Abstract
Nontypeable Haemophilus influenzae (NTHI) strains are members of the normal human nasopharyngeal flora, as well as frequent opportunistic pathogens of both the upper and lower respiratory tracts. Recently, it has been shown that NTHI can form biofilms both in vitro and in vivo. NTHI strains within in vitro-formed biofilms differentially express both epitopes of lipooligosaccharide (LOS) and the outer membrane proteins P2, P5, and P6, whereas those generated either in a 96-well plate assay in vitro or in a mammalian host have been shown to incorporate a specific glycoform of sialylated LOS within the biofilm matrix. While DNA has been identified as a key component of the biofilm matrix formed in vitro by several bacterial pathogens, here we demonstrate for the first time that in addition to sialylated LOS, the biofilm formed by NTHI in vivo contains both type IV pilin protein and a significant amount of double-stranded DNA. The DNA appeared to be arranged in a dense interlaced meshwork of fine strands as well as in individual thicker “ropes” that span water channels, suggesting that DNA could be imparting structural stability to the biofilm produced by NTHI in vivo. The presence of type IV pilin protein both appearing as small aggregates within the biofilm matrix and tracking along DNA strands supports our observations which showed that type IV pili are expressed by NTHI during experimental otitis media when these bacteria form a biofilm in the middle ear space.
Nontypeable Haemophilus influenzae (NTHI) strains are normal commensal flora of the human nasopharynx but can act as opportunistic pathogens, causing multiple upper respiratory tract illnesses, including sinusitis and acute as well as chronic and recurrent otitis media (OM), when the conditions for them to do so are optimal (3, 4). NTHI strains are also associated with diseases of the lower respiratory tract, including exacerbations of chronic obstructive pulmonary disease and bronchitis. In recent years, NTHI has been shown to be capable of forming a biofilm in vitro and in vivo (13-15, 17, 19, 24, 34, 36, 41). A biofilm is defined as a group of bacteria growing as a community and encased in a self-produced polymeric matrix (33). Residence within a biofilm matrix, wherein oxygen is limited and the metabolic rate of these sessile microbes is altered, serves several functions, including protection from environmental threats such as host immune defenses, antibiotics, and surfactants. Thus, bacteria in biofilms are characteristically highly resistant to immune-mediated clearance (7, 8, 30, 39). The biofilm matrix can also provide a scavenging system to trap and filter nutrients from the environment. The composition of this matrix can be quite diverse, depending on the organism that induced its formation and the environment in which the biofilm was produced. Based on these characteristics, the ability to form a biofilm in the airway of its mammalian host is considered to be a likely contributing factor to the recurrent and/or chronic nature of NTHI-induced diseases of the respiratory tract (10, 11, 14, 29, 34).
In an attempt to better understand the role of biofilm production in the pathogenesis of NTHI-induced disease; several groups have begun to characterize these sessile communities of bacteria in greater detail. Toward this end, we and others have investigated the biochemical nature of the NTHI-induced biofilm and have shown that lipooligosaccharide (LOS) is heavily distributed throughout biofilms formed in vitro (16) and in vivo (19). Murphy and Kirkham used a panel of monoclonal antibodies to demonstrate that middle ear isolates of NTHI resident within a biofilm produced in a 96-well plate assay show altered expression of an LOS epitope, as well as selected epitopes within the outer membrane proteins (OMPs) P2, P5, and P6, compared to planktonically grown cells (24). Gallaher and colleagues later used a proteomic approach to identify 265 proteins, including OMPs P5 and P6, in the biofilm matrix of an NTHI biofilm produced by on-filter growth (15), whereas Webster et al. used electron microscopy to similarly localize LOS to the biofilm matrix and OMP P6 to the bacterial membrane (40). The latter group also demonstrated the presence of two NTHI adhesins and immunoglobulin A1 protease within these in vitro-formed biofilm matrices. Swords et al. further showed that sialylation of NTHI LOS promotes biofilm formation by NTHI (36) and that when growing in a biofilm, the phosphorylcholine content of NTHI LOS increases, resulting in a decrease in its net bioactivity (41).
Based on our initial observations of particular sialylated glycoforms of LOS incorporated into the biofilm formed by NTHI in the middle ear of the chinchilla (19), a predominant rodent host used to study the pathogenesis and prevention of OM, here we wanted to more extensively characterize both immature and more mature in vivo-formed biofilms biochemically. Therefore, we were interested in determining, primarily via the use of a chinchilla model of NTHI-induced OM combined with immunofluorescence confocal microscopy, whether the biochemical character of LOS contained within the biofilm changed over time, whether the major subunit protein of the recently described twitching pilus of NTHI (5) could be detected within these biofilms, and whether we could now better describe what initial observations suggested might be monocyte-like host cells that appeared to be both surface associated and infiltrating the NTHI-induced biofilm matrix (19).
MATERIALS AND METHODS
Animal model.
Four adult chinchillas (Chinchilla lanigera) were each challenged transbullarly with 2,500 CFU NTHI strain 86-028NP bilaterally. At selected time points after challenge, chinchillas were euthanatized and the bullae excised. The superior bulla was removed, and the inferior bulla was rinsed with 1.0 ml sterile pyrogen-free saline. The bullae were then carefully filled with Tissue-Tek OCT embedding compound (Fisher Scientific, Pittsburgh, PA) and snap frozen over liquid nitrogen, as previously described (19). Bullae were stored at −80°C until processed. Frozen bullae were placed on a bed of dry ice, and the external bone of the inferior bulla was carefully chipped away. The resulting block was split in a plane perpendicular to the tympanic membrane and reembedded in OCT. Four-micrometer serial sections were cut on a Leica CM3050S Cryostat (Leica Microsystems, Inc., Bannockburn, IL) and placed on StarFrost Adhesive slides (Mercedes Medical, Sarasota, FL). Slides were stored at −80°C until they were examined by microscopy. Some serial sections were cut to 10-μm thickness in order to visualize biofilm structural organization in greater detail when three-dimensional confocal (stacked) images were prepared.
Lectin labeling of NTHI-induced biofilms formed in vivo.
In order both to reconfirm the presence of sialic acid in NTHI-induced biofilms formed in vivo 4 to 5 days after challenge, as we reported previously (19), and to examine the biochemical nature of the sialylated LOS present within these biofilms 16 to 17 days later (21 days after challenge), we incubated serial sections of biofilms with the fluorochrome-conjugated lectin Maackia amurensis or Sambucus nigra (EY Laboratories, San Mateo, CA). Slides were incubated either with M. amurensis conjugated to Texas Red or with S. nigra conjugated to fluorescein isothiocyanate for 30 min in a humidified chamber (both lectins were diluted 1:50 with buffer). After rinsing in buffer, slides were coverslipped and examined by fluorescence microscopy. S. nigra has specificity for terminal sialic acid in an α-2-6 linkage to galactose, while the lectin M. amurensis has specificity for terminal sialic acid in an α-2-3 linkage to galactose.
To confirm that any labeling of the biofilm sections by fluorochrome-conjugated lectins was indeed due to the presence of sialic acid, we incubated a second series of serial sections of these in vivo-formed biofilms with 0.005 U neuraminidase from Vibrio cholerae (Roche Applied Science, Indianapolis, IN) per ml of reaction buffer for 3 h at 37°C in a humidified chamber. The slides were then rinsed in buffer three times and incubated with the fluorochrome-conjugated lectins as described above.
Immunofluorescent labeling of NTHI OMPs and type IV pilin protein.
To determine the relative incorporation of NTHI OMPs in general, as well as type IV pilin protein specifically, within biofilms formed by NTHI in vivo, 10-μm serial sections were labeled for immunofluorescence imaging using a standard protocol. Briefly, slides were air dried, fixed in cold acetone, and then equilibrated in buffer. Sections were blocked with image-iT FX signal enhancer (Molecular Probes, Eugene, OR) and with background sniper (BioCare Medical, Concord, CA) as per the manufacturer's instructions. Primary antibody (rabbit polyclonal anti-soluble recombinant PilA protein [20; J.A. Jurcisek, J. E. Bookwalter, B. D. Baker, S. Fernandez, L. A. Novotny, R. S. Munson Jr., and L. O. Bakaletz, submitted for publication] or chinchilla polyclonal anti-NTHI strain 86-028NP OMP antiserum [6]) diluted 1:200 in buffer was incubated with sections for 1 h at room temperature in a humidified chamber. Slides were then rinsed five times with buffer before incubation for 30 min either with goat anti-rabbit immunoglobulin G conjugated to AlexaFluor 488 in the case of the rabbit polyclonal antibody or with protein A conjugated to AlexaFluor 546 in the case of the chinchilla-derived antibody, rinsed five times, and coverslipped using ProLong Gold antifade reagent that contained DAPI (4′,6′-diamidino-2-phenylindole) to label double-stranded DNA (dsDNA) (Molecular Probes). Naïve rabbit serum or naïve chinchilla serum was used as a negative control, as appropriate. Sections were viewed using a Zeiss LSM 510 Meta confocal system attached to a Zeiss Axiovert 200 inverted microscope (Carl Zeiss Inc., Thornwood, NY).
SEM.
NTHI strain 86-028NP was inoculated into brain heart infusion broth supplemented with 10 μg hemin/ml and 10 μg NAD/ml (both from Sigma Chemical Co., St. Louis, MO) and then incubated for 180 min prior to inoculation of normal human bronchial epithelial (NHBE) cells (Clonetics, Baltimore, MD) that had been grown as a confluent monolayer on sterile glass coverslips, with bacteria at an multiplicity of infection of 100:1 and incubation at 37°C with 5% CO2 for 24 h. The coverslips were carefully rinsed three times with phosphate-buffered saline (pH 7.4). Cells were then prefixed in 2.5% glutaraldehyde (Electron Microscopy Sciences, Fort Washington, PA) overnight at 4°C and postfixed twice in 1% osmium tetroxide solution (Electron Microscopy Sciences) for 2 h for each fixation, followed by five washes in double-distilled water. Coverslips were dehydrated through a graded series of alcohol solutions and dried with hexamethyldisilazane (HMDS) (Electron Microscopy Sciences) for 10 min. Spent HMDS was removed and fresh HMDS was added before coverslips were allowed to air dry and then adhered to stubs using a colloidal silver coat (Electron Microscopy Sciences). Stubs were air dried overnight and sputter coated with gold and palladium (using a Cressington model 108 sputter coater) prior to viewing. Mounted coverslips were viewed with a Phillips 3000 scanning electron microscope (SEM) located at The Ohio State University Campus Microscopy and Imaging Facility.
Immunofluorescent labeling of DNA present within an NTHI-induced biofilm.
In an attempt to better describe the monocyte-like host cells present within biofilms formed by NTHI in vivo, we labeled nuclei (dsDNA) using DAPI, as previously described (19). The nuclear counterstain DAPI was incorporated in the mounting medium used to coverslip the slides (Prolong Gold antifade reagent with DAPI [Molecular Probes, Eugene, OR]) following the manufacturer's protocol.
To confirm that any labeling of NTHI-induced biofilms seen with DAPI was indeed due to the presence of dsDNA and not an artifact, additional slides containing serial sections were incubated with 106 U DNase I (Ambion Inc., Austin, TX)/ml buffer for 2 h at 37°C prior to labeling with DAPI as described above. As another control to confirm that the labeling observed was due to dsDNA present in the biofilm, additional serial sections were incubated with 100 μg DNase-free RNase (Sigma, St. Louis, MO)/ml water for 2 h at 37°C prior to labeling with DAPI. The RNase was made DNase free by heating a 10-mg/ml solution to 100°C for 15 min, allowing it to slowly cool to room temperature, and then adjusting the pH to 7.4 prior to diluting to the final working concentration. The working concentration used (100 μg RNase/ml buffer) is sufficient to remove all RNA from the equivalent of 3 ml of an overnight culture of Escherichia coli, based on the manufacturer's supplied information.
Live/dead labeling.
Frozen, OCT-embedded sections were stained with the Live/Dead BacLight bacterial viability kit (Molecular Probes, Eugene, OR) as per the manufacturer's instructions, as previously described (19).
RESULTS
Immunofluorescent labeling of sialylated LOS, OMPs, and type IV pilin protein.
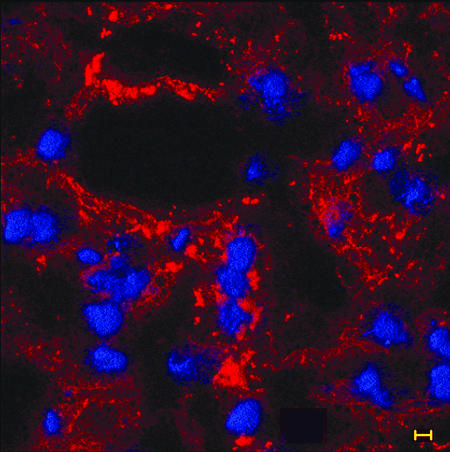
Sialylated LOS has been shown to be a major component of the biofilm formed by NTHI in vivo; however, one question yet to be answered was whether the biochemical nature of the LOS contained within the biofilm matrix changed as the biofilm matured further when resident within a mammalian host. To characterize the biofilm matrix for incorporation of both sialylated LOS and other bacterial components, serial OCT-embedded sections were incubated with several fluorochrome-conjugated antibodies or lectins. Figure 1 demonstrates labeling obtained using chinchilla polyclonal anti-NTHI strain 86-028NP OMP antiserum, which suggests the dense presence of either whole NTHI or residual OMPs throughout a 4-day biofilm matrix. DAPI labeling of what appeared to be host cells (based on size, shape, and uniformity of labeling) that had infiltrated the biofilm can be seen within this 4-day biofilm.
FIG. 1.
The distribution of NTHI (or residual OMPs) can be seen as red fluorescence throughout a 4-day biofilm recovered from the chinchilla middle ear. OCT-embedded inferior bullae were sectioned and immunofluorescently labeled with polyclonal chinchilla anti-NTHI strain 86-028NP OMP and AlexaFluor 546-conjugated protein A (red) to detect the presence of NTHI in the biofilm. DAPI, used as a counterstain, labels the nuclei of host cells (blue fluorescence) associated with, or perhaps trafficking through, the biofilm. Bar, 5 μm.
To confirm that a biofilm formed by NTHI strain 86-028NP in the middle ear of the chinchilla host incorporated sialylated LOS, both associated with the bacterial cell and within the biofilm matrix, we labeled biofilms recovered in this study with fluorochrome-conjugated lectins M. amurensis and S. nigra. The labeling obtained for the strain 86-028NP-formed biofilm was consistent with that previously observed with strain 2019 (data not shown). Briefly, the lectin S. nigra was seen heavily distributed throughout the biofilm matrix, which has been attributed (via the use of multiple NTHI LOS biosynthesis mutants [19]) to preferential binding of this lectin to 5-acetylneuraminic acid in an α-2-6 linkage to galactose that is present within the biofilm matrix itself. Whereas labeling with the fluorescent lectin M. amurensis, which has specificity for sialic acid in an α-2-3-linkage to galactose, was similarly seen distributed throughout the biofilm matrix, labeling with this lectin has been attributed to binding to 5-acetylneuraminic acid of the LOS expressed by NTHI resident within the biofilm. Serial sections that were identically lectin labeled, after treatment with neuraminidase to remove sialic acid, showed reduced labeling with both lectins. The biofilm formed 21 days after challenge of the middle ear with NTHI strain 86-028NP exhibited labeling patterns identical to those seen here when day 4 or 5 biofilms were labeled with these fluorochrome-conjugated lectins, both before and after neuraminidase treatment (data not shown).
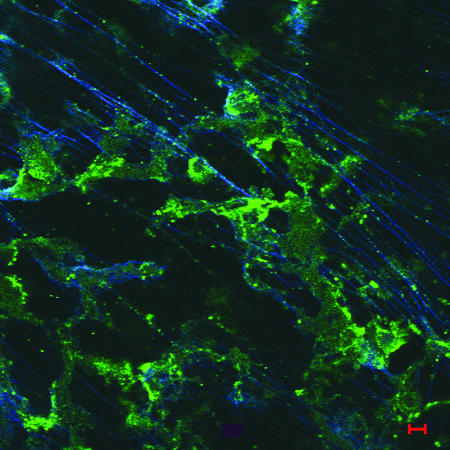
In an attempt to demonstrate the presence of type IV pili or pilin protein in the biofilm produced in the chinchilla middle ear, we labeled frozen sections with rabbit polyclonal antibody directed against recombinant soluble PilA of NTHI strain 86-028NP and detected this primary antibody using AlexaFluor 488-conjugated goat anti-rabbit serum. Figure 2 shows the labeling of NTHI pilin protein within a 4-day biofilm as evident by small aggregates of green fluorescence among as-yet-sparse strands of dsDNA (blue fluorescence). Pilin protein can also be observed as pinpoint areas of green fluorescence tracking along these fine strands of dsDNA.
FIG. 2.
Immunofluorescent image of an immature biofilm (4 days) labeled with rabbit anti-soluble recombinant PilA and AlexaFluor 488 goat anti-rabbit (green). Fine, widely spaced dsDNA strands are labeled with DAPI and appear blue in this image, whereas type IV pilin protein can be seen in both small aggregates and pinpoint spots that are colocalized along the length of the DNA strands. Bar, 5 μm.
Immunofluorescent labeling of dsDNA within biofilms formed by NTHI in vivo.
DAPI, which labels dsDNA, was originally intended for use here as a host cell nuclear counterstain; however, it became evident upon higher-magnification examination of our biofilm sections that in addition to labeling of the nuclei of host cells that appeared to be infiltrating the bacterial biofilm, there was additional labeling with DAPI that did not have the regular size and shape of a eukaryotic cell nucleus. In immature (4- to 5-day) in vivo-formed biofilms, there was clear DAPI labeling of fine strands of material present within the biofilm matrices (Fig. 2). This labeling increased significantly over time and, due to the intricate structure and dense character of the DAPI labeling, suggested that dsDNA might play a predominant role in biofilms formed by NTHI in vivo. We thereby cut thicker (10-μm) sections from a 21-day biofilm in order to allow for greater depth of field in confocal imaging and labeled theses sections both for type IV pilin protein and for dsDNA.
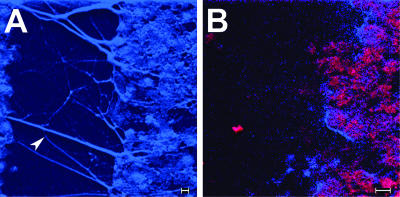
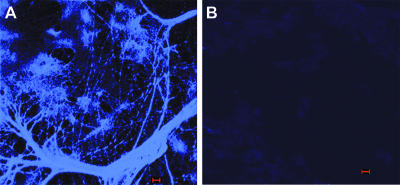
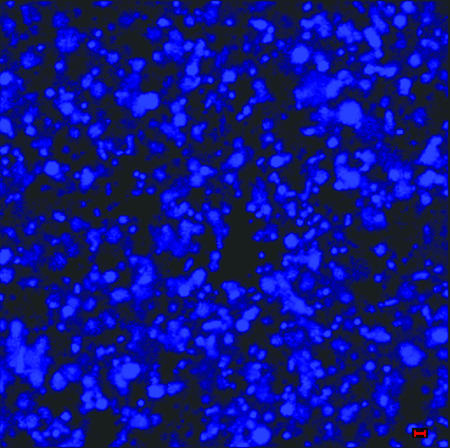
The biofilm formed by NTHI in vivo contained a significant amount of dsDNA which could be readily observed in 10-μm DAPI-stained sections. Widely spaced and thick, rope-like strands of dsDNA were seen crossing water channels (Fig. 3A) bordered by biofilm matrix material that was heavily populated with bacteria (Fig. 3B). To confirm that these strands and meshwork were indeed comprised of dsDNA, serial sections were treated with DNase prior to incubation with DAPI. DNase treatment removed all DAPI labeling (Fig. 4), whereas treatment with RNase had no effect (data not shown). To determine if the dsDNA present in the matrix of the biofilm formed by NTHI in vivo was of bacterial or eukaryotic origin, we allowed NTHI strain 86-028NP to form a biofilm in vitro, using a continuous-flow chamber and in the absence of eukaryotic cells. The matrix of this biofilm was also heavily labeled with DAPI (Fig. 5), suggesting that the bacterial cells themselves were a primary source of dsDNA observed in the matrix of an in vivo-formed NTHI biofilm.
FIG. 3.
Composite of high-resolution confocal images of a water channel present in a 21 day in vivo-formed biofilm. (A) Thick, rope-like strands of DAPI-labeled dsDNA can be seen as blue fluorescent filaments spanning the water channel (arrowhead). Also, dense clusters of what appear to be aggregates of bacteria are visible. (B) To confirm that these clusters were indeed NTHI, we dual labeled serial sections with both DAPI and polyclonal chinchilla anti-NTHI strain 86-028NP OMP antiserum that was detected with protein A-conjugated AlexaFluor 546, which imparts red fluorescence. One can observe clusters of NTHI (appearing red and fuchsia due to the heavy fluorescence in both the blue and red channels) populating the biofilm which borders the depicted water channel. Bars, 5 μm.
FIG. 4.
DNA present within a biofilm formed by NTHI strain 86-028NP in the chinchilla middle ear 21 days after challenge. The biofilm within the middle ear as well as the mucosal layer of the inferior bulla was embedded in OCT compound and snap frozen, and 10-μm sections were placed on slides. (A) dsDNA, labeled with DAPI, forms a complex network consisting of some thick strands, areas of aggregation, and thin web-like strands, giving a three-dimensional structure to the biofilm. (B) Serial section of the same biofilm after DNase treatment, confirming that the web-like mesh and strands seen in panel A were indeed comprised of dsDNA. Bars, 5 μm.
FIG. 5.
A biofilm produced in vitro by NTHI strain 86-028NP in a continuous-flow chamber was labeled with DAPI to detect the presence of dsDNA in the matrix when this organism was allowed to form a biofilm in the absence of eukaryotic cells. The image shows the presence of a significant amount of dsDNA in the biofilm matrix, as had been observed in those biofilms formed in vivo. These data suggest that the bacterial cells themselves are a primary source of dsDNA that is present in the NTHI-produced biofilm matrix. Bar, 1 μm.
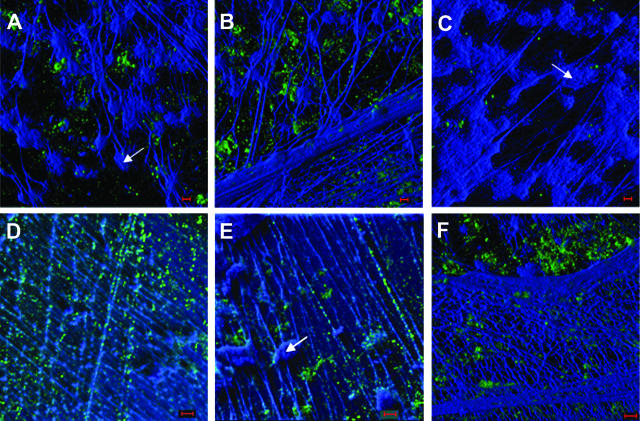
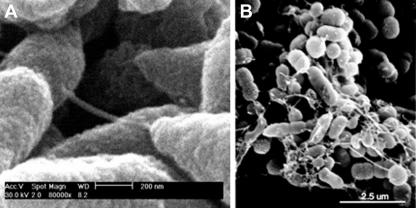
Immunolabeling further demonstrated the presence of type IV pilin protein within the biofilm matrix. In Fig. 6, among the DAPI-labeled interlaced network of thin strands of dsDNA, as well as what appears to be a heavier woven meshwork of dsDNA, pilin protein appears as the green fluorescence. The pilin protein often appeared as aggregates in the plane below the dsDNA mesh when the biofilm mass was imaged from an outermost surface (Fig. 6A, B, and F). When imaged from planes located more deeply within the biofilm matrix, where the dsDNA strands were thinner and more widely spaced, labeling of pilin protein could often be demonstrated as located on top of or tracking along the surface of many of these strands (Fig. 6D and E). To investigate whether Tfp might be providing some structural stability to the biofilm, perhaps by serving as an intrabacterial bridge, we examined NTHI that was allowed to grow on the surface of NHBE cells by SEM. We observed distinct interbacterial bridges, which were of the approximate dimensions of the type IV pilus expressed by NTHI (5), extending between individual bacterial cells present within small biofilm communities formed on the surface of respiratory epithelial cells in culture (Fig. 7).
FIG. 6.
Composite image of three-dimensional reconstructions of z-stack images from a 21-day biofilm formed in the chinchilla middle ear by NTHI strain 86-028NP and labeled for NTHI Tfp pilin protein (green fluorescence), as well as with DAPI for labeling of the dsDNA (blue fluorescence). DAPI labeling of what appear to be numerous host cell nuclei is clearly evident (A, C, and E) (arrows), as well as multiple strands forming a network of both fine, widely spaced strands (D and E) and a dense interwoven meshwork (B and F) within the biofilm. Pilin protein (or possibly NTHI that is expressing Tfp) is seen both as small aggregates (A, B and F) and tracking along fine dsDNA strand (D and E). Images are taken from different planes of sectioning within the same biofilm, where panels B and F represent the outside edge of the biofilm and panels A, C, D, and E represent more interior aspects, collectively showing the varied density but clear contribution of dsDNA to the structure of the biofilm formed by NTHI in the mammalian middle ear. Bars, 5 μm.
FIG. 7.
Composite of SEM images of biofilms formed by NTHI in in vitro cell culture systems. Structures seen extending between bacterial cells (NTHI strain 86-028NP) which formed a biofilm on NHBE cells are consistent in diameter with that reported for the type IV pilus expressed by NTHI (A). Similar structures were observed when NTHI strain 1128 was allowed to grow as a biofilm on human oropharyngeal cells (B).
Live/dead labeling.
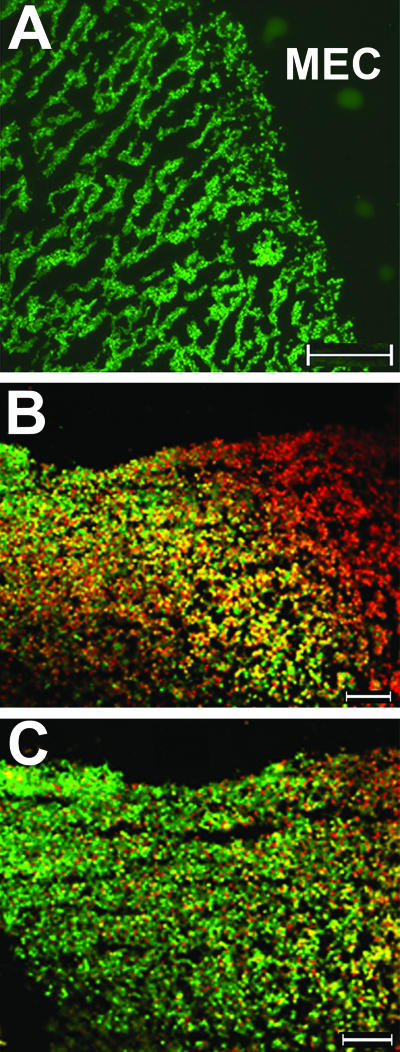
We showed previously that NTHI strain 2019 forms a well-defined, highly structured, and viable biofilm in the chinchilla middle ear as soon as 4 days after challenge (19). However, in that earlier study, we did not examine additional strains, nor did we assess the relative viability of NTHI in an in vivo-formed biofilm at later time points, where extended residence in the mammalian host may have resulted in reduced bacterial viability. Here, fluorescent vital staining confirmed that, like strain 2019, NTHI strain 86-028NP present within biofilms formed 4 to 5 days after challenge of the chinchilla host were uniformly viable (Fig. 8A). Moreover, the biofilm formed by strain 86-028NP within 5 days of challenge of the middle ear had a well-defined architecture and contained numerous water channels, again similar to that observed for biofilms formed in vivo by NTHI strain 2019.
FIG. 8.
Composite of fluorescent vital-stained images of OCT-embedded biofilms recovered from a chinchilla middle ear 5 or 21 days after challenge. Green fluorescence indicates the presence of viable bacteria, red fluorescence indicates dead bacteria, and dark areas (no bacterial cells) represent water channels within the biofilm. (A) Higher-magnification image of a 5-day middle ear biofilm (MEC, middle ear cavity). (B) Lower-magnification image of a day 21 biofilm which appears to contain mostly dead bacteria and some host cells. (C) Serial section of the day 21 biofilm shown in panel B after DNase treatment, demonstrating the presence of largely viable bacteria after extracellular DNA had been removed. Bars, 100 μm.
Imaging of a day 21 biofilm using the same fluorescent vital stain suggested to us that at this later time point the biofilm was largely nonviable (Fig. 8B). However, due to both our new understanding that there was an abundance of dsDNA present within these mature biofilms, as described above, and knowledge of the mechanism of labeling for this vital stain, we wondered if the appearance of red fluorescence might be a false readout of dead bacteria that was perhaps due to incorporation of propidium iodide by the dense network of extracellular dsDNA strands present in the mature biofilm matrix. To investigate this phenomenon further, we treated serial biofilm sections with either buffer or DNase for 2 h prior to labeling them with the vital fluorescent stain. As can be seen in Fig. 8C, removal of the extracellular dsDNA from the biofilm matrix via DNase treatment resulted in a fluorescent image that was now predominantly green, suggesting that the NTHI resident within these mature biofilms was actually as viable as its day 5 counterpart.
DISCUSSION
The literature on the role of biofilms in OM is still relatively young, as this paradigm-shifting hypothesis was originally put forth by Rayner et al. in 1998 (28). Nevertheless, data to date clearly show that NTHI, like most if not all bacteria, is readily capable of forming a biofilm both in vitro (15, 24, 41) and in vivo (13, 14, 17, 19, 34, 36). Work from several groups has now begun to both characterize the proteome of biofilm-growing NTHI (15, 40), as well as to determine that the proteins, or epitopes thereof, expressed by NTHI when grown in a sessile manner are different from those growing planktonically (24). Moreover, Greiner and colleagues (16) and later Jurcisek et al. (19) demonstrated that sialylated LOS, or endotoxin, with sialic acid in a specific linkage group to galactose was a key component of in vitro- as well as in vivo-formed biofilms, respectively. Bouchet and colleagues (9) further showed that sialylated LOS is a major virulence factor in experimental OM induced by NTHI, whereas the Swords lab has demonstrated that sialylation of LOS promotes biofilm formation by NTHI (36). By increasing the phosphorylcholine content of LOS, biofilm-growing NTHI is also capable of reducing the net bioactivity of this molecule, suggesting that by inducing less inflammation, NTHI may be promoting its ability to resist clearance (18, 41). Ehrlich and colleagues (14) were the first to show that mature NTHI-induced biofilms formed in the middle ear of the chinchilla host within 5 days of challenge, and later this group demonstrated that NTHI, as well as Streptococcus pneumoniae and Moraxella catarrhalis, had formed biofilms on middle ear mucosal samples recovered from children with recurrent and/or chronic OM (17).
In the present study, we hypothesized that due to immunological pressure put on NTHI in vivo, there may be differences in the components that comprise the biofilm matrix over time. Further, due to the fact that there appear to be multiple components, in addition to sialylated LOS, that are incorporated into the NTHI-induced matrix formed in vitro, this likely might also be true for those formed in vivo. Thus, here we attempted to expand upon our initial characterization of an in vivo-formed NTHI biofilm. To this end, we were interested in determining whether or not type IV pilin protein might be associated with, or incorporated into, the biofilms formed in vivo by NTHI, as these structures have been shown to play a key role in biofilms formed by Pseudomonas aeruginosa and Neisseria gonorrhoeae (21, 26). We also wanted to determine if the presence of sialylated LOS in specific linkage groups changed over time, and finally, we were interested in following up on a previous observation (19) which suggested that host cells (likely monocytes or polymorphonuclear leukocytes) were intimately associated with the biofilms formed by NTHI in vivo.
Immunofluorescent confocal images showed that immature (day 4 or 5), as well as mature (day 21), biofilms were highly organized and contained both viable NTHI and numerous characteristic water channels. Both day 4 (or 5) and day 21 biofilms were labeled with S. nigra and M. amurensis, indicating the presence of sialylated LOS that was removed or diminished by treatment with neuraminidase. Labeling of type IV pilin protein was also evident throughout the biofilm matrix, both as small aggregates and tracking along thin dsDNA strands. A similar phenomenon has been reported for P. aeruginosa by both Walker et al. (38), and Allesen-Holm and colleagues (2), which resulted in the latter group hypothesizing that since type IV pili are known to bind DNA (1, 12, 37), the presence of DNA in the matrix might provide a substrate on which these bacteria could migrate in order to organize into the characteristic mushroom-like structures built in a P. aeruginosa biofilm. The observation of pilin protein on top of or tracking along strands of dsDNA present in in vivo-formed biofilms here not only confirmed that Tfp were expressed by NTHI within the middle ear, as we described recently (20; Jurcisek et al., submitted for publication), but further suggested that these pili (or pilin protein) may also provide some structural stability to the biofilm, perhaps by serving as an intrabacterial bridge. This hypothesis was supported by SEM examination of NTHI growing in a biofilm community on the surface of NHBE cells. These data are additionally supported by our observation that a pilA mutant of NTHI strain 86-028NP, while able to survive in the middle ear of the chinchilla host, formed a less robust biofilm that was notably incapable of maintaining adherence to the mucosal epithelial surface lining the middle ear compared to the parental isolate (20; Jurcisek et al., submitted for publication).
Whereas we were indeed able to label what appeared (based on uniform size and shape) to be the nuclei of many host cells in association with in vivo-formed biofilms, we also observed an extensive interlaced meshwork or matrix comprised of dsDNA which was similarly labeled with DAPI. Treatment with DNase, but not RNase, removed DAPI labeling of both host cell nuclei and this intricate meshwork of dsDNA strands. The amount and arrangement of these DNA strands gave the impression not only that DNA was a key component of an NTHI-induced biofilm, shown here to notably increase in density with time in vivo, but also that this DNA-containing matrix could likely impart significant structural stability to the biofilm community. The relatively greater density of DNA at the outer edges of a biofilm mass compared to that observed within deeper aspects further suggested a compartmentalization of components within these NTHI-induced biofilms. A similar compartmental distribution of additional individual components has recently been reported for biofilms formed by NTHI in vitro (40).
Our data thus show that, like for S. pneumoniae, Streptococcus mutans, P. aeruginosa, Pseudomonas putida, Rhodococcus erythropolis, and Variovorax paradoxus, release of DNA into a biofilm appears to be a common theme among a group of bacteria (2, 31, 35) that now includes NTHI. The presence of DNA in the biofilm matrix has recently been noted to play an important role in both biofilm development and stability (2, 22, 23, 27, 31, 32, 35). In the latter regard, extracellular DNA has been shown to act as a cell-to-cell interconnecting component in biofilms produced by P. aeruginosa and S. pneumoniae (2, 23, 27, 31). However, whereas LOS in a specific linkage group as well as several outer membrane proteins, surface proteins, and adhesins (15, 16, 19, 25, 36, 40) have been identified in NTHI-produced biofilms, the presence of dsDNA in these matrices has, to the best of our knowledge, never been described. In fact, to date, studies characterizing the presence of DNA in any biofilm matrix have focused on those formed in vitro. Here, we describe the presence of DNA in the extracellular matrix of an NTHI biofilm formed in vivo and present evidence which suggests that due to the intricate arrangement and density of the dsDNA strands, this abundant DNA likely plays a key role in providing structural stability to the biofilm. Given the basket weave-like matrix of DNA, it is also possible that dsDNA contributes to providing an element of protection from phagocytic cells, as well as possibly effectors of innate and acquired immunity. Whereas the DNA present in the biofilm matrix formed by NTHI in vivo could be derived from the bacteria, from host cells, or from a combination of both, as there is evidence in the literature for each possibility (2, 31, 38), here we showed that a biofilm formed by NTHI in vitro in the absence of eukaryotic cells also contained an abundant amount of dsDNA. This observation suggested that the bacterial cells themselves served as a primary source of dsDNA present in the biofilm matrix. Understanding the mechanism(s) by which NTHI contributes dsDNA to its own biofilm matrix is currently an area of active investigation in our lab.
Here we have employed immunofluorescent labeling of cryopreserved biofilms recovered from the middle ears of chinchillas, which serve as a rodent model of experimental OM, to demonstrate that both immature and mature NTHI-induced biofilms contained sialylated LOS; OMPs, including type IV pilin protein; and a significant amount of dsDNA. The DNA present within the biofilm matrix formed a structural scaffold of sorts by assembling into a network of closely spaced interwoven strands, as well as extending as sparse but thicker rope-like strands, across water channels. Whereas no differences were seen between immature (day 4 or 5) or mature (day 21) in vivo-formed biofilms in terms of OMP content; LOS content or biochemistry, or bacterial viability, mature biofilms incorporated a notably increased amount of dsDNA that had now assumed a dense mesh or basket weave of strands. This meshwork appeared to be particularly concentrated at the outer margins of the biofilm mass, whereas there was a less dense distribution of finer strands located throughout the interior of the biofilm. NTHI could be seen heavily populating these multicomponent biofilm matrices. We are currently investigating the mechanism of dsDNA release into NTHI-formed biofilms.
Acknowledgments
This work was supported by grant R01 DC03915 to L.O.B. from NIDCD/NIH.
Footnotes
Published ahead of print on 23 February 2007.
REFERENCES
- 1.Aas, F. E., M. Wolfgang, S. Frye, S. Dunham, C. Lovold, and M. Koomey. 2002. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol. Microbiol. 46:749-760. [DOI] [PubMed] [Google Scholar]
- 2.Allesen-Holm, M., K. B. Barken, L. Yang, M. Klausen, J. S. Webb, S. Kjelleberg, S. Molin, M. Givskov, and T. Tolker-Nielsen. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59:1114-1128. [DOI] [PubMed] [Google Scholar]
- 3.Bakaletz, L. O. 2004. Developing animal models for polymicrobial diseases. Nat. Rev. Microbiol. 2:552-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakaletz, L. O. 2002. Otitis media, p. 259-298. In K. A. Brogden and J. M. Guthmiller (ed.), Polymicrobial diseases. ASM Press, Washington, DC. [PubMed]
- 5.Bakaletz, L. O., B. D. Baker, J. A. Jurcisek, A. Harrison, L. A. Novotny, J. E. Bookwalter, R. Mungur, and R. S. Munson, Jr. 2005. Demonstration of type IV pilus expression and a twitching phenotype by Haemophilus influenzae. Infect. Immun. 73:1635-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakaletz, L. O., B. J. Kennedy, L. A. Novotny, G. Duquesne, J. Cohen, and Y. Lobet. 1999. Protection against development of otitis media induced by nontypeable Haemophilus influenzae by both active and passive immunization in a chinchilla model of virus-bacterium superinfection. Infect. Immun. 67:2746-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borriello, G., L. Richards, G. D. Ehrlich, and P. S. Stewart. 2006. Arginine or nitrate enhances antibiotic susceptibility of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 50:382-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borriello, G., E. Werner, F. Roe, A. M. Kim, G. D. Ehrlich, and P. S. Stewart. 2004. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 48:2659-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouchet, V., D. W. Hood, J. Li, J. R. Brisson, G. A. Randle, A. Martin, Z. Li, R. Goldstein, E. K. Schweda, S. I. Pelton, J. C. Richards, and E. R. Moxon. 2003. Host-derived sialic acid is incorporated into Haemophilus influenzae lipopolysaccharide and is a major virulence factor in experimental otitis media. Proc. Natl. Acad. Sci. USA 100:8898-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chole, R. A., and B. T. Faddis. 2003. Anatomical evidence of microbial biofilms in tonsillar tissues: a possible mechanism to explain chronicity. Arch. Otolaryngol. Head Neck Surg. 129:634-636. [DOI] [PubMed] [Google Scholar]
- 11.Costerton, W., R. Veeh, M. Shirtliff, M. Pasmore, C. Post, and G. Ehrlich. 2003. The application of biofilm science to the study and control of chronic bacterial infections. J. Clin. Investig. 112:1466-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig, L., N. Volkmann, A. S. Arvai, M. E. Pique, M. Yeager, E. H. Egelman, and J. A. Tainer. 2006. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell 23:651-662. [DOI] [PubMed] [Google Scholar]
- 13.Daines, D. A., M. Bothwell, J. Furrer, W. Unrath, K. Nelson, J. Jarisch, N. Melrose, L. Greiner, M. Apicella, and A. L. Smith. 2005. Haemophilus influenzae luxS mutants form a biofilm and have increased virulence. Microb. Pathog. 39:87-96. [DOI] [PubMed] [Google Scholar]
- 14.Ehrlich, G. D., R. Veeh, X. Wang, J. W. Costerton, J. D. Hayes, F. Z. Hu, B. J. Daigle, M. D. Ehrlich, and J. C. Post. 2002. Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of otitis media. JAMA 287:1710-1715. [DOI] [PubMed] [Google Scholar]
- 15.Gallaher, T. K., S. Wu, P. Webster, and R. Aguilera. 2006. Identification of biofilm proteins in non-typeable Haemophilus Influenzae. BMC Microbiol. 6:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greiner, L. L., H. Watanabe, N. J. Phillips, J. Shao, A. Morgan, A. Zaleski, B. W. Gibson, and M. A. Apicella. 2004. Nontypeable Haemophilus influenzae strain 2019 produces a biofilm containing N-acetylneuraminic acid that may mimic sialylated O-linked glycans. Infect. Immun. 72:4249-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall-Stoodley, L., F. Z. Hu, A. Gieseke, L. Nistico, D. Nguyen, J. Hayes, M. Forbes, D. P. Greenberg, B. Dice, A. Burrows, P. A. Wackym, P. Stoodley, J. C. Post, G. D. Ehrlich, and J. E. Kerschner. 2006. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296:202-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong, W., K. Mason, J. Jurcisek, L. Novotny, L. O. Bakaletz, and W. E. Swords. 2007. Phosphorylcholine decreases early inflammation and promotes the establishment of stable biofilm communities of nontypeable Haemophilus influenzae strain 86-028NP in a chinchilla model of otitis media. Infect. Immun. 75:958-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jurcisek, J., L. Greiner, H. Watanabe, A. Zaleski, M. A. Apicella, and L. O. Bakaletz. 2005. Role of sialic acid and complex carbohydrate biosynthesis in biofilm formation by nontypeable Haemophilus influenzae in the chinchilla middle ear. Infect. Immun. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jurcisek, J. A., J. E. Bookwalter, R. S. Munson Jr., and L. O. Bakaletz. 2006. The type IV pilus of nontypeable Haemophilus influenzae (NTHI) plays a role in the stabilization and structure of biofilms formed both in vitro and in vivo, abstr. D-109. Abstr. 106th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 21.Lappann, M., J. A. Haagensen, H. Claus, U. Vogel, and S. Molin. 2006. Meningococcal biofilm formation: stucture, development and phenotypes in a standardized continuous flow system. Mol. Microbiol. 62:1292-1309. [DOI] [PubMed] [Google Scholar]
- 22.Molin, S., and T. Tolker-Nielsen. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14:255-261. [DOI] [PubMed] [Google Scholar]
- 23.Moscoso, M., E. Garcia, and R. Lopez. 2006. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J. Bacteriol. 188:7785-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy, T. F., and C. Kirkham. 2002. Biofilm formation by nontypeable Haemophilus influenzae: strain variability, outer membrane antigen expression and role of pili. BMC Microbiol. 2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy, T. F., C. Kirkham, S. Sethi, and A. J. Lesse. 2005. Expression of a peroxiredoxin-glutaredoxin by Haemophilus influenzae in biofilms and during human respiratory tract infection. FEMS Immunol. Med. Microbiol. 44:81-89. [DOI] [PubMed] [Google Scholar]
- 26.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 27.Petersen, F. C., L. Tao, and A. A. Scheie. 2005. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J. Bacteriol. 187:4392-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rayner, M. G., Y. Zhang, M. C. Gorry, Y. Chen, J. C. Post, and G. D. Ehrlich. 1998. Evidence of bacterial metabolic activity in culture-negative otitis media with effusion. JAMA 279:296-299. [DOI] [PubMed] [Google Scholar]
- 29.Sanderson, A. R., J. G. Leid, and D. Hunsaker. 2006. Bacterial biofilms on the sinus mucosa of human subjects with chronic rhinosinusitis. Laryngoscope 116:1121-1126. [DOI] [PubMed] [Google Scholar]
- 30.Sauer, K. 2003. The genomics and proteomics of biofilm formation. Genome Biol. 4:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spoering, A. L., and M. S. Gilmore. 2006. Quorum sensing and DNA release in bacterial biofilms. Curr. Opin. Microbiol. 9:133-137. [DOI] [PubMed] [Google Scholar]
- 32.Sriramulu, D. D., H. Lunsdorf, J. S. Lam, and U. Romling. 2005. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 54:667-676. [DOI] [PubMed] [Google Scholar]
- 33.Stanley, N. R., and B. A. Lazazzera. 2005. Defining the genetic differences between wild and domestic strains of Bacillus subtilis that affect poly-gamma-dl-glutamic acid production and biofilm formation. Mol. Microbiol. 57:1143-1158. [DOI] [PubMed] [Google Scholar]
- 34.Starner, T. D., N. Zhang, G. Kim, M. A. Apicella, and P. B. McCray, Jr. 2006. Haemophilus influenzae forms biofilms on airway epithelia: implications in cystic fibrosis. Am. J. Respir. Crit. Care Med. 174:213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinberger, R. E., and P. A. Holden. 2005. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl. Environ Microbiol. 71:5404-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swords, W. E., M. L. Moore, L. Godzicki, G. Bukofzer, M. J. Mitten, and J. VonCannon. 2004. Sialylation of lipooligosaccharides promotes biofilm formation by nontypeable Haemophilus influenzae. Infect. Immun. 72:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Schaik, E. J., C. L. Giltner, G. F. Audette, D. W. Keizer, D. L. Bautista, C. M. Slupsky, B. D. Sykes, and R. T. Irvin. 2005. DNA binding: a novel function of Pseudomonas aeruginosa type IV pili. J. Bacteriol. 187:1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker, T. S., K. L. Tomlin, G. S. Worthen, K. R. Poch, J. G. Lieber, M. T. Saavedra, M. B. Fessler, K. C. Malcolm, M. L. Vasil, and J. A. Nick. 2005. Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect. Immun. 73:3693-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walters, M. C., III, F. Roe, A. Bugnicourt, M. J. Franklin, and P. S. Stewart. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webster, P., S. Wu, G. Gomez, M. Apicella, A. G. Plaut, and J. W. St Geme, III. 2006. Distribution of bacterial proteins in biofilms formed by non-typeable Haemophilus influenzae. J. Histochem. Cytochem. 54:829-842. [DOI] [PubMed] [Google Scholar]
- 41.West-Barnette, S., A. Rockel, and W. E. Swords. 2006. Biofilm growth increases phosphorylcholine content and decreases potency of nontypeable Haemophilus influenzae endotoxins. Infect. Immun. 74:1828-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]