Abstract
The archaeon Methanocaldococcus jannaschii uses three different 2-oxoacid elongation pathways, which extend the chain length of precursors in leucine, isoleucine, and coenzyme B biosyntheses. In each of these pathways an aconitase-type hydrolyase catalyzes an hydroxyacid isomerization reaction. The genome sequence of M. jannaschii encodes two homologs of each large and small subunit that forms the hydrolyase, but the genes are not cotranscribed. The genes are more similar to each other than to previously characterized isopropylmalate isomerase or homoaconitase enzyme genes. To identify the functions of these homologs, the four combinations of subunits were heterologously expressed in Escherichia coli, purified, and reconstituted to generate the iron-sulfur center of the holoenzyme. Only the combination of MJ0499 and MJ1277 proteins catalyzed isopropylmalate and citramalate isomerization reactions. This pair also catalyzed hydration half-reactions using citraconate and maleate. Another broad-specificity enzyme, isopropylmalate dehydrogenase (MJ0720), catalyzed the oxidative decarboxylation of β-isopropylmalate, β-methylmalate, and d-malate. Combined with these results, phylogenetic analysis suggests that the pyruvate pathway to 2-oxobutyrate (an alternative to threonine dehydratase in isoleucine biosynthesis) evolved several times in bacteria and archaea. The enzymes in the isopropylmalate pathway of leucine biosynthesis facilitated the evolution of 2-oxobutyrate biosynthesis through the introduction of a citramalate synthase, either by gene recruitment or gene duplication and functional divergence.
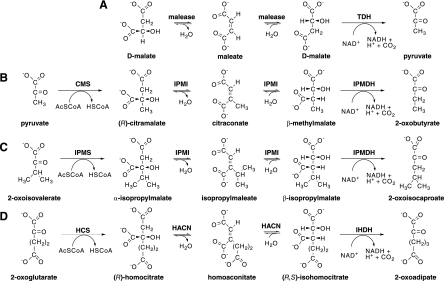
Three enzymes act in succession to extend the chain length of a 2-oxocarboxylic acid by one methylene group (Fig. 1). Using these reactions the isopropylmalate pathway in leucine biosynthesis converts 2-oxoisovalerate to 2-oxoisocaproate and the oxidative citric acid cycle converts oxaloacetate to 2-oxoglutarate. Additionally the homocitrate pathway for lysine biosynthesis converts 2-oxoglutarate to 2-oxoadipate. Three iterations of the homocitrate pathway convert 2-oxoglutarate to 2-oxosuberate in methanogenic coenzyme B biosynthesis (42).
FIG. 1.
Analogous 2-oxoacid elongation pathways. (A) Malease enzyme catalyzes the hydration of maleate to form d-malate, which can be oxidatively decarboxylated by TDH or by IPMDH. (B) The pyruvate pathway to 2-oxobutyrate for isoleucine biosynthesis requires CMS, IPMI, and IPMDH enzymes. (C) The isopropylmalate pathway for leucine biosynthesis uses IPMS and shares the IPMI and IPMDH enzymes with the pyruvate pathway. (D) The homocitrate pathway for lysine production in some organisms requires homocitrate synthase (HCS), HACN, and isohomocitrate dehydrogenase (IHDH) enzymes. Three iterations of the homocitrate pathway produce 2-oxosuberate for coenzyme B biosynthesis in methanogens. AcSCoA, acetyl-coenzyme A; HSCoA, coenzyme A.
The first reaction in each pathway is catalyzed by an acetyltransferase enzyme that condenses the acetyl group from acetyl-coenzyme A (CoA) with a 2-oxoacid to produce an α-hydroxydicarboxylate in an aldol-type reaction. Next, a hydrolyase enzyme catalyzes a dehydration reaction and the anti addition of water to a cis-unsaturated intermediate to produce a β-hydroxydicarboxylate. Finally, an NAD(P)+-dependent enzyme oxidizes the β-hydroxydicarboxylate, producing an enzyme-bound β-ketoacid intermediate, which undergoes divalent metal ion-catalyzed decarboxylation to form a new 2-oxoacid. In the leucine biosynthetic pathway, for example, the enzyme isopropylmalate synthase (IPMS) catalyzes the condensation of acetyl-CoA at the re face of 2-oxoisovalerate to produce 2S-2-isopropylmalate (α-isopropylmalate) (Fig. 1C) (8). The dehydrating isopropylmalate isomerase (IPMI) catalyzes the abstraction of water from α-isopropylmalate to give isopropylmaleate (cis-dimethylcitraconate) (17). A postulated rotation of this intermediate and the trans addition of water produces (2R,3S)-3-isopropylmalate (β-isopropylmalate) (6). Finally, the isopropylmalate dehydrogenase enzyme (IPMDH) catalyzes the NAD+-dependent oxidation and decarboxylation of β-isopropylmalate to produce 2-oxoisocaproate (5).
To make isoleucine, heterotrophic bacteria, cyanobacteria, and fungi use a threonine dehydratase enzyme to produce 2-oxobutyrate by the dehydration and deamination of l-threonine. Most methanogenic archaea lack this enzyme but still produce isoleucine. Metabolic labeling studies suggest that methanogens can assimilate exogenous propionate into isoleucine (11). In this pathway, propionyl-CoA is reductively carboxylated to produce 2-oxobutryate in a reaction that could be catalyzed by a ferredoxin-dependent oxidoreductase (3, 35). However, in the absence of exogenous propionate or isoleucine, methanogenic archaea and diverse bacteria extend the chain length of pyruvate to produce 2-oxobutyrate using a 2-oxoacid elongation pathway called either the pyruvate pathway or the citramalate pathway (Fig. 1B) (12, 13).
This pyruvate pathway to 2-oxobutyrate was first proposed to explain 14CO2 incorporation patterns in amino acids from the spirochetal bacterium Leptospira interrogans (7). The citramalate synthase enzyme (CMS) catalyzes the first step in this pathway, the condensation of acetyl-CoA with pyruvate to produce (R)-citramalate. CMS homologs from the methanogen Methanocaldococcus jannaschii (20) and L. interrogans (43) have been expressed in Escherichia coli and characterized in vitro. The isomerase and dehydrogenase enzymes that catalyze successive reactions have not been characterized from those organisms as pure proteins. Because the leucine biosynthetic IPMI from Saccharomyces cerevisiae also catalyzes the hydration of citraconate (27), we predicted that the M. jannaschii IPMI could function in both leucine and isoleucine pathways. Similarly, the M. jannaschii IPMDH homolog could also catalyze the oxidative decarboxylation of β-methylmalate to produce 2-oxobutyrate (34).
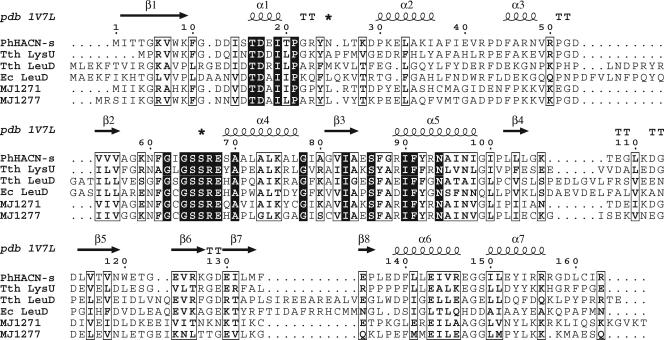
Fungal IPMI enzymes consist of a single protein with two domains. In bacteria and archaea these domains are expressed as separate protein subunits: a small subunit interacts with a large Fe4S4-containing subunit to form the active site. The genome sequence of M. jannaschii includes two homologs of each gene, encoding large and small subunits of IPMI (4). These genes are not linked on the M. jannaschii genome, and their sequences are too diverged from characterized homologs to assign specific functions. A crystal structure model of a small-subunit homolog from the archaeon Pyrococcus horikoshii suggested a highly conserved substrate-binding domain between β-strand 2 and α-helix 4 and a variable domain between α-helices 1 and 2 that could interact with the substrate's R-group side chain (Fig. 2) (45). There is no clear signature sequence in this variable domain that correlates with the enzymes' substrates; therefore, the specificity determinants for these isomerases are unknown and probably involve interactions with both subunits. Because the M. jannaschii subunits are 50% identical to each other, these proteins are all annotated as IPMI subunits in the Swiss-Prot sequence database (release 51.5, 23 January 2007). We predicted that one pair of enzymes would function as a broad-specificity isopropylmalate-citramalate isomerase and the other as the homoaconitase (HACN) in the homocitrate pathway for coenzyme B biosynthesis (42).
FIG. 2.
Sequence alignment of hydrolyase small subunits shown with the secondary structure of Pyrococcus horikoshii HACN (PhHACN-s; pdb 1V7L) (45). The additional sequences are Thermus thermophilus HACN (Tth LysU; RefSeq accession no. YP_005515.1), T. thermophilus isopropylmalate dehydratase (Tth LeuD; RefSeq accession no. YP_004837.1), Escherichia coli isopropylmalate dehydratase (Ec LeuD; Swiss-Prot accession no. P30126), Methanocaldococcus jannaschii MJ1271 (Swiss-Prot accession no. Q58667), and M. jannaschii MJ1277 (Swiss-Prot accession no. Q58673). Asterisks above the alignment indicate the loop regions that are predicted to bind substrate in the P. horikoshii HACN structure.
To identify the reactions catalyzed by the dehydratase enzymes, we heterologously expressed and purified the four combinations of large and small subunits of IPMI homologs from M. jannaschii. After reconstitution of their iron-sulfur centers, only the MJ0499/MJ1277 pair of proteins catalyzed the reversible hydration of citraconate and dehydration of (R)-citramalate. These subunits interacted to form a functional complex that efficiently catalyzes both partial reactions in the isopropylmalate pathway. Additionally, this broad-specificity enzyme had malease activity: it catalyzed the hydration of maleate to form d-malate. The M. jannaschii IPMDH homolog catalyzed the oxidative decarboxylation of β-methylmalate, as well as β-isopropylmalate and d-malate. Therefore the M. jannaschii IPMI and IPMDH proteins function in both the isopropylmalate pathway and the pyruvate pathway to 2-oxobutyrate. The latter pathway evolved by the duplication and functional divergence of an α-isopropylmalate synthase gene, with no requirement for new isomerase or dehydrogenase proteins.
MATERIALS AND METHODS
Chemicals.
(R)-Homocitrate was a gift from David Palmer (University of Saskatchewan) (29). α-Isopropylmalic acid was purchased from Aldrich. Other commercially available reagent grade chemicals were used without further purification.
Synthesis of β-isopropylmalate.
The physiologically relevant product of isopropylmalate dehydratase is (2R,3S)-3-isopropylmalate, which is commonly called β-isopropylmalate or threo-DS-3-isopropylmalate (6). Racemic dl-threo-3-isopropylmalate was synthesized as described by Yamada et al. (44), by the aldol condensation of ethyl isovalerate with veratraldehyde in the presence of hexamethylphosphoric triamide, followed by acetylation, oxidation, and acid hydrolysis reactions. The 1H nuclear magnetic resonance spectrum of the dl-threo-3-isopropylmalate product was consistent with resonances reported previously (44). dl-threo-3-isopropylmalic acid was purchased from Wako Chemicals for analytical comparison.
Cloning leuCD, aksDE, and leuB genes.
Genes were amplified from chromosomal DNA of Methanocaldococcus jannaschii JAL-1 using PCR. The large-subunit genes at loci MJ0499 (RefSeq accession no. NP_247475) and MJ1003 (NP_247997) were cloned between NdeI and KpnI sites of plasmid pCDF-Duet1 (Novagen) to create vectors pDG142 and pDG141, respectively (Table 1). The MJ1271 small-subunit gene (NP_248267) was cloned between NcoI and BamHI sites of plasmid pDG142 to create vector pDG159 and between the same sites in plasmid pDG141 to create pDG152. The MJ1277 small-subunit gene (NP_248273) was cloned between NcoI and BamHI sites of plasmid pDG142 to create pDG164 and between the same sites in plasmid pDG141 to create pDG157. The leuB gene at locus MJ0720 (NP_247705) was amplified using PCR and cloned between the NdeI and BamHI sites of plasmid pET-19b (Novagen) to create vector pMYL1. The translational start site for that protein was originally predicted to be encoded by a GTG codon, which can be recognized as an initiator codon in some archaea (20). However, there is an ATG codon downstream from the GTG codon that is closer to the predicted start site of conserved, orthologous archaeal genes. Because the GTG codon appears to be part of a ribosome binding site “GGTGAT,” we believe the ATG codon is probably the relevant initiator codon. The MJ0720 protein was expressed from this downstream start site fused to an amino-terminal polyhistidine tag in E. coli.
TABLE 1.
List of plasmids and microorganisms
| Strain or plasmid | Description | Source and/or reference |
|---|---|---|
| Methanocaldoccus jannaschii JAL-1 | Wild type | DSM 2661 |
| Escherichia coli | ||
| DH5α | General cloning host | Invitrogen, 16 |
| BL21(DE3) | Protein expression host | Novagen, 33 |
| Plasmids | ||
| pCDF-Duet1 | Protein coexpression vector | Novagen |
| pET-19b | Expression vector for proteins with an N-terminal decahistidine tag | Novagen |
| pDG141 | MJ1003 PCR product obtained using primers MJ1003Fwd and MJ1003Rev cloned into NdeI/KpnI sites of pCDF-Duet1 | This study |
| pDG142 | MJ0499 PCR product obtained using primers MJ0499Fwd and MJ0499Rev cloned into NdeI/KpnI sites of pCDF-Duet1 | This study |
| pDG152 | MJ1271 PCR product obtained using primers MJ1271Fwd and MJ1271Rev cloned into NcoI/BamHI sites of pDG141 | This study |
| pDG157 | MJ1277 PCR product obtained using primers MJ1277Fwd and MJ1277Rev cloned into NcoI/BamHI sites of pDG141 | This study |
| pDG159 | MJ1271 NcoI/BamHI fragment from pDG152 cloned into same sites of pDG142 | This study |
| pDG160 | MJ1271 PCR product cloned into NcoI/BamHI sites of pET-19b | This study |
| pDG163 | MJ1277 PCR product cloned into NcoI/BamHI sites of pET-19b | This study |
| pDG164 | MJ1277 NcoI/BamHI fragment from pDG157 cloned into same sites of pDG142 | This study |
| pDG329 | TDH NdeI/BamHI fragment from pTDH1 cloned into same site of pET-19b | This study |
| pMYL1 | MJ0720 PCR product obtained using primers MJ0720Fwd and MJ0720Rev cloned into NdeI/BamHI sites of pET-19b | This study |
| pTDH1 | Pseudomonas putida R-TDH in pET-3a | 37 |
Plasmids were propagated in E. coli DH5α (Invitrogen). Dideoxyribonucleotide sequencing confirmed the sequences of inserts in recombinant plasmids. Oligodeoxyribonucleotide primers used for PCR were as follows (restriction enzyme recognition sites used for cloning are underlined, and initiator codons are italicized): MJ1003Fwd (5′-GGTCATATGACATTGGTAGAGAAGATAC-3′), MJ1003Rev (5′-GCGGGTACCTTAATCCAATTTGTTGGT-3′), MJ0499Fwd (5′-GGTCATATGGGAATGACAATTGTAGAG-3′), MJ0499Rev (5′-GCGGGTACCTTATAAATCCCTTGGGTC-3′), MJ1271Fwd (5′-CATGCCATGGTTATTAAGGGAAGAGC-3′), MJ1271Rev (5′-CGGGATCCTTATGTTTTTACACCTTTTTTTGATTG-3′), MJ1277Fwd (5′-CATGCCATGGGAAGTATAATAAAGGGAAGAG-3′), MJ1277Rev (5′-CGGGATCCTTATTGGCTTTCAGCCATCTTTTTCTTTAAG-3′), MJ0720Fwd (5′-GCGCATATGCATAAAATATGTGTTATAG-3′), and MJ0720Rev (5′-GCGGATCCTTATTCTTCTCTTACTC-3′).
Protein expression and purification.
E. coli BL21(DE3) strains transformed with expression vectors were grown in Luria broth containing appropriate antibiotics at 37°C with shaking at 250 rpm. Cells containing pCDF-Duet1 derivatives were grown in the presence of streptomycin (50 μg ml−1). Cells containing pET-19b derivatives were grown in the presence of ampicillin (100 μg ml−1). When cultures reached an optical density at 600 nm of 0.6 to 0.8, protein expression was induced by the addition of α-d-lactose (1%, wt/vol) and shaking was continued at 20°C for 15 h. Cells were harvested by centrifugation and stored at −20°C.
Protein purification.
The MJ0499 and MJ1277 proteins were copurified from cells containing pDG164 by heat treatment, followed by anion-exchange chromatography. E. coli cells containing heterologously expressed protein were suspended in 20 mM Tris-HCl (pH 8.0), lysed by passing through a French pressure minicell at 8,000 lb/in2 (Thermo Electron), and sonicated on ice for 2 min using a Sonifier 450 with a microtip (15 W, 30% duty; Branson) to reduce viscosity. Lysates were clarified by centrifugation (14,000 × g for 10 min at 4°C), and the cell extract was heated at 70°C for 10 min. Denatured proteins were removed by centrifugation (14,000 × g for 10 min at 4°C), and the supernatant was stored on ice. This heat-stable protein was applied to a 5-ml HiTrap desalting column (GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 8.0). Fractions containing desalted protein were pooled and applied to a 5-ml HiTrap DEAE FF column (GE Healthcare) equilibrated in 20 mM Tris-HCl (pH 8.0). Chromatography was performed using an ÄKTAprime system (GE Healthcare) at a flow rate of 5 ml min−1. Protein was eluted from the column with a linear gradient to 0.5 M NaCl-20 mM Tris-HCl (pH 8.0) over 20 min. Fractions containing the target protein were identified by absorbance at 280 nm and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Fractions containing the proteins of interest were combined, loaded into SpectraPor4 dialysis tubing (molecular weight cutoff = 14,000), and dialyzed overnight in 2 liters of buffer containing 20 mM Tris-HCl (pH 8.0 at 4°C). The protein was transferred to fresh buffer and dialyzed for 5 h. Protein was concentrated inside the dialysis tubing using polyethylene glycol (20,000 Da). The MJ1003-MJ1277 and MJ1003-MJ1271 proteins were copurified from cells containing pDG141 and pDG163 and pDG160 and pDG152, respectively, as described above. Total protein concentration was determined using the Bio-Rad protein assay with bovine serum albumin as a standard.
The polyhistidine-tagged M. jannaschii IPMDH protein was purified by heat treatment followed by nickel affinity chromatography. E. coli(pMYL1) cells containing heterologously expressed protein were suspended in His tag binding buffer containing 5 mM imidazole, 500 mM sodium chloride, and 20 mM Tris-HCl (pH 7.6), and cell extracts were prepared as described above. Extracts were heated at 60°C for 10 min, and denatured protein was removed by centrifugation. The soluble protein was applied to a 5-ml HisTrap FF column (GE Healthcare) charged with NiCl2 and equilibrated in His tag binding buffer at a flow rate of 5 ml min−1. Protein was eluted from the column with a linear gradient to 0.5 M imidazole, 0.5 M sodium chloride, and 20 mM Tris-HCl (pH 7.6) over 20 min. Fractions containing the target protein were combined and concentrated in a stirred ultrafiltration cell (Amicon) with a filter having a 10-kDa molecular weight cutoff (Pall) under nitrogen gas at 4°C.
After concentration, the buffer was exchanged with 20 mM Tris-HCl (pH 7.6) in the ultrafiltration cell. The purified protein precipitated and lost activity during storage at 4°C. However, it could be stored in aliquots containing 20% glycerol, frozen at −40°C or −80°C, and thawed without significant loss of activity. Protein stored in this manner remained active for at least 2 weeks.
Analytical size exclusion chromatography.
Interactions between large and small subunits were tested by measuring apparent masses of protein pairs using size exclusion chromatography as described previously (19). The compositions of protein fractions were determined by SDS-PAGE with silver staining (Bio-Rad). Separations were performed aerobically on purified, unreconstituted apoenzymes.
Reconstitution of Fe-S centers.
Purified hydrolyase holoenzymes were reconstituted in a 2-ml glass serum vial (Wheaton) containing a magnetic stir bar and sealed with a 13-mm butyl rubber stopper. All buffers and reagents were degassed under a continuous flow of argon for at least 10 min. Anaerobic transfers were performed using gastight syringes (Hamilton). For reconstitution, purified apoprotein (0.5 to 1.0 mg ml−1) was mixed with 50 mM Tris-HCl (pH 8.0) and 3 mM dithiothreitol (2). The sealed solution was placed in an ice-water bath at 0 to 4°C, and the solution was degassed with argon for 10 min while being stirred. To the reconstitution mixture, Fe(NH4)2(SO4)2·6H2O was added to a final concentration of 0.5 mM. After 10 min, Na2S·9H2O was added dropwise to a final concentration of 0.5 mM, and the reconstitution mixture was stirred under argon until maximum activity was observed (typically 2 h). The final reconstitution volume was 1 ml. Hydrolyase activity was monitored in a standard assay with 0.3 mM citraconate as described below.
Measurement of lyase activities.
All continuous assays used a DU-800 spectrophotometer with a Peltier temperature-controlled sampler (Beckman). Reactions (1 ml) were conducted in quartz semimicrocells with screw-cap septa (Starna), including 300 mM KCl, 10 mM MgCl2, 50 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid]-KOH (pH 7.5), 2.5 or 10 μg ml−1 reconstituted protein, and various substrate concentrations. The sealed quartz cell containing buffer and substrate was degassed under argon for 10 min and equilibrated at 60°C for 10 min. Reactions were initiated by the addition of reconstituted enzyme, and the change in UV absorbance was monitored at 60°C. Initial rates were measured from the linear portion of the reaction progress curve between 5 and 60 s. For α-isopropylmalate, β-isopropylmalate, and (R)-citramalate, the formation of the reaction intermediate was observed by measuring the increase in absorbance at 235 nm, as measured in the reaction buffer at 60°C with 10 μg ml−1 of reconstituted enzyme. The molar absorption coefficient at 235 nm (ɛ235) for both citraconate and isopropylmaleate was 4,668 M−1 cm−1. The hydration of citraconate and maleate was observed as a decrease in absorbance with 2.5 μg ml−1 of reconstituted enzyme. Maleate hydration to produce malate was monitored at 235, 245, and 255 nm using molar absorption coefficients of 2,318, 1,297, and 582 M−1 cm−1, respectively, measured in the reaction buffer at 60°C. Citraconate hydration was monitored at 235 nm, with a molar absorption coefficient of 4,530 M−1 cm−1. Reaction progress curves were linear from 5 to 60 s. One unit of hydrolyase activity catalyzed the conversion of 1 μmol substrate to product per minute. Steady-state kinetic constants were estimated by nonlinear regression of initial rate data fit to the Michaelis-Menten-Henri equation using the KaleidaGraph program (Synergy Software).
Measurement of oxidative decarboxylation activities.
The rate of oxidative decarboxylation catalyzed by IPMDHMj was measured by monitoring the reduction of NAD+ to NADH at 340 nm, assuming a molar absorption coefficient for NADH of 6,220 M−1 cm−1. Assays were conducted aerobically in optical glass cuvettes (Starna) with 200 mM KCl, 10 mM MgCl2, 50 mM TAPS [Tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid]-KOH (pH 8.5), 5 mM NAD+, 2.5 μg ml−1 IPMDHMj, and various substrate concentrations. Reaction mixtures were equilibrated at 60°C for 10 min, and reactions were initiated with NAD+. Initial rates were measured using a linear portion of the reaction progress curve, from 5 to 60 seconds for β-isopropylmalate and 120 to 180 seconds for d-malate. One unit of enzymatic activity catalyzed the reduction of 1 μmol NAD+ per min in a 1-ml reaction mixture. Apparent kinetic constants were estimated as described above.
Coupled assay of hydratase activity.
The β-hydrating activity of enzymes incubated with unsaturated intermediates was measured in reaction mixtures containing IPMDHMj and NAD+ as described above. Reaction mixtures contained 300 mM KCl, 10 mM MgCl2, 50 mM TES-KOH (pH 7.5), 30 μg ml−1 IPMDHMj, 5 mM NAD+, and various substrate concentrations. Reaction mixtures were made anaerobic and preincubated at 60°C as described above, before the addition of 2.5 μg ml−1 MJ0499/MJ1277 to initiate the reaction. Initial rates were measured between 5 and 60 seconds for citraconate and between 120 and 180 seconds for maleate.
Identification of dehydratase reaction products.
Hydroxyacids and unsaturated carboxylic acids were derivatized with ethylchloroformate and analyzed by gas chromatography-mass spectrometry (GC-MS) (28). Reaction products were mixed with ethylchloroformate in a solution of water-trifluoroethanol-pyridine (60:32:8, vol/vol/vol) (40). Derivatives were analyzed on a Finnigan MAT GCQ GC-MS with a DB-5MS capillary column (0.32 mm by 29 m, 0.5-mm film; J&W Scientific) and analyzed by chemical ionization MS in the positive mode, as described previously (19). Using this method, the trifluoroethyl ester derivatives had the following retention times and mass spectral data. The molecular ion (MH+) is shown first, if observed (a minus sign indicates that MH+ was not found), followed by the base peak (italics), and characteristic fragment ions are listed in decreasing order of intensity. The derivatives were as follows: citraconate diester, 3.73 min (295, 195, 275); (R)-citramalate, 3.87 min (−, 185, 195, 275, 213); d-malate diester, 4.06 min (299, 171, 261, 279, 199); β-isopropylmalate diester, 4.61 min (−, 199, 213, 241); α-isopropylmalate diester, 4.65 min (341, 213, 195, 241, 321). In most spectra the base peak was assigned as the fragment ion following the neutral loss of CF3CH2OH or CF3CH2COOH from the molecular ion.
Alternatively, trimethylsilyl (TMS) derivatives of the hydroxycarboxylic acids were prepared by reacting dried sample with 50 μl N-methyl-N-(TMS)-trifluoroacetamide and 50 μl pyridine in a sealed vial at 60°C for 20 min. Derivatives were analyzed by GC-MS as described above. The mass range scanned was 70 to 700 atomic mass units. The derivatives were as follows: citraconate-(TMS)2, 5.54 min (275, 185, 149, 73, 259, 277, 73); citramalate-(TMS)3, 6.03 min (365, 247, 349, 321, 277, 73); β-isopropylmalate-(TMS)3, 6.30 (−, 275, 191, 377, 73); α-isopropylmalate-(TMS)3, 6.35 (393, 213, 275, 377, 73).
For quantitative analysis of reaction mixtures, carboxylic acids were separated by reversed-phase high-pressure liquid chromatography (HPLC). Reaction mixtures or standard compounds were applied to a Luna C18(2) column (4.6 by 150 mm, 5 μm; Phenomenex) with a Security Guard ODS cartridge (4 by 3 mm; Phenomenex) that was equilibrated in mobile phase containing 25 mM phosphoric acid and 2% (vol/vol) methanol (pH 2.5). Isocratic elution with this mobile phase was used at a flow rate of 1 ml min−1 at 35°C. Samples (25 μl) were injected using a Beckman 508 autosampler. Organic acids were detected using a Beckman 168 photodiode array detector. Data were collected and chromatograms were integrated using the 32 Karat software (Beckman). Under these conditions, retention factors for standard compounds were 0.63 (d-malate), 0.97 (2-oxoglutarate), 1.4 (maleate), 2.1 (homoisocitrate), 2.0 (fumarate), 2.3 (cis-aconitate), 2.7 (R-citramalate), 3.7 (trans-aconitate), and 4.7 (citraconate), with a holdup time of 1.47 min. Less-polar compounds were resolved using a mobile phase containing 25 mM phosphoric acid and 30% (vol/vol) methanol (pH 2.5). Under these conditions, retention factors for standard compounds were 0.4 (R-citramalate), 0.7 (citraconate), 2.5 (β-isopropylmalate), 2.9 (α-isopropylmalate), and 4.2 (isopropylmaleate), with a holdup time of 1.55 min. Analyte concentrations were calculated from the integrated peak areas of the chromatogram at 220-nm absorbance using the method of standard addition.
Stereochemical analysis of malate product.
Tartrate dehydrogenase (TDH) was used to confirm the stereochemistry of IPMI hydration products. Plasmid pTDH1, which encodes the TDH enzyme from Pseudomonas putida ATCC 17642, was a gift from Paul Cook (University of Oklahoma) (24). Dideoxynucleotide sequencing using T7 promoter and T7 terminator oligonucleotide primers identified several discrepancies with the originally published TDH sequence (37). The NdeI/BamHI fragment of plasmid pTDH1 was subcloned into pET-19b to produce plasmid pDG329, which encodes TDH fused to an amino-terminal decahistidine tag. The His10TDH protein was expressed from pDG329 in E. coli BL21(DE3). Protein was purified by heating the extract at 50°C for 10 min, precipitating the denatured protein, and isolating the His10TDH by nickel affinity chromatography as described above.
Reaction mixtures to determine the stereochemistry of malate produced by the malease activity of the MJ0499/MJ1277 combination contained 5 mM maleate, reconstituted MJ0499/MJ1277 protein (5 μg ml−1), and reaction buffer incubated at 60°C for 24 h. Portions of reaction product were added to an aerobic solution containing IPMI buffer salts, TDH (His10TDH; 20 μg ml−1), and 1 mM NAD+ in a volume of 1 ml. After incubation at 37°C for 1 h, the absorbance of the solution at 340 nm was measured and d-malate product concentrations were calculated according to a linear standard curve from 5 to 200 μM d-malate. Another portion of the malease reaction product was analyzed using l-malate dehydrogenase (U.S. Biochemicals) in a 1-ml reaction mixture containing 20 μg ml−1 enzyme, 100 μM phenazine methosulfate, 150 μM thiazolyl blue, and 1 mM NAD+. Reaction mixtures were incubated at room temperature for 30 min, the absorbance of the solution at 570 nm was measured, and l-malate concentrations were estimated according to a linear standard curve from 5 to 200 μM l-malate (15). The limit of detection for this method was approximately 1.5 μM l-malate.
Phylogenies of dehydratase subunits.
An alignment of 62 homologs of the large IPMI subunit was prepared using the ClustalW program (version 1.83) (36). From the full alignment 442 positions that were deemed to be confidently aligned were chosen for phylogenetic analysis. The phylogeny was inferred using the proml program or the protdist and neighbor programs (with 100 bootstrap replicates) from the Phylip package (version 3.66) (14). Both programs used the Jones-Taylor-Thornton model of amino acid changes and assumed a γ-distribution of rates (α = 2.4) approximated by three states.
Sequences of large subunit homologs (RefSeq [ref], Swiss-Prot [sp], and EMBL [emb] accession numbers [or in one case a URL] are in parentheses) were from Aquifex aeolicus (ref, NP_213641.1), Archaeoglobus fulgidus (ref, NP_070787.1 and NP_071024.1), Bacteroides thetaiotaomicron (ref, NP_810773.1), Carboxydothermus hydrogenoformans (ref, YP_359380.1 and YP_359951.1), Deinococcus radiodurans (sp, Q9RTY9 and Q9RTI6), Escherichia coli (sp, P36683 and P0A6A6), Ferroplasma acidarmanus (ref, ZP_00609074.1), Haloarcula marismortui (sp, Q5V518), Haloquadratum walsbyi (ref, YP_658418.1), Halothermothrix orenii (ref, ZP_01188446.1), Ignicoccus sp. strain Kin4-I (http://genome.jgi-psf.org), Leptospira interrogans (sp, Q8F4E6), Methanocaldococcus jannaschii (sp, P81291 and Q58409), Methanococcoides burtonii (ref, YP_566516.1 and YP_566874.1), Methanococcus maripaludis (ref, NP_988600.1 and NP_988269.1), Methanoculleus marisnigri (ref, ZP_01392213 and ZP_01390868.1), Methanopyrus kandleri (ref, NP_614723.1 and NP_614491.1), Methanosaeta thermophila (ref, YP_843415.1 and YP_843217.1), Methanosarcina acetivorans (ref, NP_617978.1 and NP_616329.1), Methanosphaera stadtmanae (ref, YP_448499.1 and YP_448126.1), Methanospirillum hungatei (ref, YP_503784.1 and YP_503240.1), Methanothermobacter thermautotrophicus (ref, NP_276743.1 and NP_276502.1), Moorella thermoacetica (ref, YP 431084.1), Natronomonas pharaonis (ref, YP_326749.1), Picrophilus torridus (ref, YP_024242 and YP_023690.1), Pyrobaculum aerophilum (ref, NP_559679.1), Pyrococcus abyssi (sp, Q9V1JO and Q9UZ07), Pyrococcus furiosus (sp, Q8U2A1 and Q8U0C0), Pyrococcus horikoshii (sp, O59391), Saccharomyces cerevisiae (ref, NP_013407.1, NP_011506.1, and NP_010520.1), Sulfolobus acidocaldarius (ref, YP_254963.1), Sulfolobus solfataricus (sp, Q97VY2; ref, NP_342564.1), Syntrophomonas wolfei (ref, ZP_00662230.1 and ZP_00664168.1), Thermoproteus tenax (emb, CAF18518.1), Thermococcus kodakarensis (ref, YP_182695.1), Thermotoga maritima (ref, NP_228103.1 and 228364.1), and Thermus thermophilus (sp, Q9ZND5; ref, YP_004349; and ref, YP_145177.1).
Nucleotide sequence accession number.
The revised sequence of the TDH gene was deposited in GenBank with accession number EF210485.
RESULTS
Expression and purification of subunit combinations.
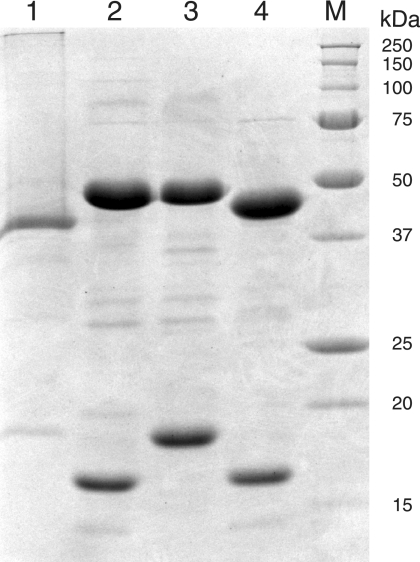
To identify protein pairs that form active hydrolyase enzymes, the four combinations of large-subunit homologs (MJ0499 or MJ1003) with small-subunit homologs (MJ1271 or MJ1277) were expressed in E. coli. All four combinations produced substantial amounts of soluble, heterologous protein. Cell extracts were heated to denature native proteins, and the target proteins were purified by anion-exchange chromatography. Combinations of MJ1003 and MJ1271 or MJ1277 copurified, as did the MJ0499 and MJ1277 combination. However, the MJ0499 and MJ1271 subunits did not copurify. Preparations of the three combinations of associated subunits were substantially pure: the two subunits comprised 84 to 93% of the total protein as judged by overloading an SDS-PAGE gel stained with Coomassie blue (Fig. 3). The apparent masses of purified proteins matched their theoretical masses: MJ0499, observed 44 kDa, expected 46.1 kDa; MJ1003, 46 kDa, 46.1 kDa; MJ1271, 18 kDa, 18.6 kDa; and MJ1277, 16 kDa, 18.4 kDa.
FIG. 3.
Purified IPMDHMj and combinations of large and small subunits of the hydrolyase apoenzymes. Lane 1, 8 μg His10MJ0720 (IPMDHMj); lane 2, 10 μg MJ1003/MJ1277; lane 3, 10 μg MJ1003/MJ1271; lane 4, 10 μg MJ0499/MJ1277; lane M, protein size markers (Bio-Rad). Proteins were separated by SDS-PAGE on a 15% acrylamide gel and stained with Coomassie blue.
Subunit interactions.
The three copurified pairs of large and small subunits also coeluted during analytical size exclusion chromatography. The MJ0499/MJ1277 pair formed a complex with a Stokes radius of 42 Å, corresponding to a 143-kDa protein, which could be the heterotetramer (MJ0499/MJ1277)2. The MJ1003/MJ1271 complex also had a Stokes radius of 42 Å and probably forms heterotetramer (MJ1003/MJ1271)2. The MJ1003/MJ1277 complex had a Stokes radius of 43 Å, corresponding to the 153-kDa complex of heterotetrameric structure (MJ1003/MJ1277)2. A mixture of purified MJ0499 and MJ1271 subunits did not form a complex; instead the proteins eluted separately as monomers.
Reconstitution of iron-sulfur centers.
The purified preparations of subunit combinations had no significant activity in assays of citraconate hydrolyase activity. Members of this protein family contain an Fe4S4 cluster with a unique iron that is readily lost during aerobic purification (18). Therefore we reconstituted the clusters under anaerobic, reducing conditions to produce maximal citraconate hydrolyase activity. The resulting protein preparation contained a broad absorbance shoulder from 400 to 450 nm, characteristic of S→Fe charge transfer bands in iron-sulfur centers (10, 23). In the absence of electron paramagnetic resonance or Mössbauer spectra we cannot conclude that all iron-sulfur centers were fully reconstituted as Fe4S42+ clusters (1), but to calculate turnover numbers each heterodimer was assumed to contain an active site.
Identification of the citramalate/isopropylmalate isomerase.
The reconstituted MJ0499/MJ1277 holoenzyme catalyzed the hydrolysis of citraconate at 60°C under anaerobic conditions with a specific activity of 15 U mg−1. The reaction product was derivatized, and citramalate or β-methylmalate was identified by GC-MS (the isomers could not be separated by this method). Both substrate and products were also resolved and detected by reversed-phase HPLC. None of the other three reconstituted protein combinations (MJ0499/MJ1271, MJ1003/MJ1271, or MJ1003/MJ1277) catalyzed this reaction, nor did a control reconstitution mixture that contained no protein.
In addition to catalyzing the hydrolysis of citraconate, the MJ0499/MJ1277 pair had malease activity: it catalyzed the hydrolysis of 2 mM maleate with a high specific activity (28 U mg−1). The enzyme catalyzed the dehydration of (R)-citramalate and α-isopropylmalate, as well as the dehydration of β-isopropylmalate product, at 10 to 25% of the rate of citraconate hydrolysis. In contrast, fumarate, mesaconate, cis-aconitate, trans-aconitate, d-tartrate, l-tartrate, l-malate, dl-isocitrate, citrate, (R)-homocitrate, and (S)-citramalate were not substrates. Therefore the enzyme is specific for cis-unsaturated intermediates and their d-α-hydroxy or β-hydroxy analogs (Fig. 1).
For optimal citraconate hydrolase activity, reactions required at least 5 mM MgCl2; the relative activity without added MgCl2 was 5% of the maximum at 10 mM MgCl2. Enzyme activity also increased at higher KCl concentrations: in the absence of added KCl, activity was 65% of the maximum (at 300 mM KCl). The enzyme demonstrated significant activity across a broad pH range, with optimal activity at pH 7.5. Maximal activity was observed at 70°C, although standard reactions were performed at 60°C for technical reasons.
To determine the equilibrium constant for the citraconate hydration reaction, vials containing 2 mM citraconate and 5 μg ml−1 reconstituted MJ0499/MJ1277 were sealed under an argon atmosphere and incubated at 60°C for 20 h. A portion of the reaction mixture was analyzed by HPLC. Concentrations of citraconate and citramalate (with β-methylmalate) were determined by standard addition: the equilibrium ratio of hydrated products to citraconate was 50:1. In similar reaction mixtures containing 2 mM β-isopropylmalate, concentrations of α-isopropylmalate and β-isopropylmalate were determined by standard addition and the isopropylmaleate concentration was estimated using maleate or citraconate as the external standard. The equilibrium ratio of α-isopropylmalate/β-isopropylmalate/isopropylmaleate was 9:30:1, averaged from two analyses. To confirm the identity of these reaction products, TMS derivatives were analyzed by GC-MS. Both α-isopropylmalate and β-isopropylmalate were detected, although the isopropylmaleate concentration was too low to detect in this analysis.
The equilibrium ratio of malate to maleate in reaction mixtures containing 2 mM maleate with 5 μg ml−1 reconstituted MJ0499/MJ1277 was 146:1. No maleate was detected in reaction mixtures containing d-malate and reconstituted enzyme due to the detection limits of this assay. Standard reaction mixtures containing enzyme, 2 mM maleate, and 5 mM d-malate exhibited no malease activity, due to product inhibition. A d-malate concentration of 0.5 mM inhibited the rate of malease activity by approximately 50%. In contrast, an l-malate concentration of 5 mM was required to reduce malease activity by approximately 50%. The much weaker binding of l-malate to the MJ0499/MJ1277 pair suggests that d-malate is the product of malease activity (38).
The enzymes l-malate dehydrogenase and TDH (which catalyzes the oxidative decarboxylation of d-malate [37]) were used to confirm the stereochemistry of malate produced by the hydration of maleate. Reaction mixtures containing 5 mM maleate and 5 μg ml−1 reconstituted MJ0499/MJ1277 were incubated for 24 h at 60°C. Enzymatic analysis identified 5.07 ± 0.05 mM d-malate and no l-malate (<8 μM) in the reaction product. Therefore the sole product of maleate hydration was d-malate.
Steady-state kinetic parameters for hydrolyase activity using substrates maleate, citraconate, (R)-citramalate, α-isopropylmalate, and β-isopropylmalate were estimated from initial rate data fit to the Michaelis-Menten-Henri equation (Table 2). The MJ0499/MJ1277 pair catalyzes the hydration of both maleate and citraconate with similar specificity constants. However, water is abstracted from the physiologically relevant substrates (R)-citramalate and α-isopropylmalate with approximately 100-fold-lower specificity constants. The Km value for α-isopropylmalate is more than double the value calculated for (R)-citramalate. However, commercially synthesized α-isopropylmalate is a racemic mixture of two enantiomers, so the Km for the relevant isomer, (2S)-2-isopropylmalate, may be lower (31). β-Isopropylmalate is also a racemic mixture, although the enzyme's Km for this racemic substrate is relatively low.
TABLE 2.
Steady-state kinetic parameters for MJ0499/MJ1277a
| Substrate | Km (μM) | Vmax (U mg−1) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| Citraconateb | 80 ± 20 | 15 ± 0.9 | 16 ± 1 | 2.0 × 105 |
| Citraconatec | 40 ± 10 | 13 ± 0.7 | 14 ± 1 | 3.5 × 105 |
| (R)-Citramalateb | 810 ± 80 | 1.5 ± 0.05 | 1.6 ± 0.1 | 2.0 × 103 |
| α-Isopropylmalateb | 1,900 ± 400 | 4.2 ± 0.3 | 4.5 ± 0.3 | 2.4 × 103 |
| β-Isopropylmalateb | 39 ± 7 | 1.8 ± 0.1 | 1.9 ± 0.1 | 5.0 × 104 |
| Maleateb | 400 ± 50 | 34 ± 2 | 36 ± 2 | 9.0 × 104 |
| Maleatec | 180 ± 30 | 19 ± 0.9 | 20 ± 1 | 1.1 × 105 |
Kinetic parameters were estimated by nonlinear regression, and the standard errors of those parameters are shown. Turnover numbers were calculated assuming one active site per MJ0499/MJ1277 pair.
Initial rates of hydrolyase activity were determined by measuring UV absorbance.
Initial rates of hydratase activity were measured in a coupled assay for NADH production using excess IPMDHMj.
Neither the MJ1003/MJ1277 nor the MJ1003/MJ1271 pair of subunits catalyzed citraconate hydrolysis or the dehydration of α-isopropylmalate or β-isopropylmalate. Therefore the MJ0499/MJ1277 pair constitutes the citramalate/isopropylmalate isomerase in M. jannaschii.
Characterizing isopropylmalate/β-methylmalate dehydrogenase.
The isopropylmalate dehydrogenase protein from M. jannaschii (MJ0720; IPMDHMj) was previously identified and characterized in cell extract from a heterologous expression system in E. coli (20). To test the substrate specificity of IPMDHMj, we expressed that protein in E. coli fused to an amino-terminal polyhistidine tag. This IPMDHMj protein was purified to homogeneity by nickel affinity chromatography, and it had an apparent mass of 40 kDa, as determined by SDS-PAGE, which is close to its expected mass of 39.1 kDa (Fig. 3). Analytical size exclusion chromatography identified a protein complex with a Stokes radius of 51 Å, corresponding to a 183-kDa native protein, which is probably a tetrameric form of the protein. Analysis of a mixture of IPMDHMj protein with the MJ0499/MJ1277 complex showed no evidence of association with the hydrolyase complex.
Reactions with IPMDHMj were initiated by the addition of NAD+, and the rate of NADH formation was determined by measuring UV absorbance at 60°C. The enzyme catalyzed the oxidative decarboxylation of β-isopropylmalate and d-malate with kinetic parameters shown in Table 3. The IPMDHMj enzyme specifically required NAD+ as reported previously: no β-isopropylmalate dehydrogenase activity was observed in the presence of NADP+ (20). This enzyme did not catalyze the oxidation of l-malate, l-tartrate, d-tartrate, dl-isocitrate, or dl-lactate.
TABLE 3.
Kinetic parameters for IPMDH homologs
| Enzyme (reference) | β-Isopropylmalate
|
d-Malate
|
||
|---|---|---|---|---|
| Km (μM) | kcat/Km (M−1 s−1) | Km (μM) | kcat/Km (M−1 s−1) | |
| IPMDHMja | 24 ± 3 | 8.8 × 104 | 410 ± 40 | 1.4 × 104 |
| Sulfolobus sp. IPMDH (34) | 1.2 | 3.0 × 106 | 390 | 1.0 × 104 |
| P. putida TDH (37) | 14 | 1.1 × 104 | 60 | 2.2 × 105 |
| T. thermophilus IPMDH (9) | 17 | 1.4 × 104 | 1,510 | 7.3 × 103 |
Data from this work. Initial rates were determined by measuring an increase in absorbance at 340 nm due to NADH in reaction mixtures incubated at 60°C.
Coupled assay for β-methylmalate oxidative decarboxylation.
We used a coupled assay with MJ0499/MJ1277 and IPMDHMj to confirm that IPMDHMj catalyzes the oxidative decarboxylation of β-methylmalate, the final step in the pyruvate pathway to 2-oxobutyrate. Reaction mixtures containing citraconate, excess NAD+, and IPMDHMj IPMDH were incubated in anaerobic vials, and reactions were initiated by the addition of reconstituted MJ0499/MJ1277. The rate of NADH production was measured at 60°C, demonstrating that IPMDHMj efficiently catalyzes the oxidative decarboxylation of β-methylmalate. Initial rates were used to estimate apparent kinetic parameters for the hydration reaction (Table 2). These parameters for β-methylmalate formation measured by this coupled assay were comparable to those estimated for the hydration of citraconate. This coupled assay was also used to measure rates of d-malate production due to hydration of maleate.
Evolution of the isopropylmalate isomerase homologs.
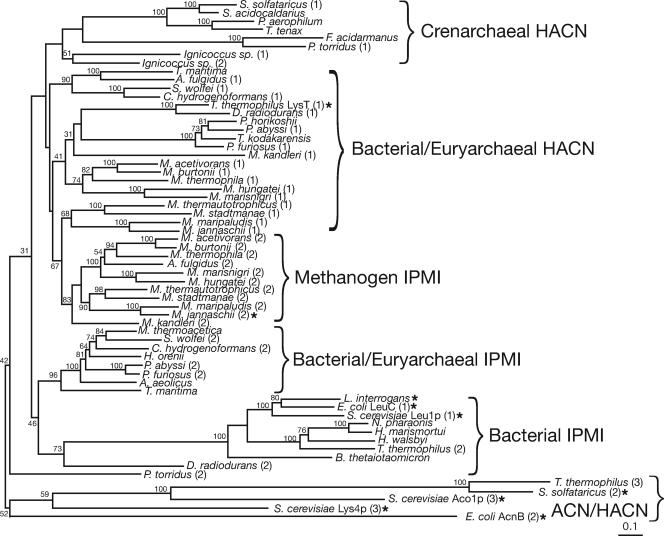
A phylogeny of IPMI large-subunit homologs was inferred, including homologous regions of aconitase (ACN) and HACN proteins, to establish the evolutionary relationship among these hydrolyases. The phylogeny of small-subunit homologs was not analyzed due to their short length and high degree of sequence similarity (Fig. 2). Phylogenetic trees were inferred by neighbor joining (Fig. 4), protein maximum likelihood, and Bayesian methods, using ACN sequences as an outgroup. Bootstrap or clade credibility values show low support for most deep branches; however, the trees have common topological features. All of the trees show clusters of methanogen IPMI sequences (orthologs of MJ0499) that share a common ancestor with a cluster of paralogous methanogen sequences predicted to encode HACNs (orthologs of MJ1003). These two paralogous groups are specifically related to putative HACNs from other euryarchaea, crenarchaea, and bacteria. IPMI sequences from bacteria, nonmethanogenic archaea, and eukaryotes are not specifically related to the methanogen IPMI sequences; therefore, either the IPMI or HACN functions must have evolved several times. IPMI and putative HACN subunits were present in the methanogen ancestor and have been vertically inherited in modern methanogenic lineages. Halophilic euryarchaea apparently recruited IPMI from bacteria. The heterotrophic euryarchaea and crenarchaea are usually cultured in complex media, so their amino acid requirements are often poorly characterized and the functions of their homologs listed in Fig. 4 are ambiguous. Additional biochemical studies are required to characterize the clusters labeled “HACN.”
FIG. 4.
Phylogram showing the evolutionary relationship between the large subunits of IPMI, ACN, and HACN proteins. The phylogeny was inferred by the neighbor joining method, and bootstrap values are shown near branches supported by a plurality of 100 trees. This tree is rooted using the ACN cluster. Related proteins predicted to catalyze the same reactions are grouped by braces. The proposed HACN function of some archaeal proteins has not yet been demonstrated. Enzymes whose functions have been experimentally confirmed are indicated by an asterisk. Full organism names and sequence accession numbers are listed in Materials and Methods. Numbers in parentheses after some species names differentiate paralogs from the same genome. The scale bar indicates 0.1 amino acid change expected per site.
DISCUSSION
The phylogram in Fig. 4 shows that the IPMI and putative HACN large subunits are paralogs that diverged early in the methanogen lineage. This underscores the difficulty in predicting enzyme function from closely related sequences and the need for experiments to assign gene functions to complex protein families. There is currently no structural model of the IPMI complex, and almost all biochemical knowledge of this protein family comes from studies on the ACN enzyme (1). Although the biochemical function of the small-subunit protein homolog from P. horikoshii is unknown, a crystal structure model suggests a substrate binding site that is shown in Fig. 2 (45). The only recognizable member of the IPMS family in P. horikoshii is a homocitrate synthase, and the only member of the IPMDH family is an isocitrate-homoisocitrate dehydrogenase (30). Therefore the crystallized P. horikoshii homolog is probably an HACN and not an IPMI subunit as proposed previously (45). An unrefined homology model of the MJ1277 protein produced by the SWISS-MODEL server (version 36.0003) (32) based on the P. horikoshii protein structure suggests that Leu27 and His71 side chains of MJ0277 form a hydrophobic pocket that could confer specificity for short alkyl-substituted hydroxy-dicarboxylates.
Bacteria that use the homoaconitate pathway for lysine biosynthesis recruited HACN from euryarchaea. This protein is only distantly related to yeast HACN. Although yeast IPMI is a cytosolic protein, IPMS is associated with mitochondria, and the yeast leucine biosynthetic proteins may have been recruited from a mitochondrial ancestor. Yeast ACN and HACN proteins are also localized to mitochondria. Therefore these iron-sulfur hydrolyases probably evolved first in bacteria and archaea that contained the apparatus for iron-sulfur cluster assembly.
The MJ0499/MJ1277 pair of subunits resembles other hydrolyases in its broad pH optimum, domain structure, and Michaelis constants for cis-unsaturated intermediates. Gel filtration results suggest that this apoprotein forms a heterotetramer, while the S. cerevisiae IPMI acts as a monomer and the Pseudomonas pseudoalcaligenes malease forms a heterodimer. Further studies will be required to determine the shape and subunit composition of the methanogen IPMI in its active, holoenzyme form. Similar to most Fe4S4-containing proteins, the methanogen IPMI is inactivated by exposure to air; both superoxide and hydrogen peroxide have been shown to oxidize IPMI metal centers (22, 41).
This enzyme's specificity constants for hydroxyacids are 10- to 100-fold lower than those for unsaturated intermediates, although the specific activity is comparable to that reported for the S. cerevisiae IPMI dehydration of α-isopropylmalate (6 U mg−1) (27). The large equilibrium constants measured for citraconate and maleate hydrolysis confirm that the hydrolysis half-reaction is favorable (ΔG′° ≈ −14 kJ mol−1 for the hydration of maleate at 60°C), consistent with the previously calculated value of −19 kJ mol−1 (at 25°C) from studies of the P. pseudoalcaligenes malease enzyme (39). Equilibrium constants measured for β-isopropylmalate isomerization show that the forward reaction (converting α-isopropylmalate to β-isopropylmalate) is slightly favored (by approximately 3 kJ mol−1 at 60°C). In the cell the isomerization reaction is probably driven by the oxidative decarboxylation reaction catalyzed by IPMDH. Continuous assays for citraconate hydration measure the production of both (R)-citramalate and β-methylmalate (Table 2). Therefore it is informative to compare those kinetic parameters with parameters measured using a coupled assay with IPMDH that monitors only β-methylmalate production. Both the Km and kcat parameters for citramalate hydration are similar for both assays, suggesting that β-methylmalate is the dominant product.
Most maleases appear to be iron-sulfur proteins that are probably homologs of IPMI (10), although no sequence information is available. However, Pseudomonas pseudoalcaligenes expresses a two-subunit maleate hydratase enzyme that is colorless and resistant to oxidation or inactivation by metal chelators (38). Therefore it may not be an iron-sulfur protein. The M. jannaschii isopropylmalate/citramalate hydratase enzyme is also proficient at catalyzing the hydration of maleate to produce d-malate. This enzyme's Km for maleate (400 μM) is similar to that reported for the P. pseudoalcaligenes protein (350 μM), as are the enzymes' Km values for citramalate (80 μM and 200 μM, respectively) (38). Both enzymes are highly stereospecific, producing only d-malate.
The MJ0499/MJ1277 pair of proteins catalyzes the isomerization of both α-isopropylmalate to β-isopropylmalate for leucine biosynthesis and (R)-citramalate to β-methylmalate for isoleucine biosynthesis. Similarly, the MJ0720 protein has both isopropylmalate and β-methylmalate dehydrogenase activities. The IPMDHMj is 44% identical to the crenarchaeal homolog from a Sulfolobus sp. (34). The two proteins have similar kinetic parameters for the oxidative decarboxylation of d-malate, although the Sulfolobus enzyme has a 20-fold-lower Km for β-isopropylmalate (Table 3). Therefore the latter two enzymes of the isopropylmalate pathway appear to be broadly specific for acids with substitutions of short-chain aliphatic compounds.
The only enzymes specific to the isopropylmalate pathway or the pyruvate pathway to 2-oxobutyrate are the IPMS and CMS. An isoleucine auxotroph (a threonine dehydratase mutant) of Serratia marcescens could be grown in minimal medium supplemented with citramalate or citraconate (25). This observation confirms the broad specificity of that bacterium's IPMI and IPMDH enzymes. The S. marcescens IPMS catalyzes citramalate synthesis with only a sevenfold-lower specificity constant than that for α-isopropylmalate synthesis (26). Therefore it is not surprising that genetic selection using the S. marcescens threonine dehydratase mutant identified a revertant strain that evolved the pyruvate pathway to 2-oxobutyrate. Compared to wild-type S. marcescens this mutant had increased IPMS activity due to reduced feedback inhibition by leucine (25). These experiments demonstrate how readily the pyruvate pathway to 2-oxobutyrate can evolve, upon mutation or duplication and functional divergence of an IPMS gene. The specialized CMS from M. jannaschii no longer catalyzes isopropylmalate synthesis, even though it is 50% identical to that organism's IPMS protein (21).
The spirochetal pathogen Leptospira interrogans uses the pyruvate pathway to 2-oxobutyrate exclusively, while saprophytic Leptospira spp. have both threonine dehydratase and citramalate synthase enzymes (7, 46). Although the presence of a CMS in L. interrogans was previously interpreted as evidence for the early divergence of the Leptospiraceae in the bacterial lineage (43), that CMS is more closely related to bacterial homologs in Bacteroides thetaiotaomicron and Flavobacterium johnsoniae than it is the methanogen CMS (data not shown). Therefore the pyruvate pathway to 2-oxobutyrate probably evolved in the Leptospiraceae by the acquisition of a CMS gene that displaced an ancestral threonine dehydratase gene. For cells without an exogenous source of threonine, the pyruvate pathway requires fewer ATP equivalents to produce 2-oxobutyrate.
Acknowledgments
This work was supported in part by grant F-1576 from the Welch Foundation and by NSF grant MCB-0425983.
We thank Smriti Canakapalli and Syed Ali Reza Hasan for experimental help with protein purification and Man-Yu Lam for cloning assistance.
Footnotes
Published ahead of print on 20 April 2007.
REFERENCES
- 1.Beinert, H., M. C. Kennedy, and C. D. Stout. 1996. Aconitase as iron-sulfur protein, enzyme, and iron-regulatory protein. Chem. Rev. 96:2335-2374. [DOI] [PubMed] [Google Scholar]
- 2.Blank, L., J. Green, and J. R. Guest. 2002. AcnC of Escherichia coli is a 2-methylcitrate dehydratase (PrpD) that can use citrate and isocitrate as substrates. Microbiology 148:133-146. [DOI] [PubMed] [Google Scholar]
- 3.Buchanan, B. B. 1969. Role of ferredoxin in the synthesis of alpha-ketobutyrate from propionyl coenzyme A and carbon dioxide by enzymes from photosynthetic and nonphotosynthetic bacteria. J. Biol. Chem. 244:4218-4223. [PubMed] [Google Scholar]
- 4.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J.-F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. M. Geoghagen, H. O. Smith, C. R. Woese, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1017-1140. [DOI] [PubMed] [Google Scholar]
- 5.Burns, R. O., H. E. Umbarger, and S. R. Gross. 1963. The biosynthesis of leucine. III. The conversion of α-hydroxy-β-carboxyisocaproate to α-ketoisocaproate. Biochemistry 2:1053-1058. [DOI] [PubMed] [Google Scholar]
- 6.Calvo, J. M., C. M. Stevens, M. G. Kalyanpur, and H. E. Umbarger. 1964. The absolute configuration of α-hydroxy-β-carboxyisocaproic acid (3-isopropylmalic acid), an intermediate in leucine biosynthesis. Biochemistry 3:2024-2027. [DOI] [PubMed] [Google Scholar]
- 7.Charon, N. W., R. C. Johnson, and D. Peterson. 1974. Amino acid biosynthesis in the spirochete Leptospira: evidence for a novel pathway of isoleucine biosynthesis. J. Bacteriol. 117:203-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, F. E., M. G. Kalyanpur, and C. M. Stevens. 1973. Absolute configuration of α-isopropylmalate and the mechanism of its conversion to β-isopropylmalate in the biosynthesis of leucine. Biochemistry 12:3346-3350. [DOI] [PubMed] [Google Scholar]
- 9.Dean, A. M., and L. Dvorak. 1995. The role of glutamate 87 in the kinetic mechanism of Thermus thermophilus isopropylmalate dehydrogenase. Protein Sci. 4:2156-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dreyer, J.-L. 1985. Isolation and biochemical characterization of maleic-acid hydratase, an iron-requiring hydro-lyase. Eur. J. Biochem. 150:145-154. [DOI] [PubMed] [Google Scholar]
- 11.Eikmanns, B., R. Jaenchen, and R. K. Thauer. 1983. Propionate assimilation by methanogenic bacteria. Arch. Microbiol. 136:106-110. [Google Scholar]
- 12.Eikmanns, B., D. Linder, and R. K. Thauer. 1983. Unusual pathway of isoleucine biosynthesis in Methanobacterium thermoautotrophicum. Arch. Microbiol. 136:111-113. [Google Scholar]
- 13.Ekiel, I., I. C. P. Smith, and G. D. Sprott. 1984. Biosynthesis of isoleucine in methanogenic bacteria: a 13C NMR study. Biochemistry 23:1683-1687. [Google Scholar]
- 14.Felsenstein, J. 2005. PHYLIP (phylogeny inference package), 3.65 ed. Department of Genetics, University of Washington, Seattle.
- 15.Graham, D. E., M. Graupner, H. Xu, and R. H. White. 2001. Identification of coenzyme M biosynthetic 2-phosphosulfolactate phosphatase: a member of a new class of Mg2+-dependent acid phosphatases. Eur. J. Biochem. 268:5176-5188. [DOI] [PubMed] [Google Scholar]
- 16.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross, S. R., R. O. Burns, and H. E. Umbarger. 1963. The biosynthesis of leucine. II. The enzymic isomerization of β-carboxy-β-hydroxyisocaproate and α-hydroxy-β-carboxyisocaproate. Biochemistry 2:1046-1052. [DOI] [PubMed] [Google Scholar]
- 18.Gruer, M. J., P. J. Artymiuk, and J. R. Guest. 1997. The aconitase family: three structural variations on a common theme. Trends Biochem. Sci. 22:3-6. [DOI] [PubMed] [Google Scholar]
- 19.Helgadóttir, S., G. Rosas-Sandoval, D. Söll, and D. E. Graham. 2007. Biosynthesis of phosphoserine in the Methanococcales. J. Bacteriol. 189:575-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howell, D. M., M. Graupner, H. Xu, and R. H. White. 2000. Identification of enzymes homologous to isocitrate dehydrogenase that are involved in coenzyme B and leucine biosynthesis in methanoarchaea. J. Bacteriol. 182:5013-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howell, D. M., H. Xu, and R. H. White. 1999. (R)-Citramalate synthase in methanogenic archaea. J. Bacteriol. 181:331-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jang, S., and J. A. Imlay. 2007. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J. Biol. Chem. 282:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordan, P. A., Y. Tang, A. J. Bradbury, A. J. Thomson, and J. R. Guest. 1999. Biochemical and spectroscopic characterization of Escherichia coli aconitases (AcnA and AcnB). Biochem. J. 344(Pt. 3):739-746. [PMC free article] [PubMed] [Google Scholar]
- 24.Karsten, W. E., P. A. Tipton, and P. F. Cook. 2002. Tartrate dehydrogenase catalyzes the stepwise oxidative decarboxylation of D-malate with both NAD and thio-NAD. Biochemistry 41:12193-12199. [DOI] [PubMed] [Google Scholar]
- 25.Kisumi, M., S. Komatsubara, and I. Chibata. 1977. Pathway for isoleucine formation from pyruvate by leucine biosynthetic enzymes in leucine-accumulating isoleucine revertants of Serratia marcescens. J. Biochem. 82:95-103. [DOI] [PubMed] [Google Scholar]
- 26.Kisumi, M., M. Sugiura, and I. Chibata. 1976. Biosynthesis of norvaline, norleucine, and homoisoleucine in Serratia marcescens. J. Biochem. 80:333-339. [DOI] [PubMed] [Google Scholar]
- 27.Kohlhaw, G. B. 1988. Isopropylmalate dehydratase from yeast. Methods Enzymol. 166:423-429. [DOI] [PubMed] [Google Scholar]
- 28.Liebich, H. M., E. Gesele, H. G. Wahl, C. Wirth, J. Wöll, and P. Hušek. 1992. Identification of side-products formed by derivatization of 2-hydroxycarboxylic acids with methyl and ethyl chloroformate. J. Chromatogr. A 626:289-293. [Google Scholar]
- 29.Ma, G. X., and D. R. J. Palmer. 2000. Improved asymmetric syntheses of (R)-(-)-homocitrate and (2R,3S)-(-)-homoisocitrate, intermediates in the α-aminoadipate pathway of fungi. Tetrahedron Lett. 41:9209-9212. [Google Scholar]
- 30.Miyazaki, K. 2005. Bifunctional isocitrate-homoisocitrate dehydrogenase: a missing link in the evolution of beta-decarboxylating dehydrogenase. Biochem. Biophys. Res. Commun. 331:341-346. [DOI] [PubMed] [Google Scholar]
- 31.Schloss, J. V., R. Magolda, and M. Emptage. 1988. Synthesis of α-isopropylmalate, β-isopropylmalate and dimethylcitraconate. Methods Enzymol. [DOI] [PubMed]
- 32.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki, T., Y. Inoki, A. Yamagishi, T. Iwasaki, T. Wakagi, and T. Oshima. 1997. Molecular and phylogenetic characterization of isopropylmalate dehydrogenase of a thermoacidophilic archaeon, Sulfolobus sp. strain 7. J. Bacteriol. 179:1174-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tersteegen, A., D. Linder, R. K. Thauer, and R. Hedderich. 1997. Structures and functions of four anabolic 2-oxoacid oxidoreductases in Methanobacterium thermoautotrophicum. Eur. J. Biochem. 244:862-868. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tipton, P. A., and B. S. Beecher. 1994. Tartrate dehydrogenase, a new member of the family of metal-dependent decarboxylating R-hydroxyacid dehydrogenases. Arch. Biochem. Biophys. 313:15-21. [DOI] [PubMed] [Google Scholar]
- 38.van der Werf, M. J., W. J. J. van den Tweel, and S. Hartmans. 1993. Purification and characterization of maleate hydratase from Pseudomonas pseudoalcaligenes. Appl. Environ. Microbiol. 59:2823-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Werf, M. J., W. J. J. van den Tweel, and S. Hartmans. 1993. Thermodynamics of the maleate and citraconate hydration reactions catalysed by malease from Pseudomonas pseudoalcaligenes. Eur. J. Biochem. 217:1011-1017. [DOI] [PubMed] [Google Scholar]
- 40.Vatankhah, M., and M. Moini. 1994. Characterization of fluorinated ethylchloroformate derivatives of protein amino acids using positive and negative chemical ionization gas chromatography/mass spectrometry. Biol. Mass Spectrom. 23:277-282. [Google Scholar]
- 41.Wallace, M. A., L.-L. Liou, J. Martins, M. H. Clement, S. Bailey, V. D. Longo, J. S. Valentine, and E. B. Gralla. 2004. Superoxide inhibits 4Fe-4S cluster enzymes involved in amino acid biosynthesis. Cross-compartment protection by CuZn-superoxide dismutase. J. Biol. Chem. 279:32055-32062. [DOI] [PubMed] [Google Scholar]
- 42.White, R. H. 1989. Steps in the conversion of α-ketosuberate to 7-mercaptoheptanoic acid in methanogenic bacteria. Biochemistry 28:9417-9423. [DOI] [PubMed] [Google Scholar]
- 43.Xu, H., Y. Zhang, X. Guo, S. Ren, A. A. Staempfli, J. Chiao, W. Jiang, and G. Zhao. 2004. Isoleucine biosynthesis in Leptospira interrogans serotype lai strain 56601 proceeds via a threonine-independent pathway. J. Bacteriol. 186:5400-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada, T., K. Kakinuma, and T. Oshima. 1987. A preparative method of DL-threo-3-isopropylmalic acid and DL-threo-[2-2H]-3-isopropylmalic acid. Chem. Lett. 9:1745-1748. [Google Scholar]
- 45.Yasutake, Y., M. Yao, N. Sakai, T. Kirita, and I. Tanaka. 2004. Crystal structure of the Pyrococcus horikoshii isopropylmalate isomerase small subunit provides insight into the dual substrate specificity of the enzyme. J. Mol. Biol. 344:325-333. [DOI] [PubMed] [Google Scholar]
- 46.Zou, Y., X. Guo, M. Picardeau, H. Xu, and G. Zhao. 2007. A comprehensive survey on isoleucine biosynthesis pathways in seven epidemic Leptospira interrogans reference strains of China. FEMS Microbiol. Lett. [DOI] [PubMed]