N., K. Ippen-Ihler, and R. A. Skurray. 1996. Structure and function of the F factor and mechanisms of conjugation, p. 2377-2401. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: molecular and cellular biology, 2nd ed. ASM Press, Washington, DC.
- 11.Frost, L., S. Lee, N. Yanchar, and W. Paranchych. 1989. finP and fisO mutations in FinP anti-sense RNA suggest a model for FinOP action in the repression of bacterial conjugation by the Flac plasmid JCFL0. Mol. Gen. Genet. 218:152-160. [DOI] [PubMed] [Google Scholar]
- 12.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu, Y. H., M. M. Tsai, Y. N. Luo, and R. C. Deonier. 1991. Deletion analysis of the F plasmid oriT locus. J. Bacteriol. 173:1012-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furste, J. P., W. Pansegrau, G. Ziegelin, M. Kroger, and E. Lanka. 1989. Conjugative transfer of promiscuous IncP plasmids: interaction of plasmid-encoded products with the transfer origin. Proc. Natl. Acad. Sci. USA 86:1771-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, Q., Y. Luo, and R. C. Deonier. 1994. Initiation and termination of DNA transfer at F plasmid oriT. Mol. Microbiol. 11:449-458. [DOI] [PubMed] [Google Scholar]
- 16.Goodrich, J. A., M. L. Schwartz, and W. R. McClure. 1990. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res. 18:4993-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howard, M. T., W. C. Nelson, and S. W. Matson. 1995. Stepwise assembly of a relaxosome at the F plasmid origin of transfer. J. Biol. Chem. 270:28381-28386. [PubMed] [Google Scholar]
- 18.Inamoto, S., Y. Yoshioka, and E. Ohtsubo. 1991. Site- and strand-specific nicking in vitro at oriT by the traY-traI endonuclease of plasmid R100. J. Biol. Chem. 266:10086-10092. [PubMed] [Google Scholar]
- 19.Lahue, E. E., and S. W. Matson. 1988. Escherichia coli DNA helicase I catalyzes a unidirectional and highly processive unwinding reaction. J. Biol. Chem. 263:3208-3215. [PubMed] [Google Scholar]
- 20.Lanka, E., and B. Wilkins. 1995. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64:141-169. [DOI] [PubMed] [Google Scholar]
- 21.Leong, J. M., S. Nunes-Duby, C. F. Lesser, P. Youderian, M. M. Susskind, and A. Landy. 1985. The phi 80 and P22 attachment sites. Primary structure and interaction with Escherichia coli integration host factor. J. Biol. Chem. 260:4468-4477. [PubMed] [Google Scholar]
- 22.Lum, P. L., M. E. Rodgers, and J. F. Schildbach. 2002. TraY DNA recognition of its two F factor binding sites. J. Mol. Biol. 321:563-578. [DOI] [PubMed] [Google Scholar]
- 23.Lum, P. L., and J. F. Schildbach. 1999. Specific DNA recognition by F Factor TraY involves beta-sheet residues. J. Biol. Chem. 274:19644-19648. [DOI] [PubMed] [Google Scholar]
- 24.Luo, Y., Q. Gao, and R. C. Deonier. 1995. Boundaries of the nicking region for the F plasmid transfer origin, oriT. Mol. Microbiol. 15:829-837. [DOI] [PubMed] [Google Scholar]
- 25.Luo, Y., Q. Gao, and R. C. Deonier. 1994. Mutational and physical analysis of F plasmid traY protein binding to oriT. Mol. Microbiol. 11:459-469. [DOI] [PubMed] [Google Scholar]
- 26.Nelson, B. D., C. Manoil, and B. Traxler. 1997. Insertion mutagenesis of the lac repressor and its implications for structure-function analysis. J. Bacteriol. 179:3721-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson, W. C., M. T. Howard, J. A. Sherman, and S. W. Matson. 1995. The traY gene product and integration host factor stimulate Escherichia coli DNA helicase I-catalyzed nicking at the F plasmid oriT. J. Biol. Chem. 270:28374-28380. [PubMed] [Google Scholar]
- 28.Nelson, W. C., B. S. Morton, E. E. Lahue, and S. W. Matson. 1993. Characterization of the Escherichia coli F factor traY gene product and its binding sites. J. Bacteriol. 175:2221-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pansegrau, W., D. Balzer, V. Kruft, R. Lurz, and E. Lanka. 1990. In vitro assembly of relaxosomes at the transfer origin of plasmid RP4. Proc. Natl. Acad. Sci. USA 87:6555-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penfold, S. S., J. Simon, and L. S. Frost. 1996. Regulation of the expression of the traM gene of the F sex factor of Escherichia coli. Mol. Microbiol. 20:549-558. [DOI] [PubMed] [Google Scholar]
- 31.Rice, P. A., S. Yang, K. Mizuuchi, and H. A. Nash. 1996. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell 87:1295-1306. [DOI] [PubMed] [Google Scholar]
- 32.Robertson, C. A., and H. A. Nash. 1988. Bending of the bacteriophage lambda attachment site by Escherichia coli integration host factor. J. Biol. Chem. 263:3554-3557. [PubMed] [Google Scholar]
- 33.Scherzinger, E., R. Lurz, S. Otto, and B. Dobrinski. 1992. In vitro cleavage of double- and single-stranded DNA by plasmid RSF1010-encoded mobilization proteins. Nucleic Acids Res. 20:41-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schildbach, J. F., C. R. Robinson, and R. T. Sauer. 1998. Biophysical characterization of the TraY protein of Escherichia coli F factor. J. Biol. Chem. 273:1329-1333. [DOI] [PubMed] [Google Scholar]
- 35.Silverman, P. M., and A. Sholl. 1996. Effect of traY amber mutations on F-plasmid traY promoter activity in vivo. J. Bacteriol. 178:5787-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stern, J. C., and J. F. Schildbach. 2001. DNA recognition by F Factor TraI36: highly sequence-specific binding of single-stranded DNA. Biochemistry 40:11586-11595. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, R., and L. Taylor. 1982. Promoter mapping and DNA sequencing of the F plasmid transfer genes traM and traJ. Mol. Gen. Genet. 188:513-518. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, R., L. Taylor, K. Kelly, R. Everett, and N. Willetts. 1984. The F plasmid origin of transfer: DNA sequence of wild-type and mutant origins and location of origin-specific nicks. EMBO J. 3:1175-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai, M. M., Y. H. Fu, and R. C. Deonier. 1990. Intrinsic bends and integration host factor binding at F plasmid oriT. J. Bacteriol. 172:4603-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, S., R. Cosstick, J. F. Gardner, and R. I. Gumport. 1995. The specific binding of Escherichia coli integration host factor involves both major and minor grooves of DNA. Biochemistry 34:13082-13090. [DOI] [PubMed] [Google Scholar]
- 41.Willetts, N., and R. Skurray. 1980. The conjugation system of F-like plasmids. Annu. Rev. Genet. 14:41-76. [DOI] [PubMed] [Google Scholar]
- 42.Williams, S. L., and J. F. Schildbach. 2006. Examination of an inverted repeat within the F factor origin of transfer: context dependence of F TraI relaxase DNA specificity. Nucleic Acids Res. 34:426-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, C.-C., and H. A. Nash. 1989. The interaction of E. coli IHF protein with its specific binding sites. Cell 57:869-880. [DOI] [PubMed] [Google Scholar]
- 44.Yang, S. W., and H. A. Nash. 1995. Comparison of protein binding to DNA in vivo and in vitro: defining an effective intracellular target. EMBO J. 14:6292-6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zechner, E. L., F. de la Cruz, R. Eisenbrandt, A. M. Grahn, G. Koraimann, E. Lanka, G. Muth, W. Pansegrau, C. M. Thomas, B. M. Wilkins, and M. Zatyka. 2000. Conjugative DNA transfer processes, p. 87-174. In C. M. Thomas (ed.), The horizontal gene pool. Harwood Academic Publishers, Amsterdam, The Netherlands.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.