Abstract
The opportunistic human pathogen bacterium Pseudomonas aeruginosa secretes various exoproteins in its surrounding environment. Protein secretion involves different secretory systems, including the type II secretion system, or T2SS, that is one of the most efficient secretory pathways of P. aeruginosa. There are two T2SS in this bacterium, the quorum-sensing-regulated Xcp system and the Hxc system, which is only present under phosphate-limiting conditions. Like T2SS of other bacteria, the Xcp T2SS is species specific, and this specificity mainly involves two proteins, XcpP (GspC family) and the secretin XcpQ (GspD family), which are the gatekeepers of the system. Interestingly, an orphan secretin, XqhA, was previously reported as being able to functionally replace the XcpQ secretin. In this study, we identified another gene, which we named xphA (xcpP homologue A), which is located next to xqhA. We showed that deletion of the xphA gene in an xcpP mutant caused the disappearance of the residual secretion observed in this mutant strain, indicating that the protein XphA plays a role in the secretion process. Our results also revealed that complementation of an xcpP/xcpQ mutant can be obtained with the gene couple xphA/xqhA. The XphA and XqhA proteins (the PAQA subunit) could thus form, together with XcpR-Z, a functional hybrid T2SS. A two-dimensional polyacrylamide gel electrophoresis analysis showed that except for the aminopeptidase PaAP, for which secretion is not restored by the PAQA subunit in the xcpP/xcpQ deletion mutant, each major Xcp-dependent exoprotein is secreted by the new hybrid machinery. Our work supports the idea that components of the GspC/GspD families, such as XphA/XqhA or XcpP/XcpQ, are assembled as a specific tandem within the T2SS. Each of these pairs may thus confer a different level of secretion specificity, as is the case with respect to PaAP. Finally, using a chromosomal xphA-lacZ fusion, we showed that the xphA-xqhA genes are transcribed from an early stage of bacterial growth. We thus suggest that the PAQA subunit might be involved in the secretion process at a different growth stage than XcpP/XcpQ.
Pseudomonas aeruginosa is an opportunistic human pathogen bacterium which secretes a wide variety of virulence factors, including hydrolytic enzymes, into its surrounding environment. Secretion of these exoproteins requires different secretion systems, defined as type I, II, III, and V (16) and recently type VI (25), showing the high secretory diversity of this bacterium. To date, type IV is the only secretory system not identified in P. aeruginosa. The type II secretion system (T2SS) is conserved and widespread among bacterial species (13). This system, which is used by exoproteins bearing a signal peptide (Sec or Tat signature), is thought to be organized as a multiprotein complex spanning the bacterial envelope (12, 13, 27, 29). In P. aeruginosa, two independent T2SS named Xcp (or secreton) and Hxc (2) coexist. The xcp genes are organized in two divergent operons containing, respectively, xcpPQ and xcpRZ genes, while the genetic organization of the hxc genes is quite different (2, 13). The two machineries consist of 11 proteins named, respectively, XcpP to -Z (13) and HxcP to -Z (2) and require the presence of the peptidase XcpA/PilD, which is also involved in the maturation of PilA, the structural unit of type IV pili (26). The Xcp machinery is involved in the secretion of some of the major hydrolytic enzymes produced by P. aeruginosa, such as elastase (LasB), exotoxin A, or phospholipase C (13). Expression of the xcp genes has been shown to be under the positive control of quorum sensing (8, 35). In contrast, the Hxc system is functional only under phosphate limitation. This phosphate-regulated system was previously described to mainly secrete one enzyme, the low-molecular-weight alkaline phosphatase LapA (2).
Although T2SS are generally conserved, heterologous secretion is mostly species dependent, and components of the machinery are not systematically exchangeable between distant organisms (13). Moreover, T2SS components homologous to XcpP (GspC family) and to the secretin XcpQ (GspD family) have been suggested to be the gatekeepers of the system and to confer specificity for substrate recognition (6, 22).
Secretins are organized as multimers of 12 to 14 subunits and are the only outer membrane components of the T2SS. They form a ring-shaped structure with a central cavity, the diameter of which varies between 50 and 90 Å according to the species and constitutes the extrusion channel of the system. Secretins are also described as bipartite proteins which consist of a C-terminal domain (or homology domain) embedded in the outer membrane and conserved in all the members of the secretin family and an N-terminal domain more species specific which extends into the periplasm (3). In Erwinia chrysanthemi, this domain was shown to interact with a T2SS-secreted enzyme (the pectate lyase PelB), suggesting that it could be the determinant of the species specificity (32). In other respects, XcpP, the partner of the secretin XcpQ, is a bitopic inner membrane protein with a large periplasmic domain (4). XcpP presents some characteristics different from its homologues of other species. It was shown to migrate as two bands in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), in agreement with the two initiation codons predicted by in silico analysis, and it contains a coiled-coil-interacting domain in contrast to the PDZ domain present in most of the XcpP homologues (5). Coiled-coil and PDZ domains exhibit the same function (protein-protein interaction) but are structurally different. Furthermore, using hybrid proteins obtained by domain swapping between XcpP and its E. chrysanthemi homologue, OutC, we identified a 35-residue region localized in the periplasmic domain of the protein which could be involved in species specificity (15). More recently, we reported that specificity could also involve the interaction of the C-terminal domain of XcpP with XcpQ and that, as previously suggested by Bleves et al. (5), such a specific interaction could promote a fine-tuning control of the secretin opening (28). These observations suggest that rather than involving one T2SS component, species specificity could depend at least on efficient interactions occurring between XcpP and XcpQ via specific domains. T2SS components cannot be exchanged in distant organisms. However, in related species such as P. aeruginosa and Pseudomonas alcaligenes, although xcpP and xcpQ genes cannot be exchanged individually, they can restore the functionality of the Xcp system when they are exchanged pairwise (9). This observation is in agreement with a species-specific interaction between XcpP and XcpQ.
Besides the organized multiproteic Xcp and Hxc secretion systems, it was found that although not required for efficient secretion in wild-type P. aeruginosa, an individual protein, XqhA, encoded by a gene isolated on the P. aeruginosa genome and highly homologous to the xcpQ gene was responsible for the residual secretion observed in a mutant of the P. aeruginosa PAK strain deleted of the xcpQ gene (23). XqhA, which belongs to the secretin family, was also shown to require Xcp components to be functional and to recognize the exoproteins of the Xcp system. The presence in P. aeruginosa of a secretin not directly associated with the Xcp system but able to independently associate with the Xcp components and to restore secretion in an xcpQ-deleted strain can argue against the hypothesis of XcpP/XcpQ pairwise specificity. However, it should be pointed out that in the study reported by Martinez et al. (23) the xqhA gene was part of a 7.5-kb DNA fragment that most probably included other genes.
In this study, we showed that upstream and adjacent to the xqhA gene, an xcpP homologue that we have annotated in the P. aeruginosa PAO1 genome as xphA (for xcpP homologue; PA1867; www.pseudomonas.com) could clearly be identified. Such a genetic organization, reminiscent of xcpP and xcpQ organized into a single operon, suggests that similarly to XcpP and XcpQ, the XphA and XqhA proteins could constitute a specific functional GspCD secretory unit involved in protein secretion. We further investigated the physiological role played by these proteins in the secretion process. Our results support the idea that XphA and XqhA can associate with the XcpR-Z proteins of the classical T2SS of P. aeruginosa to constitute a hybrid secretion machinery with its own specificity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli and P. aeruginosa strains were grown, respectively, in Luria-Bertani or tryptic soy broth medium at 37°C with agitation. When required, P. aeruginosa strains were grown in the presence of 150 μg/ml carbenicillin (Cb), 80 μg/ml tetracycline (Tc). E. coli TG1 and DH5α strains were used to propagate plasmids, and E. coli BL21(DE3) was used for gene expression. Recombinant plasmids were introduced in P. aeruginosa strains by conjugation using the conjugative properties of pRK2013. P. aeruginosa conjugants were isolated on Pseudomonas isolation agar supplemented with 300 μg/ml Cb, 200 μg/ml Tc, or 2,000 μg/ml kanamycin (Km). Deletion mutants were obtained using mutator vectors derived from pKNG101 (Table 1) as previously reported by Bleves et al. (5).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Escherichia coli strains | ||
| TG1 | supE hsdΔR thi Δ(lac-proAB) F′ (traD36 proAB+laqIqlacZΔM15) | Laboratory collection |
| 1046 | met supE hsdS recA | Laboratory collection |
| BL21(DE3) | F−hsdS gal ompT, lysogen of DE3 carrying T7 polymerase gene | 34 |
| DH5α | supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Invitrogen |
| CC118 (λpir) | Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB arg(Am) recA1 Rfr (λpir) | 17 |
| Pseudomonas aeruginosa strains | ||
| PAO1 | Prototroph, chl-2 | B. Holloway |
| PAO1ΔPQ | xcpPQ deletion mutant | 9 |
| PAO1ΔPAQA | xphA-xqhA deletion mutant | This work |
| PAO1ΔPQPAQA | xcpP xcpQ xphPAxqhQA deletion mutant | This work |
| PAO1ΔRZ | Mutant strain deleted of the xcpRZ operon | V. Chapon |
| Plasmids | ||
| pMMB190 | Broad-host-range vector, Apr, lacUV5 promoter, lacZα | 24 |
| pMMB67HE | Broad-host-range vector, Apr, tac promoter | 14 |
| pRK2013 | ColE1, Tra+ Mob+ Kmr | 10 |
| pCR2.1 | ColE1, f1 ori, Apr Kmr | Invitrogen |
| pCR2.1-PAQA | pCR2-1 bearing xphA and xqhA genes | This work |
| pKNG101 | Smr, oriR6K mobRK2 sabBR+ (suicide vector) | 19 |
| pKNΔPAQA | Mutator plasmid for xphA-xqhA deletion | This work |
| pLAFR3 | Cosmid derivative of pLAFR1, IncP1, Tcr | 33 |
| pAX24 | pLAFR3 bearing all the xcp genes | 11 |
| pMMB67D-1867 | pMMB67HE bearing the xphA gene from the gateway system with V5 and His tags | G. Ball |
| pMMB-PAQA | pMMB190 bearing xphA-xqhA genes | This work |
| Mini CTX-lacZ | Tcr; self-proficient integration vector with Tet, Ω-FRT-attP-MCS, ori, int, and oriT | 18 |
| Mini CTX-pPA-lacZ | Mini CTX-lacZ bearing a transcriptional xphA-lacZ fusion | This work |
In order to clone xphA and xqhA genes in tandem on the same plasmid, a DNA fragment containing the coding sequences of the two genes was generated by PCR using primers XpA9 (5′-ATAGGATCCCACGAAGGATCAAAG-3′) and XqA6 (5′ TATAAGCTTAAAAAGTGC-GGCTACTAA-3′) and Expand HiFi polymerase. PCR was performed in the presence of 1 M betaine (final concentration) instead of dimethyl sulfoxide (routinely used for high-GC P. aeruginosa DNA) in order to improve high-fidelity amplification. The PCR product was first cloned in pCR2.1 (giving pCR2.1-PAQA) and a BamHI/HindIII fragment subcloned in pMMB190.
Transcriptional xphA-lacZ fusion.
A 537-bp DNA fragment containing the promoter region of the xphA-xqhA operon was produced by PCR and cloned at the PstI/EcoRI sites of the mini-CTX-lacZ vector, giving mini-CTX-pPA-lacZ. This plasmid was used to generate a chromosomal xphA-lacZ fusion in the PAO1 strain by the procedure of Hoang et al. (18), and expression of the transcriptional fusion was monitored by assaying β-galactosidase activity as described by Sambrook et al. (30).
Expression of the xphA gene.
Strain PAO1ΔP bearing plasmid pMMB67D-1867, which encodes the xphA gene from a gateway library (20), was grown in tryptic soy broth medium at 37°C. Gene expression was induced with 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and XphAV5+His-tagged protein was analyzed by immunoblotting using antiserum directed to V5.
Purification of XphA.
Strain BL21(DE3) bearing plasmid pMMB67D-1867 expressing the xphA gene from a gateway library (20) was grown at 17°C for 32 h in Studier ZYP-5052 medium. Membranes were obtained after sonication of cells as previously described (29) and solubilized with 1% (wt/vol) N-dodecyl-β-d-maltopyranoside (DM). XphAV5+His was purified from the DM-soluble fraction by immobilized metal affinity chromatography (IMAC) using either His-Select columns (Sigma) or nitrilotriacetic acid-Ni2+ magnetic beads (QIAGEN) as previously described (29).
Protein analysis and immunoblotting.
Extracellular proteins were separated from the cells by centrifugation, filtered through 0.2-μm sterile Acrodisc filters (PALL), precipitated on ice using 10% (wt/vol) trichloroacetic acid, and washed twice with 90% (vol/vol) acetone before solubilization either at 95°C with SDS-PAGE buffer (21) or at 25°C with an isoelectric focusing (IEF) buffer containing 7 M urea, 2 M thiourea, 4% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate, 30 mM dithiothreitol, and 0.5% ampholytes (Biolytes 3-10). Protein concentration was adjusted to 0.04 optical density (at 600 nm) equivalent units/μl. Immobilines dry strips (pH 3 to 10; 7 cm long; Bio-Rad) were rehydrated with 125-μl protein samples for 16 to 18 h at 20°C under silicone oil and focused using a Bio-Rad IEF cell (50-μA limitation per gel) on a 250-V gradient for 15 min, 4,000-V gradient for 2 h, and 15,000-V gradient for a total of 17,000 Vh. Gel strips were then equilibrated in a buffer containing 6 M urea, 0.375 M Tris (pH 8.8), 2% (wt/vol) SDS, 20% (vol/vol) glycerol, and 2% (wt/vol) dithiothreitol for 15 min (twice) and with the SDS-PAGE running buffer for 30 min. The strips were embedded on top of 12% (wt/vol) polyacrylamide gels with 0.5% agarose in running buffer containing bromophenol blue and run at 10 mA per gel until the indicator dye was 0.5 cm from the end of the gels. Then, gels were washed with pure water for 10 min (twice) and either routinely stained with Imperial protein stain (Pierce) or, when required, with a silver staining kit (Amresco). Proteins stained with Imperial protein stain were analyzed by matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry after trypsin digestion using standard procedures (Voyager DE-RP) and identified by database screening.
Cellular and extracellular proteins were also analyzed by immunoblotting and probing with monoclonal penta-His, polyclonal V5, LasB, β-lactamase, exotoxin A, LasA, and PaAP primary antibodies. Proteins were revealed by chemiluminescence using specific horseradish peroxidase-conjugated secondary antibodies (SuperSignal West Pico luminol; Pierce).
Proteolytic activity assays on plates.
Protease activities were assayed by growing cells on tryptic soy agar (TSA) containing skim milk incubated at 37°C.
RESULTS AND DISCUSSION
In silico analysis of XphA and XqhA.
In silico analyses showed that the protein encoded by the PA1867 gene found in the PAO1 genome next to the previously identified xqhA gene has significant similarities with XcpP. We named the gene PA1867 xphA for xcpP homologue. However, the putative xphA gene product is predicted to present some differences compared to its homologue XcpP. BlastP analysis revealed 36% identity (56/154 residues aligned) and 46% homology (72/154) between the two protein homologues. Sequence alignment using Clustal W (http://www.ebi.ac.uk/clustalw/) or T-COFFEE (http://www.ch.embnet.org/software/TCoffee.html) showed that XphA lacks a C-terminal extension present in XcpP (Fig. 1). XphA has a smaller size (175 amino acid residues versus 235), a higher pI (10.8 versus 5.46), and four cystein residues compared to XcpP. The shorter size of XphA might be explained by the absence of a coiled-coil domain (ch.EMBnet.org), which is characteristic of XcpP or of a PDZ domain present in most of the XcpP homologues in other species. These domains are usually found at the C terminus of XcpP or its homologues. Since XphA is predicted to be an inner membrane protein, we used the DAS Transmembrane Prediction server (http://www.sbc.su.se/∼miklos/DAS/) to determine the transmembrane domain of the protein. A typical transmembrane domain was predicted between residues 32 and 46 of XphA, at a position rather similar to that found for XcpP (residues 34 to 48). One of the interesting features of XcpP is that the gene which encodes this protein contains two initiation codons, giving two products of different molecular weights and which migrate as a doublet in SDS-PAGE (5). Cloning and expression of the two forms of the xcpP gene led to products that behave similarly in the secretion process and that do not appear to play a different physiological role (5). Analysis of the xphA DNA sequence revealed that, as for the xcpP gene, the xphA gene could encode two potential products (Fig. 1). The first one starts from the first base pair and stops at the end of the sequence (bp 528), giving a 175-amino-acid product, while the second might extend from bp 37 of the DNA sequence to bp 528 (163-amino-acid residue) (ATGpr server at www.hri.co.jp).
FIG. 1.
Sequence alignment of XcpP and XphA. The schematic representation of the proteins was drawn from sequence alignment using the Clustal W program. Numbers correspond to the amino acid position in the sequence. Arrows indicate the starts of the protein sequences. TM (gray), transmembrane domain; CC (black), coiled-coil domain.
BlastP searches established that XqhA and XcpQ present 63% identity (387/605 aligned residues) and 80% homology (488/605). The XqhA protein (776 amino acid residues; 83 kDa) is predicted to be larger than XcpQ (658 amino acid residues; 70 kDa), and sequence alignment using Clustal W and T-COFFEE predicted a large C-terminal extension in XqhA which is absent in XcpQ (data not shown). Furthermore, as for XphA and XcpP, the pI of XqhA is also predicted to be higher than that of XcpQ (7.3 compared to 5.78).
Therefore, XphA and XqhA present some differences compared to their respective homologues, which could have a physiological significance and be related to peculiar functions played by these proteins. On the one hand, sequence alignment of XcpP and XphA showed the presence of a C-terminal extension of XcpP that was absent for XphA. We have previously reported that the lack of the C-terminal coiled-coil domain in XcpP does not prevent the protein from assembling into a functional T2SS. It is thus a possibility that a shorter version of a GspC protein, such as XphA, may still be a functional protein. On the other hand, a C-terminal extension is present in XqhA but absent in XcpQ. The presence of such an extension could have a physiological significance and might compensate for the short size of XphA. These differences might indicate the high specificity which drives the interaction between XcpP and XcpQ or XphA and XqhA and the requirement of pairwise replacement.
Functionality and characteristics of XphA.
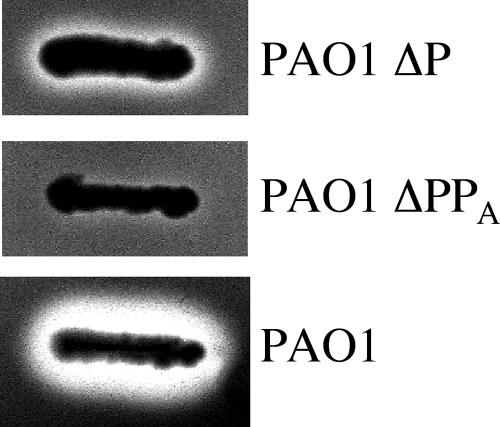
In contrast to XqhA, the presence of which was already reported by Martinez et al. (23), the XphA protein has never been described in previous studies. One of the objectives of this work was to show that this protein is produced in P. aeruginosa and that it plays a physiological role in vivo. The functionality of the secretion process was studied by assaying proteolytic activity on milk plates. Deletion of the xcpP gene was shown to drastically affect protease secretion compared to the control strain (Fig. 2). However, similar to the observation previously reported in an xcpQ mutant (23), a slight hydrolysis halo corresponding to a residual secretion was still observed in an xcpP deletion mutant strain, suggesting that although secretion was seriously affected, it was not completely abolished (Fig. 2). This residual secretion appeared to be dependent on XphA, since deletion of the xphA gene resulted in the disappearance of the proteolytic halo in the xcpP mutant (Fig. 2). Therefore, it seems likely that like XqhA, XphA plays a physiological role in P. aeruginosa and that it could contribute to the secretion process.
FIG. 2.
Functionality of XphA in P. aeruginosa. Wild-type and mutant strains deleted of the xcpP gene or of both the xcpP and the xphA genes were streaked on TSA-skim milk plates to assay secreted proteolytic activities. ΔP, xcpP-deleted mutant; ΔPPA, mutant deleted of both xcpP and xphA genes.
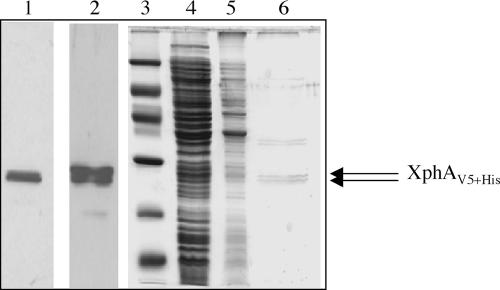
The xphA gene (PA1867) encoding a histidine (six-His)- and V5-tagged version of the XphA protein was cloned from a gateway library (20) in the expression vector pMMB67HE, giving pMMB67D-1867. Gene expression was carried out in an xcpP deletion mutant of P. aeruginosa. Interestingly, immunoblotting experiments using a V5 antiserum showed for the first time that, like the xcpP gene, xphA was expressed in P. aeruginosa as a protein doublet (XphAV5+His), exhibiting a molecular mass in SDS-PAGE close to 25 kDa (Fig. 3, lane 1). The xphA gene was also expressed in the E. coli BL21(DE3) strain for purification attempts. As depicted on the immunoblot probed with penta-His antiserum (Fig. 3, lane 2), XphA was produced in cells as a protein doublet, as previously observed in P. aeruginosa extracts (Fig. 3, lane 1). Detergent screening for membrane solubilization showed that DM was found to be more efficient than Triton X-100 or N-octyl-β-d-glucopyranoside for XphA solubilization. DM-soluble proteins were purified on nitrilotriacetic acid-Ni2+ magnetic agarose beads, and proteins were analyzed by SDS-PAGE and Western blotting (Fig. 3, lanes 4 to 6). Coomassie blue staining revealed that XphA was mainly recovered in the elution fraction in the presence of imidazole (Fig. 3, lane 6). As expected, purified XphA was shown to migrate like its homologue XcpP as a protein doublet corresponding to products of close molecular mass (around 25 kDa). Such a characteristic was confirmed by immunoblotting experiments with the purified fraction (data not shown).
FIG. 3.
Characteristics of the XphA protein. The xphA gene cloned on plasmid pMMB67D-1867 was expressed in an xcpP mutant of P. aeruginosa (lane 1) or in E. coli BL21(DE3) (lane 2). Proteins were analyzed by immunoblotting, probed either with V5 (lane 1) or penta-His (lane 2) antisera, and revealed by chemiluminescence using horseradish peroxidase-conjugated antibodies (Pierce). The apparent molecular mass of XphA was estimated from the position of protein standards (lane 3), from the top to the bottom, 97, 66, 45, 30, 20, and 14.4 kDa. IMAC purification of XphA (lanes 4 to 6) was carried out from protein extracts obtained from E. coli BL21 bearing pMMB67D-1867 as indicated in Materials and Methods. Lane 4, DM-soluble proteins; lane 5, flowthrough fraction; lane 6, purified fraction eluted with imidazole. Proteins from lanes 3 to 6 were stained with Imperial protein stain (Pierce).
XphA and XqhA can constitute a functional secretory subunit.
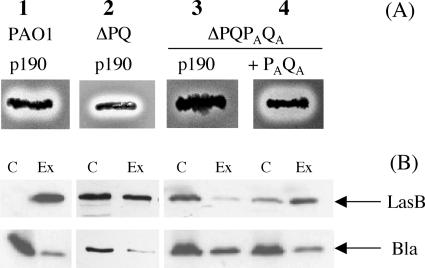
The secretion process of wild-type P. aeruginosa was characterized both by a proteolytic halo on TSA-milk plates and by immunodetection of secreted enzymes such as LasB (Fig. 4A and B, lane 1). In a mutant deleted of xcpP and xcpQ genes, a residual proteolysis halo was observed on the plates (Fig. 4A, lane 2), in agreement with the partial recovery of LasB in the extracellular medium (Fig. 4B, lane 2). This result suggested that the absence of XcpP and XcpQ (the GspC and -D components of the Xcp system) could be at least partially compensated by homologous components such as XphA and XqhA. Indeed, in a quadruple mutant deleted of xcpP, xcpQ, xphA, and xqhA genes, no proteolysis halo was observed on the plate (Fig. 4A, lane 3), while LasB was found to have accumulated intracellularly (Fig. 4B, lane 3). These results indicated that XphA and XqhA are responsible for the residual secretion observed in the xcpP/xcpQ double mutant (Fig. 4, lane 2). GspC and GspD components of the T2SS are known to require pairwise exchange to promote an efficient secretion in heterologous species-related hosts (9, 22). As GspC and GspD homologues are able to constitute like XcpP and XcpQ a GspCD pair, XphA and XqhA have thus to be produced together in order to check their role in the secretion process. To this goal, xphA and xqhA genes were cloned in tandem on the same plasmid and expressed in the quadruple mutant. As shown in Fig. 4A (lane 4), the complemented mutant strain was characterized by a clear proteolysis halo on the plate, while LasB secretion was restored (Fig. 4B, lane 4). Moreover, the secretory activity of the XphA/XqhA pair was not restricted to LasB, since exotoxin A and LasA secretion were also restored in the same strain (data not shown). Thus, it can be concluded that the XphA/XqhA pair substitutes for the XcpP/XcpQ pair and constitutes a secretion subunit (the PAQA subunit) not genetically linked to the classical Xcp system.
FIG. 4.
Functionality of XphA and XqhA in the secretory process. (A) Proteolytic activities of strains derived from ΔPQ and ΔPQPAQA mutant strains on TSA-milk plates. (B) Immunoblotting of protein extracts from the strains described for panel A bearing either the empty vector pMMB190 (p190) or the pMMB-PAQA plasmid (PAQA) expressing xphA and xqhA genes in tandem. PAO1, wild-type strain; ΔPQ, PAO1 mutant deleted of xcpP and xcpQ; ΔPQPAQA, PAO1 mutant deleted of xcpP, xcpQ, xphA, and xqhA genes. Proteins were probed with antisera directed to LasB or β-lactamase (Bla) and revealed by chemiluminescence. C, cellular proteins; Ex, extracellular proteins.
Deletion of the xcpRZ operon abolishes the secretory activity of the PAQA subunit.
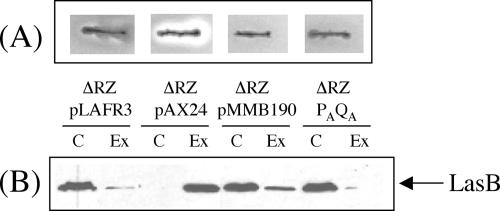
In order to check whether the secretion observed with the couple XphA/XqhA is only dependent on these proteins or whether it requires additional secretion factors, the xphA and xqhA genes were overexpressed in a mutant strain deleted of the xcpRZ operon. In this genetic background, the PAQA secretion subunit was unable to restore secretion, as shown by proteolytic assay on milk plates (Fig. 5A). However, complementation of the xcpRZ deletion by a plasmid bearing all the xcp genes caused the appearance of a clear hydrolysis halo as expected for a functional secretory process. This observation was confirmed by immunoblotting experiments showing that the XphA/XqhA pair is not able alone to promote secretion of LasB (Fig. 5B). Therefore, these results show that the PAQA subunit requires the presence of XcpR-Z as additional secretion components to constitute a functional hybrid T2SS secretory apparatus.
FIG. 5.
Effect of xcpRZ deletion on the secretory activity of PAQA. The secretory activity of the PAQA subunit was studied in a mutant strain deleted of the xcpRZ operon (ΔRZ). (A) Proteolytic activity, on a TSA-milk plate containing 2 mM IPTG, of the mutant strain bearing pLAFR3, pAX24 (xcp gene cluster), pMMB190, or pMMB-PAQA (PAQA). (B) Immunoblot of cellular and extracellular protein extracts from the same strains described for panel A and probed with LasB antiserum. C, cellular proteins; Ex, extracellular proteins.
Substrate specificity of the PAQA secretion subunit.
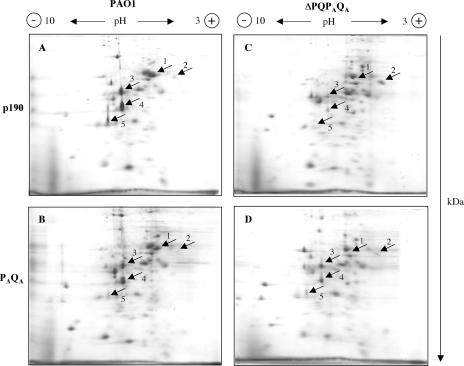
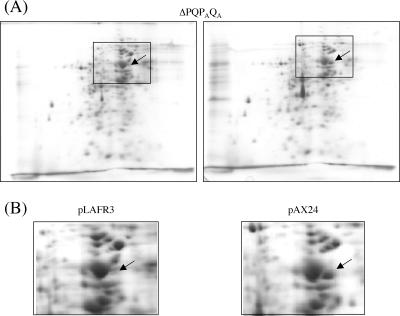
The PAQA secretion subunit appeared to work in the Xcp secreton, albeit less efficiently than XcpPQ regarding the diversity of substrates secreted by this T2SS. Thus, we investigated the possibility for the PAQA subunit to have a specific set of substrates by comparing the secretomes of the different strains used in this work. Extracellular proteins secreted during growth were separated by two-dimensional (2D)-PAGE and identified after Imperial protein staining by MALDI-TOF mass spectrometry. Protein spots 1 to 5 (Fig. 6A) were reproducibly observed in the parental strain PAO1 grown under our standard culture conditions and were identified by mass spectrometry as the aminopeptidase PaAP (PA2939) for spot 1 (7), the alkaline protease AprA (PA1249) for spot 2, the chitin binding protein CbpD (PA0852) for spot 3, the elastase LasB (PA3724) for spot 4, and the protease PrpL (PA4175) for spot 5. Except for AprA, which is secreted by the T1SS, all the other proteins identified are suggested to be T2SS substrates. The secretome of the quadruple mutant ΔPQPAQA showed a drastic decrease of the four T2SS-dependent proteins, PaAP, CbpD, LasB, and PrpL, showing as expected an alteration of secretion via T2SS (Fig. 6A and C). Overexpression of the xphA and xqhA genes in this mutant strain restored at least partially the secretion of three out of the four T2SS-dependent exoproteins that were affected for secretion, confirming that although apparently less efficient than their respective Xcp homologues, XphA and XqhA can reconstitute in association with XcpR-Z a functional hybrid T2SS (Fig. 6C and D). Interestingly, the aminopeptidase PaAP (spot 1) was not recovered after complementation of the xphA/xqhA deletion under the growth conditions tested, suggesting that its secretion requires a higher degree of specificity than for the other T2SS substrates (Fig. 6B and D). These results were confirmed by SDS-PAGE and immunoblotting of cellular and extracellular protein extracts probed with PaAP antiserum. PaAP was only detected in the cell fraction of the quadruple mutant bearing either the empty vector or expressing xphA/xqhA, showing that the protein was normally produced but not secreted (data not shown). Control experiments using the ΔPQPAQA mutant strain bearing the empty vector pLAFR3 or the plasmid pAX24 containing all the xcp genes showed that expression of the xcpP and xcpQ genes restored the secretion of PaAP in the mutant strain (Fig. 7A and B). Therefore, it seems likely that although supporting the secretion of a majority of T2SS substrates, the PAQA subunit cannot promote the secretion of the aminopeptidase and that secretion of this protein is particularly XcpPQ specific. This interesting observation lends support to the idea that substrate recognition could imply different levels of specificity in direct relation not only with the XcpP/XcpQ pair but also with the XphA/XqhA pair.
FIG. 6.
Substrate specificity of the PAQA subunit. Extracellular proteins from PAO1 and the mutant strain ΔPQPAQA carrying the empty vector pMMB190 (p190) or pMMB-PAQA (PAQA) were analyzed by 2D-PAGE as described in Materials and Methods. Polarity of the IEF is indicated at the top of the gels, and the second-dimension migration is shown on the right (in kDa). Identified protein spots are indicated by arrows. Spot 1, PaAP; spot 2, AprA; spot 3, CbpD; spot 4, LasB; spot 5, PrpL.
FIG. 7.
Secretion specificity of PaAP. Deletion of xcpP/xcpQ in the ΔPQPAQA mutant strain was complemented by introduction of pAX24 bearing all the xcp genes. Extracellular proteins were analyzed by 2D-PAGE under the same conditions as for Fig. 6. (A) Global protein profile of the secretome of the ΔPQPAQA strain carrying either pLAFR3 (empty vector) or pAX24. Squares highlight a small gel area containing PaAP. (B) Enlarged pictures of the squares shown in panel A. The arrow indicates the position of PaAP.
Expression of xphA and xqhA genes.
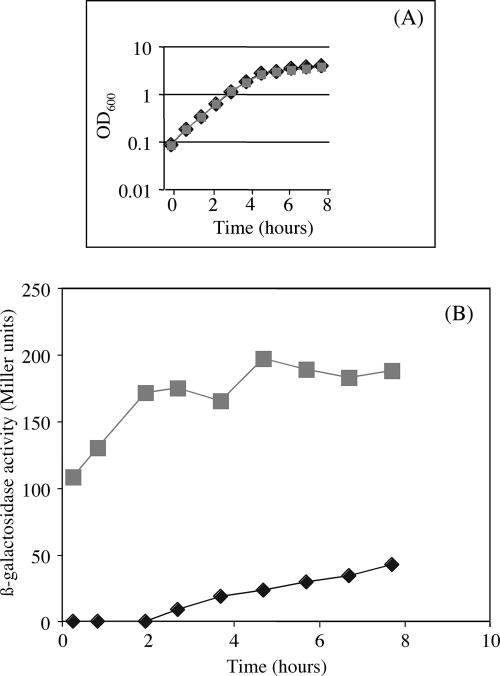
The xcpP and xcpQ genes of the classical T2SS of P. aeruginosa are known to be organized in an operon (1). It is likely that their homologues xphA and xqhA could also present the same genetic organization. Indeed, in silico analyses revealed an overlapping of the xphA and xqhA gene sequences. Moreover, a search for promoters using the BPROM software (www.softberry.com) indicated −10 and −35 boxes, respectively, 392 bp and 416 bp upstream of the initiation codon of xphA, and no additional such boxes could be found upstream of the xqhA gene. These observations strongly suggest the presence of only one putative promoter for the two genes and thus that, similarly to the xcpP and xcpQ genes, xphA and xqhA could be organized in an operon. However, nothing is known yet about the expression of these genes. Therefore, since xphA and xqhA genes are thought to be organized in an operon with a promoter localized upstream of xphA, an xphA-lacZ transcriptional fusion was constructed and integrated in the PAO1 chromosome, and its expression was studied by assaying the β-galactosidase activity under standard culture conditions. As shown in Fig. 8, the xphA-lacZ fusion was continuously expressed over the bacterial growth cycle, suggesting that XphA and XqhA proteins are produced from early stages of growth. This result is in agreement with microarray analyses, which did not detect xphA and xqhA as quorum-sensing-controlled genes (31, 35). Moreover, it has also been shown that expression of most of the genes encoding T2SS substrates is under the control of quorum sensing (31, 35). From these microarray studies, it should be emphasized that PaAP is likely to be produced later compared to LasB during P. aeruginosa growth. It should also be pointed out that expression of the xcpRZ operon is induced earlier than the xcpPQ one (35). Therefore, since the putative xphA-xqhA operon was shown to be activated at the onset of the growth, it can be hypothesized that in early growth stages, xphA and xqhA genes are expressed together with the xcpRZ operon and trigger assembly of a hybrid secretion machinery able to secrete LasB but unable to secrete PaAP. As cells grow and enter the stationary growth phase, expression of the xcpPQ operon is stimulated and XcpP and XcpQ preferentially associate with the XcpR-Z components to constitute a fully functional T2SS machinery that promotes secretion of PaAP. Delay in the expression of the genes encoding PaAP, XcpP, and XcpQ should have a physiological significance.
FIG. 8.
Expression of the xphA-xqhA genes in PAO1. A transcriptional xphA-lacZ fusion was integrated in the chromosome of PAO1 as described in Materials and Methods. The strain expressing the transcriptional lacZ fusion and the control strain were grown in Luria-Bertani medium at 37°C, and samples were withdrawn at intervals and assayed for β-galactosidase activity. (A) Growth curve. (B) β-galactosidase activity of the xphA-lacZ fusion. Results are expressed in Miller units and are means of duplicate samples from two independent experiments. Gray squares, gene fusion; black diamonds, promoterless control.
Conclusions.
P. aeruginosa is characterized by the existence of two independent T2SS, the Xcp and the Hxc systems, which are controlled, respectively, by quorum sensing (8) and by the phosphate concentration of the culture medium (2). The existence of the XqhA secretin and its involvement in the residual secretion observed in an xcpQ mutant was already reported by Martinez et al. (23). However, the presence of the XcpP homologue XphA and its role in the secretion process had not been described. Our results show that XphA is a newly described secretion protein that is normally produced by P. aeruginosa. Furthermore, the results also show that XphA works together with XqhA and further support the idea that these two proteins form an independent subunit within the T2SS that exerts various degrees of specificity towards a range of substrates. Regarding the species specificity of the T2SS, which has not been elucidated, the presence in the same bacterial species of proteins playing the same function although encoded by genes which are found at distant loci on the chromosome could contribute to additional insights on the determinants of the specificity of the T2SS.
However, the significance of having two independent GspCD subunits in P. aeruginosa (XcpP/XcpQ and XphA/XqhA) remains unclear. Therefore, several hypotheses can be proposed concerning the physiological role of the PAQA subunit in P. aeruginosa. A specific substrate, PaAP, has been found for XcpP/XcpQ, but no PAQA-specific substrate has been discovered so far. It can thus be proposed that this secretory subunit is mainly involved in the secretion of substrates, which are produced under peculiar culture conditions (such as growth on solid medium or biofilms, for instance) or in response to an unknown signal occurring during host infection. Alternatively, since it is not quorum sensing regulated, the PAQA subunit, in contrast to the XcpPQ one, can be involved at early growth stages by associating specifically with the Xcp proteins already produced in the exponential growth phase. Such an association leads to a hybrid secretory system which could secrete virulence factors at an early growth stage and help the establishment of P. aeruginosa in the host.
Acknowledgments
This work was supported by the Bettencourt-Schueller foundation and by Fondation Recherche Médicale. Eric Durand was supported by a fellowship from Fondation Recherche Médicale.
We thank Sabrina Lignon (IBSM, Plate-forme Proteome) for performing mass spectrometry analyses, Genevieve Ball for providing plasmid pMMB67D-1867, Dennis Ohman and Efrat Kessler for kindly providing us with LasA and PaAP antibodies, and Christophe Bordi and Christophe Bernard for helpful discussions.
Footnotes
Published ahead of print on 9 March 2007.
REFERENCES
- 1.Akrim, M., M. Bally, G. Ball, J. Tommassen, H. Teerink, A. Filloux, and A. Lazdunski. 1993. Xcp-mediated protein secretion in Pseudomonas aeruginosa: identification of two additional genes and evidence for regulation of xcp gene expression. Mol. Microbiol. 10:431-443. [DOI] [PubMed] [Google Scholar]
- 2.Ball, G., E. Durand, A. Lazdunski, and A. Filloux. 2002. A novel type II secretion system in Pseudomonas aeruginosa. Mol. Microbiol. 43:475-485. [DOI] [PubMed] [Google Scholar]
- 3.Bitter, W. 2003. Secretins of Pseudomonas aeruginosa: large holes in the outer membrane. Arch. Microbiol. 179:307-314. [DOI] [PubMed] [Google Scholar]
- 4.Bleves, S., A. Lazdunski, and A. Filloux. 1996. Membrane topology of three Xcp proteins involved in exoprotein transport by Pseudomonas aeruginosa. J. Bacteriol. 178:4297-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bleves, S., M. Gerard-Vincent, A. Lazdunski, and A. Filloux. 1999. Structure-function analysis of XcpP, a component involved in general secretory pathway-dependent protein secretion in Pseudomonas aeruginosa. J. Bacteriol. 181:4012-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouley, J., G. Condemine, and V. E. Shevchik. 2001. The PDZ domain of OutC and the N-terminal region of OutD determine the secretion specificity of the type II Out pathway of Erwinia chrysanthemi. J. Mol. Biol. 308:205-219. [DOI] [PubMed] [Google Scholar]
- 7.Cahan, R., I. Axelrad, M. Safrin, D. E. Ohman, and E. Kessler. 2001. A secreted aminopeptidase of Pseudomonas aeruginosa. Identification, primary structure, and relationship to other aminopeptidases. J. Biol. Chem. 276:43645-43652. [DOI] [PubMed] [Google Scholar]
- 8.Chapon-Herve, V., M. Akrim, A. Latifi, P. Williams, A. Lazdunski, and M. Bally. 1997. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol. Microbiol. 24:1169-1178. [DOI] [PubMed] [Google Scholar]
- 9.de Groot, A., M. Koster, M. Gerard-Vincent, G. Gerritse, A. Lazdunski, J. Tommassen, and A. Filloux. 2001. Exchange of Xcp (Gsp) secretion machineries between Pseudomonas aeruginosa and Pseudomonas alcaligenes: species specificity unrelated to substrate recognition. J. Bacteriol. 183:959-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filloux, A., M. Bally, M. Murgier, B. Wretlind, and A. Lazdunski. 1989. Cloning of xcp genes located at the 55 min region of the chromosome and involved in protein secretion in Pseudomonas aeruginosa. Mol. Microbiol. 3:261-265. [DOI] [PubMed] [Google Scholar]
- 12.Filloux, A., G. Michel, and M. Bally. 1998. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol. Rev. 22:177-198. [DOI] [PubMed] [Google Scholar]
- 13.Filloux, A. 2004. The underlying mechanisms of type II protein secretion. Biochim. Biophys. Acta 1694:163-179. [DOI] [PubMed] [Google Scholar]
- 14.Furste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 15.Gerard-Vincent, M., V. Robert, G. Ball, S. Bleves, G. P. Michel, A. Lazdunski, and A. Filloux. 2002. Identification of XcpP domains that confer functionality and specificity to the Pseudomonas aeruginosa type II secretion apparatus. Mol. Microbiol. 44:1651-1665. [DOI] [PubMed] [Google Scholar]
- 16.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoang, T., T. Kutchma, A. J. Becher, and H. P. Schweizer. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59-72. [DOI] [PubMed] [Google Scholar]
- 19.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 20.Labaer, J., Q. Qiu, A. Anumanthan, W. Mar, D. Zuo, T. V. Murthy, H. Taycher, A. Halleck, E. Hainsworth, S. Lory, and L. Brizuela. 2004. The Pseudomonas aeruginosa PAO1 gene collection. Genome Res. 14:2190-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lindeberg, M., G. P. Salmond, and A. Collmer. 1996. Complementation of deletion mutations in a cloned functional cluster of Erwinia chrysanthemi out genes with Erwinia carotovora out homologues reveals OutC and OutD as candidate gatekeepers of species-specific secretion of proteins via the type II pathway. Mol. Microbiol. 20:175-190. [DOI] [PubMed] [Google Scholar]
- 23.Martinez, A., P. Ostrovsky, and D. N. Nunn. 1998. Identification of an additional member of the secretin superfamily of proteins in Pseudomonas aeruginosa that is able to function in type II protein secretion. Mol. Microbiol. 28:1235-1246. [DOI] [PubMed] [Google Scholar]
- 24.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 25.Mougous, J. D., M. E. Cuff, S. Raunser, A. Shen, M. Zhou, C. A. Gifford, A. L. Goodman, G. Joachimiak, C. L. Ordonez, S. Lory, T. Walz, A. Joachimiak, and J. J. Mekalanos. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nunn, D. N., and S. Lory. 1991. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc. Natl. Acad. Sci. USA 88:3281-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Py, B., L. Loiseau, and F. Barras. 2001. An inner membrane platform in the type II secretion machinery of gram-negative bacteria. EMBO Rep. 2:244-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robert, V., A. Filloux, and G. P. Michel. 2005. Role of XcpP in the functionality of the Pseudomonas aeruginosa secreton. Res. Microbiol. 156:880-886. [DOI] [PubMed] [Google Scholar]
- 29.Robert, V., A. Filloux., and G. P. F. Michel. 2005. Subcomplexes from the Xcp secretion system of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 252:43-50. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shevchik, V. E., J. Robert-Baudouy, and G. Condemine. 1997. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 16:3007-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 35.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]