Abstract
In an age of comparative microbial genomics, knowledge of the near-native architecture of microorganisms is essential for achieving an integrative understanding of physiology and function. We characterized and compared the three-dimensional architecture of the ecologically important cyanobacterium Prochlorococcus in a near-native state using cryo-electron tomography and found that closely related strains have diverged substantially in cellular organization and structure. By visualizing native, hydrated structures within cells, we discovered that the MED4 strain, which possesses one of the smallest genomes (1.66 Mbp) of any known photosynthetic organism, has evolved a comparatively streamlined cellular architecture. This strain possesses a smaller cell volume, an attenuated cell wall, and less extensive intracytoplasmic (photosynthetic) membrane system compared to the more deeply branched MIT9313 strain. Comparative genomic analyses indicate that differences have evolved in key structural genes, including those encoding enzymes involved in cell wall peptidoglycan biosynthesis. Although both strains possess carboxysomes that are polygonal and cluster in the central cytoplasm, the carboxysomes of MED4 are smaller. A streamlined cellular structure could be advantageous to microorganisms thriving in the low-nutrient conditions characteristic of large regions of the open ocean and thus have consequences for ecological niche differentiation. Through cryo-electron tomography we visualized, for the first time, the three-dimensional structure of the extensive network of photosynthetic lamellae within Prochlorococcus and the potential pathways for intracellular and intermembrane movement of molecules. Comparative information on the near-native structure of microorganisms is an important and necessary component of exploring microbial diversity and understanding its consequences for function and ecology.
Environmental and culture-based comparative genomic analyses have established that closely related bacteria can exhibit considerable genetic diversity (2, 38, 39, 48). Information on the comparative, near-native cellular structures of closely related microorganisms is an important component of understanding the functional consequences and significance of their genomic diversity. Although this has been studied intensively in heterotrophic bacteria, much less is known in this regard about the ecologically significant group of microbial phototrophs that play key roles in oxygen production, carbon fixation, and global biogeochemical cycles. Prochlorococcus is a globally important cyanobacterium that contributes nearly half of the net primary production in certain open ocean regions (3, 7, 14, 18, 35, 50, 52). Using cryo-electron tomography, we characterized and compared the three-dimensional (3D) structure of two Prochlorococcus strains in a near-native state. The two strains selected for this study represent the major Prochlorococcus ecotypes, eMIT9313 and eMED4 (18). MIT9313 belongs to a deeply branched clade and possesses a genome size (2.41 Mbp) comparable to that of the closely related marine cyanobacterium Synechococcus sp. strain WH8102 (2.43 Mbp) (39). Members of eMIT9313 have been found to be particularly abundant in deeper (>50 m) subtropical and tropical waters (18). In contrast, MED4 belongs to a large clade comprised of recently derived lineages and possesses one of the smallest genomes (1.66 Mbp) of any known photosynthetic organism (39). Cells of eMED4 are relatively numerous in deeper (>50 m) waters but have been found to be most abundant in surface and near-surface waters (18).
Although our understanding of Prochlorococcus genomics, physiology, and ecology has advanced significantly, our knowledge of the architecture of this ecologically important cyanobacterium is incomplete. Traditional chemical fixation techniques for preserving bacterial cells for conventional transmission electron microscopy have proven unsatisfactory for the preservation of major cellular structures in Prochlorococcus, including the cell wall and intracytoplasmic (photosynthetic) membranes (24). It is unknown how Prochlorococcus strains, which possess dissimilar genome sizes and thus genetic potential, differ in their cellular architecture and respond at the structural level to abiotic stress. This knowledge is essential for an integrative understanding of the molecular physiology and ecology of Prochlorococcus.
One major advantage of cryo-electron tomography is that cells can be preserved in the absence of chemical fixatives and visualized in a near-native state. Specimens with a thickness of 1 μm or less can be well preserved by plunge freezing and are suitable for studies aimed at characterizing 3D cellular organization (9, 46). Resolution of macromolecular complexes requires thinner specimens (100 nm to 200 nm), which can be achieved through cryoultramicrotomy.
In recent years, cryo-electron tomography has provided detailed information on the near-native, 3D structures and organizations of a range of specimens, including mammalian tissue, eukaryotic and prokaryotic microorganisms, viruses, organelles, and bacterial microcompartments (9, 12, 13, 17, 25, 27, 32, 41, 46, 53). Macromolecular complexes that have been visualized successfully include flagellum-related structures (33, 47), the cytoskeleton (22, 27), and retrovirus envelope protein complexes (8). This high-resolution technique has permitted us to resolve key differences between Prochlorococcus strains that were undetectable by conventional chemical fixation and transmission electron microscopy techniques. Our cryo-electron tomography data indicate that Prochlorococcus strains, which differ by less than 3% in their 16S rRNA gene sequences, have diverged substantially in key structural characteristics, including size, shape, cell wall structure, and the extent of their intracytoplasmic (photosynthetic) membrane system. The evolution of a streamlined cellular architecture, characteristic of the MED4 strain, is expected to have physiological consequences for niche differentiation in nutrient-limited open oceans.
MATERIALS AND METHODS
Cell culturing and preparation for cryo-electron tomography.
Prochlorococcus strains MIT9313 and MED4 were cultured under identical conditions in the laboratory. Both strains were grown in batch cultures in an artificial seawater medium at 21 ± 1°C and 10 μmol photons m−2 s−1. Illumination was provided by “cool white” fluorescent lights on a 14-h-light/10-h-dark cycle.
For plunge freezing, cells collected by centrifugation were resuspended in artificial seawater containing 20-nm colloidal gold particles, which served as alignment fiducial markers in the tomographic tilt series. A 5-μl aliquot of the specimen was applied to a glow-discharged 200-mesh Cu R3.5/1 Quantifoil grid (Quantifoil Microtools, Jena, Germany). Prior to plunge freezing in liquid ethane, each grid was blotted simultaneously from both sides with Whatman no. 1 filter paper. This procedure is known as “double blotting” (6). Chemical cryoprotectants, fixatives, or stains were not added to the samples at any point during preparation. Vitrified specimens were transferred immediately to liquid nitrogen for storage.
For frozen-hydrated cryoultramicrotome sections, cells collected by centrifugation were resuspended in dextran (molecular weight, 43,000; 20% final concentration), and an aliquot (∼1 μl) of the suspension was frozen using an HPM 010 high-pressure freezer (Bal-Tec, Balzers, Liechtenstein). The sample carrier utilized in this procedure was a 3-mm by 0.5-mm aluminum platelet, with a 2-mm cavity diameter and 0.2-mm cavity depth. Following high-pressure freezing, samples were stored under liquid nitrogen. Frozen-hydrated sections were cut at −160°C using a UCT ultramicrotome with an EM-FCS cryokit (Leica, Vienna, Austria) (16). The frozen-hydrated sections (80 nm to 200 nm) were collected on 200-mesh molybdenum Quantifoil grids that had been coated first with a 10-nm-thick continuous carbon film and then with colloidal gold particles.
Cryoelectron tomography.
Specimens were transferred from liquid nitrogen storage to a model 626 cryotransfer specimen holder (Gatan, Pleasanton, CA) and imaged at below −176°C with a JEM-4000FX electron microscope (JEOL, Tokyo, Japan) equipped with a Gatan GIF 2002 energy filter, operated in zero-loss mode with a slit width of 15 eV. In order to optimize contrast at the expected tomographic resolution, images were recorded at a 15-μm underfocus; at this defocus, the first minimum of the contrast transfer function corresponds to a spacing of 5 nm. All tomograms were recorded with a 1.8-nm pixel size relative to the specimen, but some additional images were recorded with a 1.0-nm pixel size. Specimens were tilted about a single axis in 1° angular increments over a 120° range. The total electron dose for a tilt series was 50 e−/Å2 for the frozen-hydrated sections and 70 e−/Å2 for the plunge-frozen whole cells. The resolution of these single-tilt tomograms is anisotropic. A rough estimate can be obtained from the formula d = πD/n (4), where D is the thickness of the cell suspension layer or the frozen-hydrated section and n is the number of projection images (120 in our case). The resolution along the tilt axis (y) is better than this estimate, and that along the z axis (depth) is worse, due to incomplete angular sampling (37). Thus, the “average” 3D resolution varies from about 6 nm for 170-nm-thick frozen-hydrated sections to about 20 nm for the 800-nm-thick whole MIT9313 cells.
Image processing, analysis, and 3D modeling.
Alignment of the two-dimensional projection images of a tilt series was accomplished utilizing the colloidal gold particles as fiducial markers (36). The SPIDER program (10) was used to calculate the 3D reconstructions by weighted back-projection. For MIT9313, the tomogram modeled contained 330 reconstruction slices and represented one of six reconstructed MIT9313 tomograms (number of reconstruction slices ranging from 290 to 360). For MED4, the tomogram modeled contained 200 reconstruction slices and represented one of six reconstructed MED4 tomograms (number of reconstruction slices ranging from 130 to 230). In addition to these reconstructed tomograms, approximately 35 and 82 projection views were collected for MIT9313 and MED4, respectively, with many of the individual projection views containing images of more than one cell. Tomograms were displayed and analyzed using ImageJ (http://rsb.info.nih.gov/ij/). Cell volumes were determined based on equations for a sphere (4/3πr3; r is the radius) for MED4 and a prolate spheroid (4/3πab2; a and b are the semimajor and semiminor axes, respectively) for MIT9313. For visualization, surface and volume rendering were accomplished with Amira 3D (Mercury Computer Systems, San Diego, CA).
Comparative genomic analyses.
Sequences used in the comparative genomic analyses were obtained from the complete Prochlorococcus MIT9313 and MED4 and marine Synechococcus WH8102 genomes (http://www.kazusa.or.jp/cyano/cyano.html) and from GenBank. Predicted amino acid sequences were aligned using the CLUSTALW program with the BLOSUM matrix. The identities summarized in Table 1 were calculated from pairwise sequence alignments in EMBOSS using the BLOSUM40 matrix (gap open = 10; gap extend = 0.2). Database accession numbers of the protein sequences presented in Fig. 4C and Table 1 are as follows: PMM0549 (MED4, CsoS1-1), PMM0022 (MED4, MurC), PMM0610 (MED4, MurF), PMM0197 (MED4, MurG), PMM0617 (MED4, MurI), PMM1709 (MED4, MraY), PMT1199 (MIT9313, CsoS1-2), PMT1206 (MIT9313, CsoS1-1), PMT0027 (MIT9313, MurC), PMT0401(MIT9313, MurF), PMT2101 (MIT9313, MurG), PMT0393 (MIT9313, MurI), PMT2264 (MIT9313, MraY), SYNW0029 (WH8102, MurC), SYNW1002 (WH8102, MurF), SYNW2333 (WH8102, MurG), SYNW1010 (WH8102, MurI), and SYNW2516 (WH8102, MraY).
TABLE 1.
Percent identities of the amino acid sequences of five putative peptidoglycan biosynthesis gene products in Prochlorococcus MED4 and MIT9313 and the marine Synechococcus strain WH8102
| Prochlorococcus strain | % Identity
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MurC
|
MurF
|
MurG
|
MurI
|
MraY
|
||||||
| MIT9313 | WH8102 | MIT9313 | WH8102 | MIT9313 | WH8102 | MIT9313 | WH8102 | MIT9313 | WH8102 | |
| MED4 | 37.5 | 35.8 | 34.8 | 36.2 | 37.6 | 35.7 | 36.9 | 34.9 | 36.1 | 35.7 |
| MIT9313 | 61.4 | 62 | 71.5 | 67.2 | 70.7 | |||||
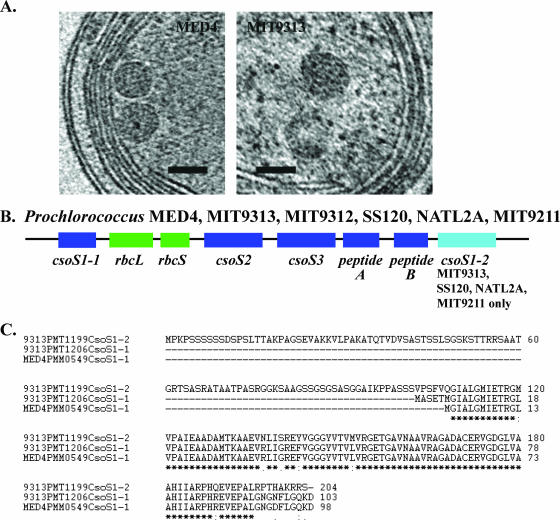
FIG. 4.
The near-native structure of Prochlorococcus α-carboxysomes and the overall genomic organization of carboxysome shell polypeptide genes are highly conserved. (A) Carboxysomes of MED4 and MIT9313 are located typically in the central cytoplasmic space and are polygonal. Depicted are 1.8-nm-thick slices from a tomogram of a frozen-hydrated MED4 cell and a frozen-hydrated cryoultramicrotome section of a high-pressure-frozen MIT9313 cell. Bars = 100 nm. (B) The overall genomic organization of genes (csoS1-1, csoS2, csoS3, peptide A gene, peptide B gene) encoding putative carboxysome shell polypeptides is highly conserved in Prochlorococcus strains MED4, MIT9313, MIT9312, SS120, NATL2A, and MIT9211. Although the csoS1-2 gene is present in the genomes of strains (MIT9313, SS120, NATL2A, MIT9211) belonging to low-light-adapted Prochlorococcus clades, it is missing from the genomes of strains (MED4, MIT9312) that are members of high-light-adapted clades. rbcL and rbcS encode the large and small subunits of RuBisCO, respectively. (C) Alignment of the predicted amino acid sequence of CsoS1-1 from MED4 with CsoS1-1 and CsoS1-2 from MIT9313. Although these amino acid sequences are highly conserved (an asterisk indicates identical amino acid residues, a semicolon indicates conserved residues, and a period indicates semiconserved residues), CsoS1-2 possesses an extended N-terminal region.
RESULTS AND DISCUSSION
Significance of divergent cell geometries in closely related strains.
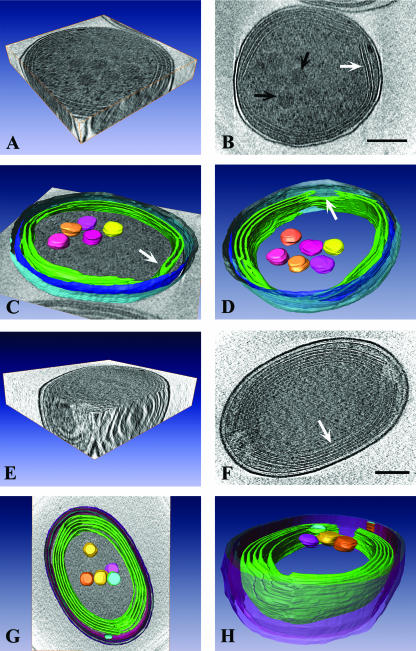
Our cryo-electron tomography-based determination of Prochlorococcus dimensions is the first reported for these cells preserved in a near-native state. Although Prochlorococcus is one of the smallest known photosynthetic organisms, differences in cell size and geometry distinguish strains cultured under identical conditions. MED4 cells are roughly spherical and are approximately 0.7 μm in diameter (Fig. 1A to D and 2A). In contrast, although MIT9313 grows slightly faster under the irradiance (10 μmol photons m−2 s−1) and temperature (21°C) conditions used in our experiments (doubling times of 57 h for MIT9313 and 72 h for MED4), MIT9313 cells are larger and more oval and are approximately 0.8 by 1.2 μm (Fig. 1E to H and 2D). The volume of MED4, which is approximately 0.2 μm3, is about two times less than the volume of MIT9313 (approximately 0.44 μm3).
FIG. 1.
Striking differences exist in the near-native cellular architecture of closely related Prochlorococcus strains. (A) 3D view of a single cell of Prochlorococcus MED4. (B) A 1.8-nm-thick tomographic slice of a MED4 cell. Two to three bands of intracytoplasmic (photosynthetic) lamellae are visible near the cell periphery (white arrow). Several polygonal structures resembling carboxysomes are clustered in the central cytoplasmic space (black arrows). (C) Surface-rendered model of a MED4 cell superimposed on a slice from a tomogram. Modeled 3D structures include the outer membrane (light blue), inner membrane (dark blue), intracytoplasmic lamellae (green), and carboxysomes (yellow, pink, orange). A distinct junction between two lamellae is visible (white arrow). (D) Surface-rendered model of MED4 showing that there are specific regions (white arrow) where the intracytoplasmic membranes (green) terminate, resulting in the formation of large gaps in the membrane bands. Note that an additional carboxysome is visible when the MED4 cell is viewed from this perspective. Color coding is as in panel C. (E) 3D view of a frozen-hydrated Prochlorococcus MIT9313 cell. (F) A 1.8-nm-thick tomographic slice of a MIT9313 cell. Several bands of intracytoplasmic lamellae are located near the cell membrane (white arrow). (G) Surface-rendered model of MIT9313 superimposed on a slice from a tomogram. Structures depicted include the cell wall (purple, pink, blue), extensive intracytoplasmic membrane system (green), and carboxysomes (yellow, pink, orange, blue). (H) Side view of a surface-rendered model of MIT9313, in which the regions where the intracytoplasmic membranes (green) terminate at the cell poles are visible. Note that these fenestrations in the intracytoplasmic membranes create areas of direct contact between the central cytoplasmic space and the region between the cell membrane and outermost intracytoplasmic membrane. Bars = 200 nm.
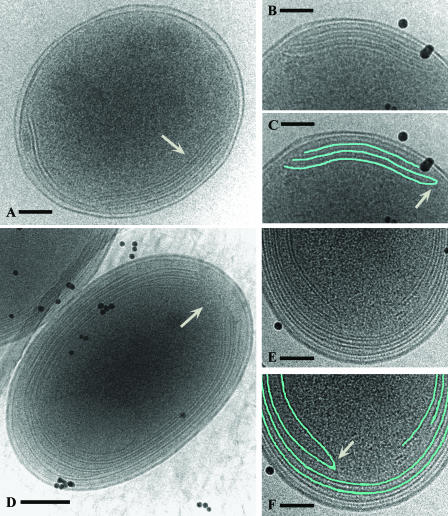
FIG. 2.
Prochlorococcus MED4 and MIT9313 cell structure as seen in frozen-hydrated whole mounts. In MED4, the intracytoplasmic membranes are typically apposed to one or two regions of the cell membrane (A, white arrow). (B and C) Higher magnifications of the internal membranes and a tracing (in blue) of their structure. A distinct junction between two intracytoplasmic membranes is shown (white arrow). (D, E, F) In MIT9313, the cell pole is the primary location for termination of the intracytoplasmic membranes and this results in prominent fenestrations (D, white arrow). (E) Structure of the intracytoplasmic membranes at a cell pole at a higher magnification; (F) tracing (in blue) of the same membranes. Note the distinct junction between two membranes (white arrow). In these images (B to F), the black particles are colloidal gold particles that were added to each sample and which serve as alignment fiducial markers. Bars = 100 nm (A, B, C, E, F) and 200 nm (D).
Differences in cell size between closely related bacteria are often correlated with differences in other cellular attributes. Previous studies using scanning electron microscopy have suggested that within the Prochlorococcus lineage, there exists a positive correlation between cell size and genome size and thus genetic potential (49). The results of the current study, where cells were preserved in a near-native state, support these previous observations. MIT9313, which possesses a genome size of 2.41 Mbp, was found to possess a larger cell size than MED4, which has the smallest genome (1.66 Mbp) of any known oxygen-evolving photosynthetic organism. Compared to MED4, the MIT9313 genome contains 243 additional unique genes (39).
The small size and roughly spherical geometry of MED4 have been reported for other Prochlorococcus strains, including SS120. The latter strain has been sized by electronic (Coulter) methods (approximately 0.6 μm in diameter) (30) and visualized by scanning electron microscopy (approximately 0.7 μm in diameter) (49). Differences in size, shape, and volume between these strains and MIT9313 are expected to have an impact on physiology, as several processes exhibit potential size and/or surface area dependence, including cellular metabolic requirements and rates, metabolite leakage rates, nutrient uptake rates per unit biomass, and interactions with light. As these processes are also affected by other cellular attributes, it will be important to determine how they differ between strains. It is well known that small spherical cells, such as MED4, possess very high ratios of surface area to volume and that the rate at which external materials and metabolites diffuse passively into a cell is enhanced as the cell surface area is maximized (34). However, the exchange of specific metabolites could be facilitated by the presence of transporters in the cell envelope. Our comparative genomic analyses indicate that the larger MIT9313 genome contains a greater proportion of transporters than the MED4 genome (39). Additional work is necessary in order to establish how these differences at the genetic level translate into dissimilarities in metabolite exchange rates in MIT9313 and MED4 under different nutrient conditions.
Furthermore, although the absorption of visible wavelengths of light is optimal theoretically for small spherical cells (34), it is critical to understand how other cellular properties, such as pigment concentration and intracytoplasmic membrane content, collectively affect the light absorption and photosynthetic characteristics of Prochlorococcus. Early work comparing roughly spherical Prochlorococcus (MED4/SS120-like cells) and rod-shaped Synechococcus cells suggested that Prochlorococcus cells possess a higher probability of absorbing, rather than scattering, light (30). This was attributed in part to their high photosynthetic pigment concentration and tiny size compared to Synechococcus (30). However, recent studies have suggested that distinct differences exist between Prochlorococcus strains in their light absorption characteristics and thus in their relative photosynthetic capacity. When Prochlorococcus MED4 and MIT9313 are cultured under low growth irradiance levels (<20 μmol photons m−2 s−1), MIT9313 possesses a higher growth rate, maximum quantum yield of photosynthesis, and maximum rate of photosynthesis than MED4 (29). These data indicate that the larger, oval MIT9313 cells are better adapted for photosynthetic growth at low irradiance levels than the smaller, spherical MED4 cells, and differences in several factors, including light-harvesting pigment concentrations and intracytoplasmic membrane content, must play a role.
Recent field studies on the distribution and abundance of MIT9313 and MED4 ecotypes (eMIT9313 and eMED4) in the Atlantic Ocean indicate that while eMIT9313 is most abundant deep in the water column (50 to 200 m), eMED4 is actually found throughout the water column (surface to 200 m) and occurs at particularly high cell densities from the surface to about 125 m (18). Thus, in the natural environment, eMED4 is not restricted in its distribution to surface and near-surface waters but is capable of thriving under a variety of irradiance conditions. In the oceans, several environmental factors interact to influence the overall distribution and abundance of Prochlorococcus ecotypes. Although light is a key variable, other factors, including nutrient concentrations, are expected to affect the population dynamics of Prochlorococcus ecotypes (18).
Evolution of a reduced cell wall in Prochlorococcus MED4.
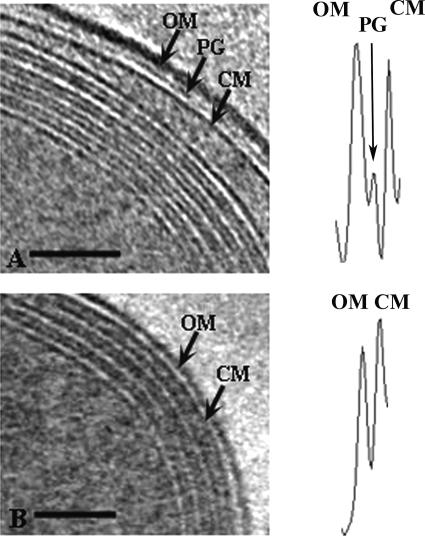
Striking differences exist in the cell wall ultrastructure of the Prochlorococcus MED4 strain. These differences, which have been undetectable using conventional transmission electron microscopy (24), were prominent in our tomograms of frozen hydrated Prochlorococcus cells. Cyanobacterial cell walls are similar structurally to those of gram-negative bacteria but share some characteristics with those of gram-positive bacteria (15). In MIT9313, the cell wall is approximately 34 nm thick and is composed of an outer membrane, a distinct peptidoglycan layer, and an inner membrane (Fig. 3A). The peptidoglycan layer in MIT9313 is approximately 4 nm thick. This is considerably less than the approximately 16-nm thickness we observed for the peptidoglycan layer in the marine cyanobacterium Synechococcus WH8102 (C. Ting, unpublished data) and less than the thickness reported for other cyanobacteria (15). Notably, the cell wall of the MED4 strain is significantly reduced, and at 19 nm it is only about half the width of the MIT9313 cell wall (Fig. 3B). This difference is due in part to a significantly thinner outer membrane, which is approximately 6 nm thick in MED4 and approximately 12 nm thick in MIT9313 (Fig. 3). Furthermore, the cell wall of MED4 is characterized by a reduced periplasmic space and the absence of a prominent peptidoglycan layer. When visible, the peptidoglycan layer of MED4 was less dense and narrower. Density profiles of the cell walls underscore these differences between MIT9313 and MED4. The MIT9313 cell wall density profile has an additional peak, which is associated with the prominent peptidoglycan layer, and the peak for the outer membrane is wider than in MED4 (Fig. 3).
FIG. 3.
Prochlorococcus MED4 possesses a minimal cell wall architecture compared to MIT9313. Shown are 1.8-nm-thick slices from tomograms of frozen-hydrated MIT9313 (A) and MED4 (B) cells, with the cell wall area depicted. Cell wall structures indicated include the outer membrane (OM), peptidoglycan layer (PG), and cell membrane (CM). Note the layers of intracytoplasmic lamellae adjacent to the cell membrane in both cells. Density profiles of the wall region are shown adjacent to the corresponding cell images. The cell wall structure represented by a particular density peak is indicated, and these density profiles emphasize the differences in cell wall structure between MED4 and MIT9313. Bars = 100 nm.
Although the cell walls in both MIT9313 (Fig. 1F, 2D to F, and 3A) and MED4 (Fig. 1B, 2A to C, and 3B) are quite uniform at different positions in a single cell, a few regions could be identified in the MED4 cell wall where the periplasm appears to be a bit wider. This could be due in part to structural dissimilarities between the MED4 and MIT9313 cell walls that in turn affect the extent to which the protoplast can move within the periplasmic space. A degree of variability in the width of the cell walls of the gram-negative bacteria Escherichia coli and Pseudomonas aeruginosa was reported by Matias et al. (26), who attributed this to protoplast movement. These authors also suggested that the width of the cell envelope could be affected by the presence of efflux transporter complexes that are typically anchored in the cell membrane and span the width of the periplasm to associate with channels in the outer membrane (26). Although several of such efflux transporters have been identified in the MIT9313 and MED4 genomes (39), their biochemical properties and localization in the cell wall are currently unknown.
Bacterial cell walls form the barrier between the cytoplasm and the external environment and have a key role in several functions, including the movement of nutrients and other compounds into and out of the cell, the preservation of cell shape, and protection against environmental stresses (5, 15, 20, 54). In addition, the wall counters intracellular osmotic pressure and thus assists in preventing the rupture of the cytoplasmic membrane and cell death. The strength of the cell wall is attributable in large part to the peptidoglycan layer (5). The striking differences between MED4 and MIT9313 in the extent of their peptidoglycan layer and in the overall thickness of their cell wall suggest that dissimilarities in the overall robustness of their walls must exist.
However, the synthesis of a prominent peptidoglycan layer could be costly to a cell, in part because nitrogen is an important structural component of the peptide cross-links and N-acetyl groups. Bacteria capable of surviving with a significantly reduced peptidoglycan layer might have a competitive advantage under nutrient-limited conditions, such as those found in large regions of the open oceans that are dominated by Prochlorococcus. While in certain habitats (i.e., coastal regions or freshwater lakes), a prominent peptidoglycan layer might be necessary because it provides mechanical stability to the wall and resistance to chemical substances (15, 40), areas such as the open ocean are relatively more stable and pristine environments in which the evolution of a reduced cell wall might confer a selective advantage.
Our comparative genomic analyses of MIT9313 and MED4 indicate that several of the genes involved in peptidoglycan biosynthesis possess low sequence identities and might be diverging. The two strains possess similar complements of putative peptidoglycan biosynthesis genes, including murA, murB, murC, murD, murE, murF, murG, murI, mraY, ddl, and pbp. However, the amino acid sequence identities for the predicted products of these genes are not very high (<52%) between MED4 and MIT9313. Notably, the particularly low level of sequence conservation (35% to 38% sequence identity) for MurC, MurF, MurG, MurI, and MraY suggests that the functions of these proteins might be diverging (Table 1). In contrast, comparisons between MIT9313 and marine Synechococcus WH8102 indicate that these same gene products (MurC, MurF, MurG, MurI, and MraY) share 61% to 72% amino acid identity (Table 1).
Near-native structure and organization of α-carboxysomes.
Through their associated carbonic anhydrase activity and their compartmentalization of ribulose bisphosphate carboxylase oxygenase (RuBisCO), carboxysomes contribute to the optimization of cellular CO2 fixation (19). Two different types of carboxysomes have been identified, and those found in form 1A and form 1B RuBisCO-containing organisms are α-carboxysomes and β-carboxysomes, respectively. Prochlorococcus possesses form 1A RuBisCO and thus α-carboxysomes, and it has been grouped with the α-cyanobacteria (14, 42). At present, detailed information on the structure of α-carboxysomes from α-cyanobacteria is unavailable (1), and our study is the first to report on their near-native structure in Prochlorococcus.
The carboxysomes of MED4 and MIT9313 are polygonal (Fig. 4A) and appear to be similar in size and shape to the β-carboxysomes characterized in Synechocystis sp. strain PCC 6803 (19). Previous work in Prochlorococcus employing immunogold labeling and conventional transmission electron microscopy demonstrated that RuBisCO is localized in these structures (24). Our tomograms indicate that the carboxysomes of Prochlorococcus MED4 and MIT9313 tend to cluster in groups in the central cytoplasmic space (Fig. 1B, D, and G and 4A). This clustering of the carboxysomes might further facilitate carbon fixation, as CO2 leaked from any carboxysome could enter and be fixed directly in an adjacent one. The carboxysomes of MED4 are smaller than those of MIT9313, with diameters of approximately 90 nm and 130 nm, respectively.
Carboxysomes are associated with a polypeptide shell that serves not only to sequester the enzymes involved in carbon fixation but also to regulate metabolite flow (19). Our comparative genomic analyses indicate that differences in the carboxysome polypeptide shells of MED4 and MIT9313 are likely to exist. The MIT9313 genome contains a gene encoding an additional carboxysome shell protein (CsoS1-2) that is not present in the MED4 genome (Fig. 4B). The predicted amino acid sequence of CsoS1-2 shares high (78%) identity with CsoS1-1 of MIT9313 but is nearly twice the length (204 residues) of the MIT9313 CsoS1-1 (103 residues) due to an extended N-terminal region (Fig. 4C). Notably, the gene encoding CsoS1-2 is also present in the genomes of three other strains (SS120, NATL2A, and MIT9211), which are similar to MIT9313 in that they belong to low-light-adapted Prochlorococcus clades and grow particularly well at low irradiance levels. All of these strains contain both the CsoS1-1 and CsoS1-2 polypeptide genes (Fig. 4B). In contrast, the gene encoding CsoS1-2 is absent from the genomes of MED4 and MIT9312, both of which belong to the high-light-adapted Prochlorococcus clade. As the overall chromosomal organization of carboxysome shell polypeptide genes is highly conserved in the genomes of these six Prochlorococcus strains (Fig. 4B), a potential role for the CsoS1-2 polypeptide in facilitating carbon fixation under low-irradiance conditions needs to be addressed.
Comparative 3D architecture of the intracytoplasmic lamellae.
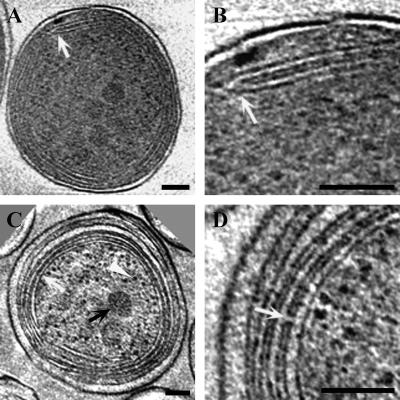
A key structure in the Prochlorococcus cell is the intracytoplasmic membrane system. In cyanobacteria, these membranes are critical for photosynthesis as well as other fundamental metabolic processes (11, 23, 28, 31, 44, 45, 51). When grown under identical conditions of irradiance level (10 μmol photons m−2 s−1), temperature (21°C), and nutrients, MIT9313 synthesizes a more extensive network of internal membranes than MED4. MIT9313 cells possess three to five distinct layers of intracytoplasmic membranes, which are arranged concentrically near the cell periphery and which occupy a significant fraction of the cytoplasmic space (Fig. 1E to H, 2D to F, and 5C). These intracytoplasmic membranes are well ordered and are tightly appressed at the cell sides and less tightly appressed near or at the cell poles, where they are often associated with a larger luminal space. The lumen is consistently less dense than the cytoplasmic space. Although this tight packing of the internal membranes is possible because Prochlorococcus lacks phycobilisomes, the structure and organization of these membranes are distinct from those of other chlorophyll a/b-containing cyanobacteria (11, 28) in that the membrane layers are arranged uniformly and are not interspersed throughout the central cytoplasm. In contrast, MED4 possesses a less extensive internal membrane system, with only two to four membrane bands apposed to the cell membrane in one or two regions of the cell and sometimes extending around the entire cell periphery (Fig. 1A to D, 2A to C, and 5A and B). Previous work has established that at the growth irradiance levels used in our studies, MIT9313 possesses a significantly greater maximum quantum yield of photosynthesis and chlorophyll b/a ratio than MED4 (29). Since the light-harvesting and reaction center complex polypeptides involved in photosynthesis are associated with the intracytoplasmic membranes (24), differences in the organization and abundance of these internal membranes must have consequences for the photosynthetic physiology of these strains.
FIG. 5.
Prochlorococcus intracytoplasmic lamellae are connected at distinct junctions and by membrane-lined channels. (A and B) Slices (1.8 nm thick) from a tomogram of a frozen-hydrated MED4 cell. The region in panel A at the white arrow is depicted at a higher magnification in panel B. Note the distinct junction between two intracytoplasmic membranes (B, white arrow). (C and D) Two different 1.8-nm tomographic slices of frozen-hydrated cryoultramicrotome sections of a high-pressure-frozen MIT9313 cell. The defocus (15 μm) and tilt angle (120°) were the same as those used for the frozen-hydrated whole-mount samples. Structures visible in panel C include carboxysomes (black arrow), ribosomes (white arrowhead), and tightly appressed intracytoplasmic lamellae (white arrow). Note that the luminal space of a lamella is less dense than the surrounding cytoplasm. A channel connecting adjacent intracytoplasmic lamellae is shown in panel D (white arrow). The apparent differences in the protein density of the cytoplasm between MED4 (A and B) and MIT9313 (C and D) reflect differences in contrast in the projection images of these samples. Comparisons of MED4 and MIT9313 cells that were prepared using the same technique of rapid freezing in liquid ethane followed by whole-mount imaging of frozen-hydrated samples indicate that the density of the cytoplasm is similar in these strains. Bars = 100 nm.
Through cryoelectron tomography we have established that the internal membranes of Prochlorococcus form an extensive interconnected network. Notably, at the cell poles in MIT9313, the intracytoplasmic lamellae interconnect with other lamellae or possess prominent fenestrations (Fig. 1F to H and 2D to F). Distinct membrane junctions are also observed in MED4 (Fig. 1C, 2B and C, and 5A and B). Thus, in Prochlorococcus, the luminal space of the intracytoplasmic membranes intercommunicates to some degree. Similar interconnections among the internal membranes have not been observed in cryo-electron tomography studies on Synechocystis sp. strain PCC 6803 (51) or the marine Synechococcus strain WH8102 (Ting, unpublished). In MIT9313, the persistence of the intracytoplasmic membrane fenestrations at the cell poles (Fig. 1F to H and 2D) strongly suggests that they have a cellular function. One possibility involves the intracellular movement of compounds. The tight packing of the membranes at the cell sides could preclude efficient intracellular trafficking of macromolecules, smaller organic molecules, and other cytoplasmic components. In MIT9313, these components might move directly to and from the central cytoplasmic space at the cell pole regions, where these distinct fenestrations exist in the intracytoplasmic membranes. For both MIT9313 and MED4, our tomographic data do not provide evidence for physical continuity between the intracytoplasmic membranes and cell membrane. Although the internal membranes sometime extend quite close to the cell membrane, as reported recently for Synechocystis sp. strain PCC 6803 (23, 51), the region was not resolved sufficiently to allow unambiguous confirmation of contact.
Tomography of cryoultramicrotome sections of frozen-hydrated cells provided higher resolution of the extensive intracytoplasmic membrane system of MIT9313 (Fig. 5C and D). We were able to visualize for the first time distinct membrane-lined connections between the lumens of the tightly appressed membranes at the cell periphery (Fig. 5D). These membrane-lined channels were approximately 10 nm in length and 6 nm in diameter. Because these channels linking the luminal space of adjacent intracytoplasmic membranes are not abundant, it is unlikely that their major function involves facilitating the diffusion of solutes between adjacent compartments. However, they could contribute to maintaining a uniform electrochemical potential across the intracytoplasmic lamellae. Membrane connections between individual granum layers, as revealed in tomographic studies of higher plant chloroplasts, are thought to have a similar function (43). Maintenance and regulation of the electrochemical potential across photosynthetic membranes is critical for several key processes, including synthesizing ATP and preserving a luminal pH optimal for enzyme activity (21).
In an age of comparative genomics, high-resolution information on the native architecture of cells is essential for achieving an integrative understanding of physiology and function. We have used cryo-electron tomography to characterize the comparative 3D structure of Prochlorococcus strains in a near-native state, and we have discovered a pronounced streamlining of cellular architecture in a strain that belongs to a large clade comprised of recently differentiated lineages. Structural dissimilarities between closely related microorganisms can reflect differences in their potential to thrive under particular environmental conditions and have consequences for microbial niche differentiation.
Acknowledgments
This work was supported by the National Science Foundation (award no. MCB-0615680 to C.S.T.), Williams College (C.S.T., S.S.), the Woodrow Wilson National Fellowship Foundation (C.S.T.), an American Society of Plant Biologists Summer Undergraduate Research Fellowship (S.S.), and NIH-NCRR grant P41 RR01219 (P. I. J. Frank) that supports the Wadsworth Center's Resource for the Visualization of Biological Complexity as a National Biotechnological Resource (C.H., C.M., M.M.).
We thank the three anonymous reviewers of the manuscript for their insightful comments and helpful suggestions.
Footnotes
Published ahead of print on 20 April 2007.
REFERENCES
- 1.Badger, M. R., and G. D. Price. 2003. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J. Exp. Bot. 54:609-622. [DOI] [PubMed] [Google Scholar]
- 2.Bergthorsson, U., and H. Ochman. 1998. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol. Biol. Evol. 15:9-16. [DOI] [PubMed] [Google Scholar]
- 3.Campbell, L., H. B. Liu, H. A. Nolla, and D. Vaulot. 1997. Annual variability of phytoplankton and bacteria in the subtropical North Pacific Ocean at Station ALOHA during the 1991-1994 ENSO event. Deep-Sea Res. Part I 44:167. [Google Scholar]
- 4.Crowther, R. A., D. J. DeRosier, and A. Klug. 1970. The reconstruction of a three-dimensional structure from its projections and its application to electron microscopy. Proc. R. Soc. Lond. A 317:319-340. [Google Scholar]
- 5.Dmitriev, B., F. Toukach, and S. Ehlers. 2005. Towards a comprehensive view of the bacterial cell wall. Trends Microbiol. 13:569-574. [DOI] [PubMed] [Google Scholar]
- 6.Dubochet, J., M. Adrian, J. J. Chang, J. C. Homo, J. Lepault, A. W. McDowall, and P. Schultz. 1988. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21:129-228. [DOI] [PubMed] [Google Scholar]
- 7.DuRand, M. D., R. J. Olson, and S. W. Chisholm. 2001. Phytoplankton population dynamics at the Bermuda Atlantic Time-series station in the Sargasso Sea. Deep-Sea Res. Part II 48:1983-2003. [Google Scholar]
- 8.Forster, F., O. Medalia, N. Zauberman, W. Baumeister, and D. Fass. 2005. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc. Natl. Acad. Sci. USA 102:4729-4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank, J., T. Wagenknecht, B. F. McEwen, M. Marko, C. E. Hsieh, and C. A. Mannella. 2002. Three-dimensional imaging of biological complexity. J. Struct. Biol. 138:85-91. [DOI] [PubMed] [Google Scholar]
- 10.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 11.Giddings, T. H., N. W. Withers, and L. A. Staehelin. 1980. Supramolecular structure of the stacked and unstacked regions of the photosynthetic membranes of Prochloron sp., a prokaryote. Proc. Natl. Acad. Sci. USA 77:352-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimm, R., H. Singh, R. Rachel, D. Typke, W. Zillig, and W. Baumeister. 1998. Electron tomography of ice-embedded prokaryotic cells. Biophys. J. 74:1031-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grunewald, K., P. Desai, D. C. Winkler, J. B. Heymann, D. M. Belnap, W. Baumeister, and A. C. Steven. 2003. Three-dimensional structure of the herpes simplex virus from cryo-electron tomography. Science 302:1396-1398. [DOI] [PubMed] [Google Scholar]
- 14.Hess, W. R., G. Rocap, C. S. Ting, F. Larimer, S. Stilwagen, J. Lamerdin, and S. W. Chisholm. 2001. The photosynthetic apparatus of Prochlorococcus: insights through comparative genomics. Photosynthesis Res. 70:53-71. [DOI] [PubMed] [Google Scholar]
- 15.Hoiczyk, E., and A. Hansel. 2000. Cyanobacterial cell walls: news from an unusual prokaryotic envelope. J. Bacteriol. 182:1191-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsieh, C. E., A. Lieth, C. A. Mannella, J. Frank, and M. Marko. 2006. Towards high-resolution three-dimensional imaging of native mammalian tissue: electron tomography of frozen-hydrated rat liver sections. J. Struct. Biol. 153:1-13. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh, C. E., M. Marko, J. Frank, and C. A. Mannella. 2002. Electron tomographic analysis of frozen-hydrated tissue sections. J. Struct. Biol. 138:63-73. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, Z. I., E. R. Zinser, A. Coe, N. P. McNulty, E. M. S. Woodward, and S. W. Chisholm. 2006. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311:1737-1740. [DOI] [PubMed] [Google Scholar]
- 19.Kerfeld, C. A., M. R. Sawaya, S. Tanaka, C. V. Nguyen, M. Phillips, M. Beeby, and T. O. Yeates. 2005. Protein structures forming the shell of primitive bacterial organelles. Science 309:936-938. [DOI] [PubMed] [Google Scholar]
- 20.Koch, A. L. 1988. Biophysics of bacterial walls viewed as stress-bearing fabric. Microbiol. Rev. 52:337-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramer, D. M., J. A. Cruz, and A. Kanazawa. 2003. Balancing the central roles of the thylakoid proton gradient. Trends Plant Sci. 8:27-32. [DOI] [PubMed] [Google Scholar]
- 22.Kurner, J., A. S. Frangakis, and W. Baumeister. 2005. Cryo-electron tomography reveals the cytoskeletal structure of Spiroplasma melliferum. Science 307:436-438. [DOI] [PubMed] [Google Scholar]
- 23.Liberton, M., H. Berg, J. Heuser, R. Roth, and H. B. Pakrasi. 2006. Ultrastructure of the membrane systems in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. Protoplasma 227:129-138. [DOI] [PubMed] [Google Scholar]
- 24.Lichtlé, C., J. C. Thomas, and A. Spilar. 1995. Immunological and ultrastructural characterization of the photosynthetic complexes of the prochlorophyte Prochlorococcus (Oxychlorobacteria). J. Phycol. 31:934-941. [Google Scholar]
- 25.Mannella, C. A. 2006. Structure and dynamics of the mitochondrial inner membrane cristae. Biochim. Biophys. Acta 1763:542-548. [DOI] [PubMed] [Google Scholar]
- 26.Matias, V. R. F., A. Al-Amoudi, J. Dubochet, and T. J. Beveridge. 2003. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J. Bacteriol. 185:6112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medalia, O., I. Weber, A. S. Frangakis, D. Nicastro, G. Gerisch, and W. Baumeister. 2002. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science 298:1209-1213. [DOI] [PubMed] [Google Scholar]
- 28.Miller, K. R., J. S. Jacob, T. Burger-Wiersma, and H. C. Matthijs. 1988. Supramolecular structure of the thylakoid membrane of Prochlorothrix hollandica: a chlorophyll b-containing prokaryote. J. Cell Sci. 91:577-586. [DOI] [PubMed] [Google Scholar]
- 29.Moore, L. R., and S. W. Chisholm. 1999. Photophysiology of the marine cyanobacterium Prochlorococcus: ecotypic differences among cultured isolates. Limnol. Oceanogr. 44:628-638. [Google Scholar]
- 30.Morel, A., Y. H. Ahn, F. Partensky, D. Vaulot, and H. Claustre. 1993. Prochlorococcus and Synechococcus: A comparative study of their optical properties in relation to their size and pigmentation. J. Mar. Res. 51:617-649. [Google Scholar]
- 31.Mullineaux, C. W. 1999. The thylakoid membranes of cyanobacteria: structure, dynamics and function. Aust. J. Plant Physiol. 26:671-677. [Google Scholar]
- 32.Nicastro, D., A. S. Frangakis, D. Typke, and W. Baumeister. 2000. Cryo-electron tomography of neurospora mitochondria. J. Struct. Biol. 129:48-56. [DOI] [PubMed] [Google Scholar]
- 33.Nicastro, D., J. R. McIntosh, and W. Baumeister. 2005. 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc. Natl. Acad. Sci. USA 102:15889-15894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niklas, K. J. 1997. The evolutionary biology of plants, p. 219-244. University of Chicago Press, Chicago, IL.
- 35.Partensky, F., W. R. Hess, and D. Vaulot. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63:106-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penczek, P., M. Marko, K. Buttle, and J. Frank. 1995. Double-title electron tomography. Ultramicroscopy 60:393-410. [DOI] [PubMed] [Google Scholar]
- 37.Radermacher, M., and W. Hoppe. 1980. Properties of three-dimensionally reconstructed objects from projections by conical tilting compared to single axis tilting. Proc. 7th Eur. Congr. Electron Microsc. Den Haag 1:132-133. [Google Scholar]
- 38.Rappé, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 39.Rocap, G., F. W. Larimer, J. Lamerdin, S. Malfatti, P. Chain, N. A. Ahlgren, A. Arellano, M. Coleman, L. Hauser, W. R. Hess, Z. I. Johnson, M. Land, D. Lindell, A. F. Post, W. Regala, M. Shah, S. L. Shaw, C. Steglich, M. B. Sullivan, C. S. Ting, A. Tolonen, E. A. Webb, E. R. Zinser, and S. W. Chisholm. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042-1047. [DOI] [PubMed] [Google Scholar]
- 40.Rogers, H. J., H. R. Perkins, and J. B. Ward. 1980. Microbial cell walls and membranes. Chapman & Hall, London, United Kingdom.
- 41.Schmid, M. F., A. M. Paredes, H. A. Khant, F. Soyer, H. C. Aldrich, W. Chiu, and J. M. Shively. 2006. Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J. Mol. Biol. 364:526-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimada, A., S. Kanai, and T. Maruyama. 1995. Partial sequence of ribulose-1,5-bisphosphate carboxylase/oxygenase and the phylogeny of Prochloron and Prochlorococcus (Prochlorales). J. Mol. Evol. 40:671-677. [DOI] [PubMed] [Google Scholar]
- 43.Shimoni, E., O. Rav-Hon, I. Ohad, V. Brumfeld, and Z. Reich. 2005. The three-dimensional organization of higher-plant chloroplast thylakoid membranes revealed by electron tomography. Plant Cell 17:2580-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanier, G. 1988. Fine structure of cyanobacteria. Methods Enzymol. 167:157-172. [Google Scholar]
- 45.Stanier, R. Y., and G. Cohen-Bazire. 1977. Photosynthetic prokaryotes: the cyanobacteria. Annu. Rev. Microbiol. 31:225-274. [DOI] [PubMed] [Google Scholar]
- 46.Steven, A. C., and U. Aebi. 2003. The next ice age: cryo-electron tomography of intact cells. Trends Cell Biol. 13:107-110. [DOI] [PubMed] [Google Scholar]
- 47.Sui, H., and K. H. Downing. 2006. Molecular architecture of axonemal microtubule doublets revealed by cryo-electron tomography. Nature 442:475-478. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J. R., S. Pacocha, C. Pharino, V. Klepac-Ceraj, D. E. Hunt, J. Benoit, R. Sarma-Rupavtarm, D. Distel, and M. F. Polz. 2005. Genotypic diversity within a natural coastal bacterioplankton population. Science 307:1311-1313. [DOI] [PubMed] [Google Scholar]
- 49.Ting, C. S., E. Westly, and E. Russell-Roy. 2005. Genome diversification in marine cyanobacteria: implications for photosynthetic physiology and environmental stress response mechanisms, p. 614-616. In A. van der Est and D. Bruce (ed.), Photosynthesis: fundamental aspects to global perspectives. Alliance Communications Group, Lawrence, KS.
- 50.Ting, C. S., G. Rocap, J. King, and S. W. Chisholm. 2002. Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies. Trends Microbiol. 10:134-142. [DOI] [PubMed] [Google Scholar]
- 51.van de Meene, A. M. L., M. F. Hohmann-Marriott, W. F. Vermaas, and R. W. Roberson. 2006. The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch. Microbiol. 184:259-270. [DOI] [PubMed] [Google Scholar]
- 52.Vaulot, D., D. Marie, R. J. Olson, and S. W. Chisholm. 1995. Growth of Prochlorococcus, a photosynthetic prokaryote, in the equatorial Pacific Ocean. Science 268:1480-1482. [DOI] [PubMed] [Google Scholar]
- 53.Wagenknecht, T., C. E. Hsieh, B. K. Rath, S. Fleischer, and M. Marko. 2002. Electron tomography of frozen-hydrated isolated triad junctions. Biophys. J. 83:2491-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young, K. D. 2003. Bacterial shape. Mol. Microbiol. 49:571-580. [DOI] [PubMed] [Google Scholar]