Abstract
Hormogonia are the infective agents in many cyanobacterium-plant symbioses. Pilus-like appendages are expressed on the hormogonium surface, and mutations in pil-like genes altered surface piliation and reduced symbiotic competency. This is the first molecular evidence that pilus biogenesis in a filamentous cyanobacterium requires a type IV pilus system.
Cyanobacteria, such as Nostoc spp., differentiate specialized filaments, known as hormogonia, which serve as the infective units in the establishment of symbioses with plants (1, 31) and exhibit a surface-dependent form of motility known as gliding. Social gliding of myxobacteria (30) and the twitching motility observed in a wide range of gram-negative bacteria, including the unicellular cyanobacterium Synechocystis sp. strain PCC 6803 (7, 9, 48, 63), are associated with surface-expressed, retractile appendages known as type IV pili (Tfp). While there is evidence that hormogonia possess pilus-like structures that are expressed during their formation (14) and transcripts hybridizing to the pilus retraction gene, pilT, increase in abundance within the first few hours of hormogonium formation (16), direct evidence that these structures underlie their motility is still lacking. Similarly, questions remain over the contribution of pili to establishment of cyanobacterium-plant symbioses. Dick and Stewart (15) noted that pilus-like structures were important in the specificity of the Nostoc-Peltigera symbioses, but Johansson and Bergman (20) noted that they appeared to be of little significance in the establishment of Nostoc-Gunnera symbioses.
Now that the genome sequence for Nostoc punctiforme ATCC 29133 (PCC 73102) is available (http://genome.jgi-psf.org/finished_microbes/nospu/nospu.home.html), it is clear that it contains open reading frames (ORFs) that are predicted to encode proteins with similarity to Tfp biogenesis proteins. To address the possibility that hormogonia require a Tfp system, we inactivated six of the pil-like ORFs and examined the mutant phenotypes.
Identification of N. punctiforme pil-like genes.
Of 15 ORFs identified (Table 1), 6 were chosen for disruption analysis. The predicted amino acid sequence of the protein encoded by the pilT-like ORF NpR0117 is most similar (75% identity; 88% similarity) to PilT1 from Synechocystis strain PCC 6803 (Slr0161) and shares 54% identity (69% similarity) with PilT from Pseudomonas aeruginosa (GenBank accession no. M55524). The PilT-like NpF2507 protein mostly resembles PilT2 of Synechocystis (74% identity; 86% similarity). Both PilT-like proteins possess the conserved Walker box motifs characteristic of nucleotide binding proteins (Fig. 1). Walker box A, in ATP/GTP-binding consensus sequence, [AG]-X4-G-K-[ST], where X is a variable (PROSITE accession no. PDOC00017), is glycine rich (54, 57). Walker box B is also present in some of the bacterial type II secretion pathway (GSP) proteins (40, 54). The NpF2507 protein has a proline-rich N-terminal extension of about 74 amino acids that is also present in PilT2 of Synechocystis strain PCC 6803 (7). pilT-like NpR0117 is clustered with two other Nostoc pil-like genes pilB (NpR0118) and pilC (NpR0116), in the order pilB, pilT, and pilC. In Synechocystis strain PCC 6803, pilC is clustered with pilT but not with a pilB homologue (63), while pilC is downstream of pilB, and pilT is found at a different locus in P. aeruginosa (29).
TABLE 1.
List of putative pilus biogenesis genes in the Nostoc punctiforme ATCC 29133 genomea
| N. punctiforme gene | No. of aa residues encoded | Annotation (Nostoc genome) | Homologous gene in P. aeruginosa/Synechocystis strain PCC 6803b | Proposed function/productc | Mutant phenotypec | References |
|---|---|---|---|---|---|---|
| NpR0117* | 375 | pilT | pilT* | Pilus retraction | Hyperpiliated, nonmotile | 7, 8, 39, 40, 47, 57 |
| NpF2507* | 425 | pilT | pilT* | Pilus retraction/phototaxisd | Hyperpiliated, nonmotile/negative phototaxisd | 7, 8, 39, 40, 47, 57 |
| NpR2800* | 293 | Prepilin peptidase gene pulO | pilD* | Prepilin peptidase | Loss of motility and Tfp | 3, 7, 25, 38, 49, 50, 51, 63 |
| NpF0676* | 188 | Tfp assembly protein pilE | pilA | Prepilin | Loss of motility and Tfp | 6, 7, 28, 42, 47, 63 |
| NpF0067 | 226 | pulG pseudopilin | pilA | Prepilin | Loss of motility and Tfp | 6, 7, 28, 42, 47, 63 |
| NpF0068* | 322 | Hypothetical | pilA* | Prepilin | Loss of motility and Tfp | 6, 7, 28, 42, 47, 63 |
| NpF0069* | 232 | Tfp assembly protein FimT gene | pilA* | Prepilin | Loss of motility and Tfp | 6, 7, 28, 42, 47, 63 |
| NpF0073 | 185 | pulG pseudopilin gene | pilA* | Prepilin | Loss of motility and Tfp | 6, 7, 28, 42, 47, 63 |
| NpR0118 | 668 | pilB | pilB | Pilus extension | Loss of motility and Tfp | 28, 36, 52, 57, 63 |
| NpF2747 | 371 | pilB | pilB | Pilus extension | Loss of motility and Tfp | 28, 36, 52, 57, 63 |
| NpR0116 | 411 | GSP member pulF | pilC | Counteracts PilT-mediated pilus retraction | Loss of motility and Tfp | 36, 39 |
| NpF5005 | 368 | pilM | PilM | Unknown | Loss of motility and Tfp | 27, 63 |
| NpF5006 | 255 | pilN | pilN | Localization/stability of Tfp | Loss of motility and Tfp | 27, 63 |
| NpF5007 | 258 | pilO | pilO | Localization/stability of Tfp | Loss of motility and Tfp | 27, 63 |
| NpF5008 | 792 | pilQ | pilQ | Pore-forming secretin | Loss of motility and Tfp | 26, 63 |
*, ORFs chosen for characterization in the present study.
Nostoc pil-related genes were identified by searching the protein database derived from the genome of N. punctiforme with deduced amino acid sequences of the known P. aeruginosa and Synechocystis strain PCC 6803 pil proteins.
The proposed functions and mutant phenotypes are based primarily on that established for P. aeruginosa but is also based on that for Synechocystis, unless otherwise noted.
Phototaxis is a proposed function/phenotype for Synechocystis only.
FIG. 1.
Conserved motifs of the N. punctiforme PilT proteins (NpR0117 and NpF2507 proteins), P. aeruginosa PilT (Pseu), Synechocystis strain PCC 6803 PilT1, and Synechocystis strain PCC 6803 PilT2. Black boxes indicate identical or conserved residues in all five of the sequences. Protein sequences were aligned using the program ClustalX. The Walker box motifs (thick lines), the Asp box and the His box (asterisks) are indicated. Note that only the central regions of the PilT sequences are shown.
The NpR2800 protein resembles prepilin peptidases (PilD proteins), which process pilin and a number of other prepilin-like proteins (3, 38) (data not shown). NpR2800 shares most sequence similarity (48%; 63% identity) with the Synechocystis strain PCC 6803 prepilin peptidase (Slr1120) and contains at least six transmembrane helices, suggesting that the protein may be an integral membrane protein. The highly conserved two-pair cysteine signature, which is required for both proteolysis and methylation activities (25), is also present (data not shown).
The N. punctiforme genome has at least five ORFs encoding proteins containing features that characterize the pilus subunit (PilA), a unique leader peptide sequence, and high sequence conservation in the N-terminal region of the mature protein (data not shown). Other significant similarities include conserved Gly residue at position −1 that is essential for cleavage of the leader sequence, a conserved Phe (+1 position of the mature protein) and a Glu residue (+5 position of the mature protein). It is worth noting that pseudopilins, which are components of the GSP (18, 21, 37, 45, 51), are also synthesized as precursors and share extensive sequence similarity with PilA (18, 25).
Mutants were constructed by the insertion of an omega neomycin phosphotransferase gene into each ORF and the introduction of inactivated alleles into wild-type N. punctiforme as described previously (13). Complete segregation was achieved with all the mutants, except pilA-like ORFs NpF0068 and NpF0676.
Pilus structure on the cell surface of hormogonia.
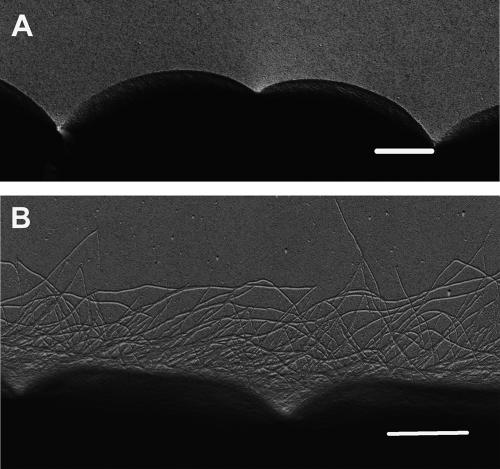
In agreement with previous observations (15, 20, 53), the surface of wild-type N. punctiforme hormogonia possesses peritrichously arranged pilus-like structures (Fig. 2B). Individually, these structures have a fairly uniform diameter of 7 to 10 nm, whereas the length is variable, extending up to 10 μm from the cell surface. We were able to discern only one Nostoc pilus morphotype, whereas others have determined that Synechocystis strain PCC 6803 has two morphologically distinct pilus types (7, 63). By contrast, the vegetative trichomes of the immotile parent filaments are largely devoid of pili (Fig. 2A). The pili of the pilA-like NpF0069 mutant were, in terms of abundance, distribution, and structure, indistinguishable from those expressed on the surface of wild-type hormogonia, implying that the gene is not responsible for the structural monomer of the Nostoc pilus-like appendages (data not shown).
FIG. 2.
Comparison of the cell surfaces of wild-type vegetative filaments and hormogonia. (A) Wild-type vegetative filaments; (B) wild-type hormogonia. Scale bars represent 1 μm. For electron microscopy, platinum wire (2 cm by 0.2 mm) was evaporated onto the surface of each sample by using an Edwards 306A high-vacuum coating unit and samples were viewed on a JEOL1200EX transmission electron microscope at 80 kV.
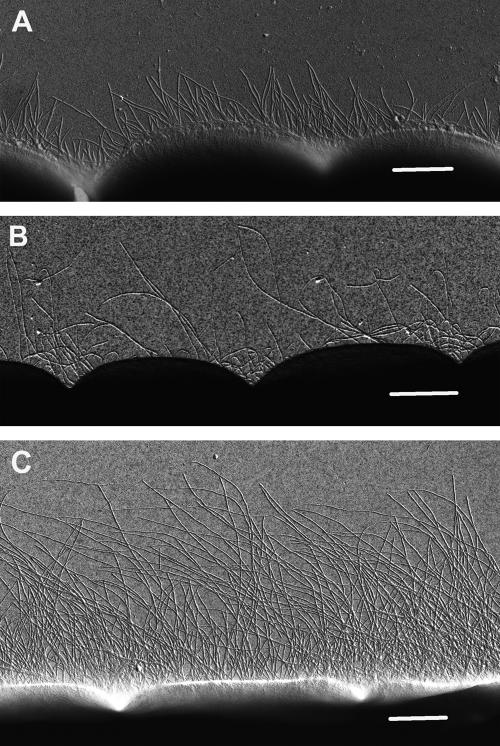
The phenotype of the pilT-like NpR0117 mutant is unmistakably the hyperpiliated phenotype (i.e., overproducing pili) (Fig. 3C) that is a typical result of mutations within pilT in other bacteria (7, 8, 32, 57, 59, 61). The mutation of the second pilT-like NpF2507 mutant resulted in pili that were slightly less abundant than those in the wild type, although their structure, in terms of length and diameter, appeared to be similar to that of the wild type (Fig. 3B). Remarkably, disruption of pilD-like NpR2800 gave rise to much shorter pili (up to 2.5 μm in length) than those of the wild type (up to 10 μm in length), although the pilus diameter (7 to 10 nm) was similar to that of the wild type (Fig. 3A). In other bacteria, mutations in pilD abolish motility and produce a nonpiliated phenotype (see, e.g., references 7, 19, and 25). The bald phenotype of the Synechocystis strain PCC 6803 pilD mutant is of special interest in that it implies that both of its pilus morphotypes are processed by the same prepilin peptidase (7). Searches of the N. punctiforme genomic sequences did not reveal any other PilD-like prepilin peptidases. The possibility of functional homologs, however, cannot be excluded. The truncated pilus phenotype reported here is unusual and may represent partially retracted or partially extended pili.
FIG. 3.
Presence of pilus-like appendages on the cell surface of hormogonia produced by pil mutants of N. punctiforme. (A) Hormogonia of the NpR2800 mutant (pilD homologue); (B) hormogonia of the NpF2507 mutant (pilT homologue); (C) hormogonia of the NpR0117 mutant (pilT homologue). Scale bars represent 1 μm. At least 50 filaments were observed for each mutant phenotype, and the electron micrographs shown are representative of each strain.
Symbiotic competencies of wild-type and mutant N. punctiforme strains.
The importance of cyanobacteria in their associations with plants is evidenced by the fact that host plants synthesize hormogonium-inducing factors (4, 10, 12, 22, 46, 55) and that both natural host plants and nonhost plants induce chemoattractive behavior in hormogonia (22, 34, 55). There is evidence, however, that hormogonium production per se is not sufficient for the establishment of symbiosis, because Johansson and Bergman (20) noted that noninfective Nostoc strains were also capable of forming motile hormogonia in the presence of the angiosperm Gunnera spp. (see also references 17 and 46). Fast hormogonium formation and rapid migration of these filaments appear critical to the successful establishment of artificial associations (33). Several factors therefore contribute to symbiotic competency. Tfp also function in adhesion (44). Indeed, adhesion, rather than motility, has been shown to be necessary for the initiation of biofilm formation in P. aeruginosa (11). Our wild-type hormogonia glided at rates of 0.7 μm s−1 to 1.7 μm s−1 (although it should be noted that motility was not always present in the wild type). Slight twitching of filaments, without forward progression, was detected by light microscopy in the NpF0069 and NpF2507 mutants. Motility in strains with NpR2800 and NpR0117 inactivated was not observed. To investigate the importance of functional pili in the establishment of symbiosis, the frequency of infection was estimated after coculture with the host bryophyte Blasia pusilla (Table 2) (22). Reconstitution of symbiotic associations was performed as described previously (60).
TABLE 2.
Mean infection frequencies of wild-type and mutant strains of N. punctiforme estimated after 14 and 28 days of coculture with Blasia
| N. punctiforme strain (putative homologue)a | 14 days
|
28 days
|
||||
|---|---|---|---|---|---|---|
| No. of coculture replicates | Infection frequency (%)b
|
No. of coculture replicates | Infection frequency (%)b
|
|||
| Mean | SD | Mean | SD | |||
| Wild-type N. unctiforme | 28 | 12.03 | 7.8 | 32 | 15.95 | 12 |
| NpF0069 mutant (pilA) | 7 | 3.49 | 1.12 | 8 | 6.00 | 2.72 |
| NpF2507 mutant (pilT) | 23 | 6.00 | 4.35 | 18 | 14.13 | 8.35 |
| NpR0117 mutant (pilT) | 17 | 0.48 | 0.56 | 18 | 1.52 | 1.03 |
| NpR2800 mutant (pilD) | 20 | 0.68 | 0.4 | 20 | 0.39 | 0.37 |
The putative homologue of each ORF is indicated in parentheses.
Values are expressed as the mean number of infected auricles (the symbiotic cavities of Blasia) expressed as a percentage of the total number of auricles counted for each determination (a minimum of 400 auricles were counted for each determination).
The wild-type-piliation phenotype observed with the pilA-like NpF0069 mutant implies that the gene is not required for the biosynthesis of the pilus-like appendages. It was therefore surprising that the NpF0069 mutant infected plant tissue at frequencies lower than those of the wild type, both during the early stages of the infection process and after a protracted period of coculture (Table 2). Some mutations in the P. aeruginosa pilA gene give rise to strains that are capable of assembling surface pili but have impaired motility and adherence (reviewed in reference 9). Further work is therefore required to fully discount the possibility that NpF0069 encodes the Nostoc pilin monomer. Alternatively, the protein encoded by pilA-like NpF0069 may represent a minor pilin (2, 58) that may support the activity of the major pilin in some aspect of motility and/or adhesion and therefore contribute to symbiotic competency. Synechocystis strain PCC 6803 has at least 10 pilA-like genes (5, 7, 62, 63), some of which are critical for motility and therefore probably represent components of the Tfp machinery.
For the pilD-like NpR2800 mutant, the frequency of infection after 14 days was 0.68%, decreasing to 0.31% after 28 days. Many of the symbiotic filaments were brown, implying that they were dead or dying and that the mutant was unable to grow effectively once it had reached the host symbiotic cavity. It is well established that prepilin peptidases, as well as processing pilin precursor, are also involved in the extracellular secretion of proteins (23, 24, 41, 50). pilD mutants are therefore pleiotrophic. Indeed, there is evidence to suggest that the Synechocystis strain PCC 6803 pilD gene is also critical for normal growth (63). The death of symbiotically associated Nostoc NpR2800 filaments opens the intriguing possibility that the Nostoc pilD-like gene may be required for some aspect of symbiotic growth.
Because wild-type hormogonia were occasionally immotile, we were unable to demonstrate irrefutably that the apparent loss of motility associated with the NpR0117 mutant resulted from mutation in the pilT-like ORF. Questions therefore remain as to whether motility, adhesion, or both function in the establishment of symbioses. Nevertheless, the NpR0117 (hyperpiliated) mutant infected Blasia tissue at a considerably lower frequency than the wild type (Table 2), implying that normal surface piliation is required for effective establishment of symbioses. Both N. punctiforme PilT-like proteins resemble other PilT proteins and share sequence homology with components of the GSP found in gram-negative bacteria (37, 43). Both proteins, however, lack the tetracysteine motif that is characteristic of GSP members and is also found in the DNA competence PilF protein (56) and the Tfp biogenesis protein PilB (35), indicating that the Nostoc PilT-like proteins can be included in the PilT family. Because the NpR0117 product resembles other PilT proteins and inactivation of the gene resulted in a hyperpiliated strain with a reduced capacity for infection, we suggest that NpR0117 at least is a component of the Nostoc piliation system and encodes the functional homologue of the pilus retraction protein PilT. The role of pilT-like NpF2507 is less clear. The NpF2507 protein mostly resembles PilT2 from Synechocystis strain PCC 6803, which is dispensable for motility but is believed to function in a signaling pathway that regulates the positive phototaxis of this cyanobacterium (7). The NpF2507 mutant infected Blasia tissue at an initial frequency 50% that of the wild type but reached a level equivalent to that of the wild type after 28 days (Table 2), implying a possible role during the early stages of the infection process. Whether NpF2507 is required for the direction of hormogonia motility (possibly involving chemotaxis-like regulatory elements), as may be the case with Synechocystis PilT2, remains to be investigated. Surprisingly, many of the plant structures that host cyanobacterial colonies are in regions that receive little or no light, suggesting that the chemoattractants released by the host plant override the elements that regulate phototactic behavior.
In conclusion, we report that N. punctiforme has a type IV piliation system and that inactivation of the pilT- and pilD-like components (NpR0117 and NpR2800, respectively) specifically altered the surface piliation levels and reduced symbiotic competency. We speculate that hormogonia motility is driven by type IV pili and that these structures contribute to the establishment of Nostoc-Blasia symbioses.
Acknowledgments
The financial support of BBSRC (grant 24/C14515) is gratefully acknowledged.
We also thank Adrian Hick for electron microscopy.
Footnotes
Published ahead of print on 6 April 2007.
REFERENCES
- 1.Adams, D. G. 2000. Symbiotic interactions, p. 523-561. In B. Whitton and M. Potts (ed.), Ecology of cyanobacteria: their diversity in time and space. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 2.Alm, R. A., and J. S. Mattick. 1996. Identification of two genes with prepilin-like leader sequences involved in type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 178:3809-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alm, R. A., and J. S. Mattick. 1997. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene 192:89-98. [DOI] [PubMed] [Google Scholar]
- 4.Bergman, B., A. Matveyev, and U. Rasmussen. 1996. Chemical signalling in cyanobacterial-plant symbioses. Trends Plant Sci. 1:191-197. [Google Scholar]
- 5.Bhaya, D., A. Takahashi, P. Shahi, and A. R. Grossman. 2001. Novel motility mutants of Synechocystis strain PCC 6803 generated by in vitro transposon mutagenesis. J. Bacteriol. 183:6140-6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhaya, D., N. Watanabe, T. Ogawa, and A. R. Grossman. 1999. The role of an alternative sigma factor in motility and pilus formation in the cyanobacterium Synechocystis PCC 6803. Proc. Natl. Acad. Sci. USA 96:3188-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhaya, D., N. R. Bianco, D. Bryant, and A. Grossman. 2000. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC 6803. Mol. Microbiol. 37:941-951. [DOI] [PubMed] [Google Scholar]
- 8.Bradley, D. E. 1980. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can. J. Microbiol. 26:146-154. [DOI] [PubMed] [Google Scholar]
- 9.Burrows, L. L. 2005. Weapons of mass retraction. Mol. Microbiol. 57:878-888. [DOI] [PubMed] [Google Scholar]
- 10.Campbell, E. L., and J. C. Meeks. 1989. Characteristics of hormogonia formation by symbiotic Nostoc spp. in response to the presence of Anthoceros punctatus or its extracellular products. Appl. Environ. Microbiol. 55:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang, P., and L. L. Burrows. 2003. Biofilm formation by hyperpiliated mutants of Pseudomonas aeruginosa. J. Bacteriol. 185:2374-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen, M. F., and J. C. Meeks. 1997. A hormogonium regulating locus, hrmUA, of the cyanobacterium Nostoc punctiforme strain ATCC 29133 and its response to an extract of a symbiotic plant partner Anthoceros punctatus. Mol. Plant-Microbe Interact. 10:280-289. [DOI] [PubMed] [Google Scholar]
- 13.Cohen, M. F., J. G. Wallis, E. L. Campbell, and J. C. Meeks. 1994. Transposon mutagenesis of Nostoc sp. strain ATCC 29133, a filamentous cyanobacterium with multiple differentiation alternatives. Microbiology 140:3233-3240. [DOI] [PubMed] [Google Scholar]
- 14.Damerval, T., G. Guglielmi, J. Houmard, and N. Tandeau de Marsac. 1991. Hormogonium differentiation in the cyanobacterium Calothrix: a photoregulated developmental process. Plant Cell 3:191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dick, H., and W. D. P. Stewart. 1980. The occurrence of fimbriae on a N2-fixing cyanobacterium which occurs in lichen symbioses. Arch. Microbiol. 124:107-109. [Google Scholar]
- 16.Doherty, H. M., and D. G. Adams. 1999. The organization and control of cell division genes expressed during differentiation in cyanobacteria, p. 453-461. In G. A. Peschek, W. Loffelhardt, and G. Schmetterer (ed.), The phototrophic prokaryotes. Kluwer Academic/Plenum Publishers, New York, NY.
- 17.Enderlin, C. S., and J. C. Meeks. 1983. Pure culture and reconstitution of the Anthoceros-Nostoc symbiotic association. Planta 158:157-165. [DOI] [PubMed] [Google Scholar]
- 18.Filloux, A., G. Michel, and M. Bally. 1998. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol. Rev. 22:177-198. [DOI] [PubMed] [Google Scholar]
- 19.Friedrich, A., C. Prust, T. Hartsch, A. Henne, and B. Averhoff. 2002. Molecular analyses of the natural transformation machinery and identification of pilus structures in the extremely thermophilic bacterium Thermus thermophilus strain HB27. Appl. Environ. Microbiol. 68:745-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansson, C., and B. Bergman. 1994. Reconstitution of the Gunnera manicata Linde symbioses: cyanobacterial specificity. New Phytol. 126:643-652. [Google Scholar]
- 21.Johnson, T. L., J. Abendroth, W. G. Hol, and M. Sandkvist. 2006. Type II secretion: from structure to function. FEMS Microbiol. Lett. 255:175-186. [DOI] [PubMed] [Google Scholar]
- 22.Knight, C. D., and D. G. Adams. 1996. A method for studying chemotaxis in nitrogen fixing cyanobacterium-plant symbiosis. Physiol. Mol. Plant Pathol. 49:73-77. [Google Scholar]
- 23.Lammertyn, E., and J. Anne. 2004. Protein secretion in Legionella pneumophila and its relation to virulence. FEMS Microbiol. Lett. 238:273-279. [DOI] [PubMed] [Google Scholar]
- 24.Liu, H., Y. Kang, S. Genin, M. A. Schell, and T. P. Denny. 2001. Twitching motility of Ralstonia solanacearum requires a type IV pilus system. Microbiology 147:3215-3229. [DOI] [PubMed] [Google Scholar]
- 25.Lory, S., and M. S. Strom. 1997. Structure-function relationship of type-IV prepilin peptidase of Pseudomonas aeruginosa—a review. Gene 192:117-121. [DOI] [PubMed] [Google Scholar]
- 26.Martin, P. R., M. Hobbs, P. D. Free, Y. Jeske, and J. S. Mattick. 1993. Characterization of pilQ, a new gene required for the biogenesis of type 4 fimbriae in Pseudomonas aeruginosa. Mol. Microbiol. 9:857-868. [DOI] [PubMed] [Google Scholar]
- 27.Martin, P. R., A. A. Watson, T. F. McCaul, and J. S. Mattick. 1995. Characterization of a five-gene cluster required for the biogenesis of type 4 fimbriae in Pseudomonas aeruginosa. Mol. Microbiol. 16:497-508. [DOI] [PubMed] [Google Scholar]
- 28.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 29.Mattick, J. S., C. B. Whitchurch, and R. A. Alm. 1996. The molecular genetics of type-4 fimbriae in Pseudomonas aeruginosa—a review. Gene 179:147-155. [DOI] [PubMed] [Google Scholar]
- 30.McBride, M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 31.Meeks, J. C. 1998. Symbiosis between nitrogen-fixing cyanobacteria and plants. BioScience 48:266-276. [Google Scholar]
- 32.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 33.Nilsson, M., U. Rasmussen, and B. Bergman. 2005. Competition among symbiotic cyanobacterial Nostoc strains forming artificial associations with rice (Oryza sativa). FEMS Microbiol. Lett. 245:139-144. [DOI] [PubMed] [Google Scholar]
- 34.Nilsson, M., U. Rasmussen, and B. Bergman. 2006. Cyanobacterial chemotaxis to extracts of host and nonhost plants. FEMS Microbiol. Ecol. 55:382-390. [DOI] [PubMed] [Google Scholar]
- 35.Nudleman, E., and D. Kaiser. 2004. Pulling together with type IV pili. J. Mol. Microbiol. Biotechnol. 7:52-62. [DOI] [PubMed] [Google Scholar]
- 36.Nunn, D., S. Bergman, and S. Lory. 1990. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J. Bacteriol. 172:2911-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nunn, D. N. 1999. Bacterial type II protein export and pilus biogenesis: more than just homologies? Trends Cell Biol. 9:402-408. [DOI] [PubMed] [Google Scholar]
- 38.Nunn, D. N., and S. Lory. 1991. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc. Natl. Acad. Sci. USA 88:3281-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okamoto, S., and M. Ohmori. 1999. Analysis of cyanobacterial motility in Synechocystis sp. PCC6803. Plant Cell Physiol. Suppl. 40:135. [Google Scholar]
- 40.Okamoto, S., and M. Ohmori. 2002. The cyanobacterial PilT protein responsible for cell motility and transformation hydrolyzes ATP. Plant Cell Physiol. 43:1127-1136. [DOI] [PubMed] [Google Scholar]
- 41.Paranjpye, R. N., J. C. Lara, J. C. Pepe, C. M. Pepe, and M. S. Strom. 1998. The type IV leader peptidase/N-methyltransferase of Vibrio vulnificus controls factors required for adherence to HEp-2 cells and virulence in iron-overloaded mice. Infect. Immun. 66:5659-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pasloske, B. L., and W. Paranchych. 1988. The expression of mutants pilins in Pseudomonas aeruginosa: fifth position glutamate affects pilin methylation. Mol. Microbiol. 2:489-495. [DOI] [PubMed] [Google Scholar]
- 43.Peabody, C. R., Y. J. Chung, M. R. Yen, D. Vidal-Ingigliardi, A. P. Pugsley, and M. H. Saier, Jr. 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149:3051-3072. [DOI] [PubMed] [Google Scholar]
- 44.Pizarro-Cerdá, J., and P. Cossart. 2006. Bacterial adhesion and entry into host cells. Cell 124:715-727. [DOI] [PubMed] [Google Scholar]
- 45.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rasmussen, U., C. Johansson, and B. Bergman. 1994. Early communication in the Gunnera-Nostoc symbiosis: plant-induced cell differentiation and protein synthesis in the cyanobacterium. Mol. Plant-Microbe Interact. 6:696-702. [Google Scholar]
- 47.Sastry, P. A., B. B. Finlay, B. L. Pasloske, W. Paranchych, J. R. Pearlstone, and L. B. Smillie. 1985. Comparative studies of the amino acid and nucleotide sequences of pilin derived from Pseudomonas aeruginosa PAK and PAO. J. Bacteriol. 164:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strom, M. S., and S. Lory. 1993. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47:565-596. [DOI] [PubMed] [Google Scholar]
- 50.Strom, M. S., D. N. Nunn, and S. Lory. 1991. Multiple roles of the pilus biogenesis protein pilD: involvement of pilD in excretion of enzymes from Pseudomonas aeruginosa. J. Bacteriol. 173:1175-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strom, M. S., D. N. Nunn, and S. Lory. 1994. Posttranslational processing of type IV prepilin and homologs by PilD of Pseudomonas aeruginosa. Methods Enzymol. 235:527-540. [DOI] [PubMed] [Google Scholar]
- 52.Turner, L. R., J. C. Lara, D. N. Nunn, and S. Lory. 1993. Mutations in the consensus ATP-binding sites of XcpR and PilB eliminate extracellular protein secretion and pilus biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 175:4962-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaara, T., and M. Vaara. 1988. Cyanobacterial fimbriae, p. 189-195. In L. Packer and A. N. Glazer (ed.), Methods in enzymology, vol. 167. Academic Press Inc., San Diego, CA. [Google Scholar]
- 54.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watts, S. D., C. D. Knight, and D. G. Adams. 1999. Characterisation of plant exudates inducing chemotaxis in nitrogen-fixing cyanobacteria, p. 679-684. In G. A. Peschek, W. Löffenhardt, and G. Schmetter (ed.), The phototrophic prokaryotes. Kluwer Academic/Plenum Publishers, New York, NY.
- 56.Watson, A. A., R. A. Alm, and J. S. Mattick. 1996. Identification of a gene, pilF, required for type 4 fimbrial biogenesis and twitching motility in Pseudomonas aeruginosa. Gene 180:49-56. [DOI] [PubMed] [Google Scholar]
- 57.Whitchurch, C. B., M. Hobbs, S. P. Livingston, V. Krishnapillai, and J. S. Mattick. 1991. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene 101:33-44. [DOI] [PubMed] [Google Scholar]
- 58.Winther-Larsen, H. C., M. Wolfgang, S. Dunham, J. P. van Putten, D. Dorward, C. Lovold, F. E. Aas, and M. Koomey. 2005. A conserved set of pilin-like molecules controls type IV pilus dynamics and organelle-associated functions in Neisseria gonorrhoeae. Mol. Microbiol. 56:903-917. [DOI] [PubMed] [Google Scholar]
- 59.Wolfgang, M., H. S. Park, S. F. Hayes, J. P van Putten, and M Koomey. 1998. Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 95:14973-14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong, F. C., and J. C. Meeks. 2002. Establishment of a functional symbiosis between the cyanobacterium Nostoc punctiforme and the bryophyte Anthoceros punctatus requires genes involved in nitrogen control and initiation of heterocyst differentiation. Microbiology 148:315-323. [DOI] [PubMed] [Google Scholar]
- 61.Wu, S. S., J. Wu, and D. Kaiser. 1997. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23:109-121. [DOI] [PubMed] [Google Scholar]
- 62.Yoshihara, S., X. Geng, and M. Ikeuchi. 2002. pilG Gene cluster and split pilL genes involved in pilus biogenesis, motility and genetic transformation in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 43:513-521. [DOI] [PubMed] [Google Scholar]
- 63.Yoshihara, S., X. Geng, S. Okamoto, K. Yura, T. Murata, M. Go, M. Ohmori, and M. Ikeuchi. 2001. Mutational analysis of genes involved in pilus structure, motility and transformation competency in the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 42:63-73. [DOI] [PubMed] [Google Scholar]