Abstract
Degradation of the cspA mRNA in vivo is very rapid at temperatures greater than 30°C and is moderately dependent on RNase E. Investigations in vitro show that degradosomes prepared from normal or cold-shocked cultures cleave the cspA mRNA preferentially at a single site in vitro between two stem-loops ∼24 residues 3′ to the termination codon and ∼31 residues from the 3′ end. The site of cleavage is independent of the temperature and largely independent of the phosphorylation status of the 5′ end of cspA mRNA. A 5′ stem-loop, potential occlusion of the initiation and termination codons, temperature-dependent translational efficiency, and the position of the RNase E cleavage site can explain the differential stability of the cspA mRNA.
Exposure of Escherichia coli or other mesophilic microorganisms to temperatures below 20°C induces an immediate growth arrest (“cold shock”) which is followed by a period of adaptation and subsequent resumption of growth (17, 30, 31, 35). A set of at least 26 proteins is induced immediately following the shift to low temperature (17). Among these is a family of low-molecular-weight proteins including CspA (16). CspA contains an OB-fold domain (1, 29), which is typical of many RNA binding proteins (34). Indeed, various roles in RNA metabolism have been attributed to CspA and other related cold shock proteins, including chaperoning RNAs (17, 19, 31). Although its expression can be detected at 37°C, CspA becomes one of the mostly highly expressed proteins following a shift to low temperature (4, 12). The induction of CspA at low temperature is largely independent of transcription and is dependent primarily on posttranscriptional controls, including enhanced translation (4, 14, 15, 17). In particular, its mRNA is selectively stabilized after the onset of cold shock (4, 12-15, 36). In fact, the half-life of cspA mRNA during exponential growth at 37°C is only 10 to 12 s, but it increases to 10 min when the organism is grown continuously at 15°C (12, 15). Moreover, mutations which stabilize the cspA mRNA permit it to be expressed constitutively, independent of a shift to 15°C (12). Finally, cspA mRNA expressed from a plasmid is stabilized in a strain carrying a temperature-sensitive RNase E (rne-50; G66S) but not sufficiently to permit detection of CspA (12).
Details of how cspA mRNA can be recognized very efficiently by the mRNA decay apparatus at 37°C but can evade degradation at 15°C are largely unknown. A potential stem-loop structure at the 5′ terminus of the cspA mRNA is critical for its stabilization following cold shock (12-15, 36). This finding, as well as circumstantial evidence for stabilization of cspA mRNA by rne-50 (12), points to a role for RNase E in regulating the expression of CspA. It has been suggested that a pathway of mRNA decay may be transiently inactivated during the initial phases of cold shock, somehow sparing cspA mRNA (15). In this regard, the finding that the RNA degradosome isolated from cold-shocked cells is modified by the presence of the DEAD box helicase, CsdA (32), raises the possibility that cspA mRNA might be differentially susceptible to degradosomes at 37°C. Alternatively, it is possible that efficient translation and possibly CspA itself may contribute to the stabilization of cspA mRNA during cold shock (15).
In this communication we show directly that cspA mRNA, which is normally quite labile at temperatures of 30°C or higher, is susceptible to RNase E in vivo and in vitro. We have determined the secondary structure of potential regulatory features within the cspA mRNA and have mapped the primary site of endonucleolytic cleavage to the 3′ untranslated region (UTR). Our data suggest a model for the regulation of expression of CspA that rationalizes the roles of secondary structure and RNase E.
MATERIALS AND METHODS
Strains and plasmids.
All strains are derivatives of E. coli K-12. CF881 [F− Δlac argA trp recB1009 (ΔxthA-pnc) Δrna] was obtained from Michael Cashel (NIH, Bethesda, MD). A set of isogenic strains including MG1693 (thi rph), SK5665 (thi rph rne-1 [rneG66S] [28]), SK5691 (thi rph pnp-7), and SK7988 (thi rph ΔpcnB) was obtained from Sidney Kushner, University of Georgia, Athens, GA. Strain KCB1008 (thi rph rne-131 [rneΔ585-1061] [21]) was constructed from MG1693 by P1-mediated transduction (K. E. Baker and G. A. Mackie, unpublished). To construct plasmid pCspA-1, DNA corresponding to a T7 promoter, the 5′ UTR, coding sequence, and 3′ UTR of cspA was amplified from E. coli DNA using primers CSPA-F and CSPA-R (Table 1). The product was purified, digested with EcoRI and BamHI, and ligated into pUC18 digested with the same enzymes. Following transformation of strain DH5α, a recombinant plasmid, pCspA-1, from a single clone was purified, and its authenticity was verified by DNA sequencing.
TABLE 1.
Sequences of oligonucleotides
| Oligonucleotide | Sequence (5′→3′) |
|---|---|
| CSPA-F | CGCAGAATTCTAATACGACTCACTATAGGGTTTGACGTACAGACC |
| CSPA-R | GGACGGATCCTGAAAACATTTAAAAAAATCCCCGC |
| CSPA-PE1 | CGATATGGCGTGCTTTAC |
| CSPA-PE2 | GAACACATCTTTAGAGCC |
| CSPA-PE3 | TAAGCAGAGATTACAGGC |
Enzymes and assays.
“Standard” degradosomes were purified from strain CF881 grown in LB medium with vigorous aeration at 37°C as described previously (26). “Cold shock” degradosomes were purified from the same strain grown at 30°C in LB medium to an A600 of 0.4 and then mixed with an equal volume of ice-cold LB and shifted to a water bath at 15°C. Growth was continued to saturation prior to harvest. Purification was performed as described previously (32). The compositions of both preparations were checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and by Western blotting to verify the presence of CsdA in the cold shock degradosomes (32). Both preparations were assayed against 9S pre-rRNA; their relative activities were directly proportional to the amount of RNase E determined by Western blotting (data not shown). Assays were performed in a final volume of 30 μl at 30°C as described previously (9), using 400 to 1,000 ng degradosomes and 20 nM RNA substrate. Samples were removed at timed intervals, denatured in buffered 90% formamide, boiled for 45 s, and separated on polyacrylamide gels containing 8 M urea.
Preparation and analysis of RNA.
Internally labeled CspA mRNA substrate was prepared from pCspA-1 linearized by cleavage with DraI and transcribed by T7 RNA polymerase, using [α-32P]CTP (NEN) as a label with a kit from Ambion, Inc. To prepare 3′-labeled RNA, unlabeled cspA mRNA was labeled with [32P]pCp (Amersham) and T4 RNA ligase under conditions recommended by the manufacturer (Fermentas). Following extraction with phenol-chloroform, the recovered RNA was purified on a 6% polyacrylamide gel containing 8 M urea, detected by staining with ethidium bromide, and eluted from a macerated gel slice with 0.5 M NH4-acetate, 10 mM Tris-HCl (pH 7.7), 1 mM EDTA, 0.1% sodium dodecyl sulfate, and 10 μg/ml carrier tRNA. Structure mapping experiments were generally performed as previously described (2) with the exception noted below. Unlabeled cspA mRNA was incubated with a dilution of T1 RNase sufficient to cleave less than 50% of the input RNA. The primers (CSPA-PE [Table 1]) were unlabeled, and [α-32P]dCTP was included in the primer extension step. Extraction of RNA from exponentially growing cultures and Northern blotting were performed as described previously (2, 24). Primer extension (see above) was used for half-life determination, using CSPA-PE1 (Table 1).
RESULTS
Effect of rne alleles on the stability of cspA mRNA in vivo.
As an initial step in determining the basis for the lability of the cspA mRNA at normal growth temperatures, we measured its half-life in vivo to determine whether it was sensitive to RNase E. Cultures of the isogenic strains MG1693 (wild type), SK5665 (rne-1), and KCB 1008 (rne-131) were grown at permissive temperature (30°C) and shifted to either 42°C (nonpermissive) or 15°C prior to extraction of RNA (see Materials and Methods). Half-lives of the cspA mRNAs from different samples were initially determined by Northern blotting. However, to obtain sufficient sensitivity, we subsequently resorted to primer extension. A summary of the data obtained is shown in Table 2. We could not reliably detect the cspA mRNA in the total RNA extracted from cultures shifted to 42°C, as the band corresponding to full-length cspA RNA was diffuse (data not shown). Moreover, no signal could be detected after treatment with rifampin. Taken together, these observations suggest that the cspA mRNA is very labile at elevated temperature. Others have reported that the half-life of the cspA mRNA is only 10 s at 37°C (15). In contrast, cspA mRNA could be detected readily from cultures grown at 30°C. Its half-life was 47 s in the wild-type strain and increased to 80 s in both strains carrying rne alleles (Table 2). As expected, the abundance and stability of the cspA mRNA increased significantly in cultures subjected to cold shock for 60 min (i.e., shifted from 30°C to 15°C). Table 2 shows that the half-life of the cspA mRNA in MG1693 was at least 600 s. The mRNA was also equally if not more stable in the two rne alleles. More accurate estimates could not be obtained, as no significant decay occurred during the time of sampling.
TABLE 2.
Half-lives of cspA mRNA in different strains and conditions
| Strain | Allele | Half-life (s)a
|
||
|---|---|---|---|---|
| 42°C | 30°C | 15°C | ||
| MG1693 | Wild type | NDb | 47 | >600 |
| SK5665 | rne-1 | ND | 80 | >600 |
| KCB1008 | rne-131 | ND | 80 | >600 |
Cultures were grown in LB at 30°C and then shifted to 42°C for 20 min or to 15°C for 60 min prior to addition of rifampin. Samples were withdrawn at 45-s intervals (42°C and 30°C) or at 2.5-min intervals (15°C) and RNA extracted immediately. Half-lives were determined by primer extension using CSPA-PE1 (Table 1). The values given represent the averages of duplicate determinations.
ND, not detectable. The cspA mRNA extracted from cultures shifted to 42°C was badly smeared, even in the time zero sample (see the text).
Endonucleolytic cleavage of cspA mRNA in vitro.
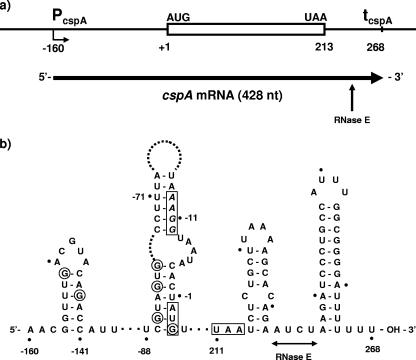
Examination of the DNA sequence of the cspA gene suggests that it should encode an mRNA of 428 nucleotides (nt) (Fig. 1a). A combination of inspection, computer-assisted folding (27), and structure mapping with T1 RNase was used to assess the potential secondary structure of the cspA mRNA, particularly at its extremities. In agreement with previous work (36), the data summarized in Fig. 1b show that this mRNA contains a moderately stable 5′ stem-loop (ΔG = −3.4 kcal/mol), a feature which is normally associated with enhanced mRNA stability (2, 3, 7, 11, 18, 23, 33) and the cold shock response (36). The 3′ end of the mRNA contains a predicted highly stable terminator stem-loop (ΔG = −14.3 kcal/mol). In addition, we predict that the AUG codon and Shine-Dalgarno sequence can be occluded by base pairing with sequences further upstream in the 5′ UTR. We presume that cold shock releases this (or other) putative structural barrier to translation, consistent with cold shock sensitivity residing in the 5′ UTR (12-15, 36).
FIG. 1.
Structure of the cspA gene and its mRNA. (a) Diagram of the cspA gene and its mRNA. The sites of initiation (−160) and termination (+268) of transcription are inferred from previous work (12, 13, 36) and inspection of the sequence. The in vitro transcript used in this work extends from position −158 (changing C to G) to +268 (see the text and the legend to Fig. 2). The vertical arrow shows the site of a major RNase E cleavage (see the text). (b) Model for portions of the secondary structure of the cspA mRNA, based on partial enzymatic digestion with T1 RNase and computer-assisted folding (RNAdraw [27]). Informative data were obtained for the 5′ UTR and the first 30 residues of the coding sequence. Residues are numbered relative to the A of the AUG initiation codon. The initiation and termination codons are boxed; the likely Shine-Dalgarno sequence (AAGG) is italicized. G residues that were clearly resistant to T1 RNase are circled. The horizontal line indicates the region in which RNase E cleavage occurs (see the text). Based on the minimum size of the cleavage products, the sites of cleavage are 3′ to residues 233 and 234. The extent of the horizontal line indicates that the precise sites of cleavage are uncertain.
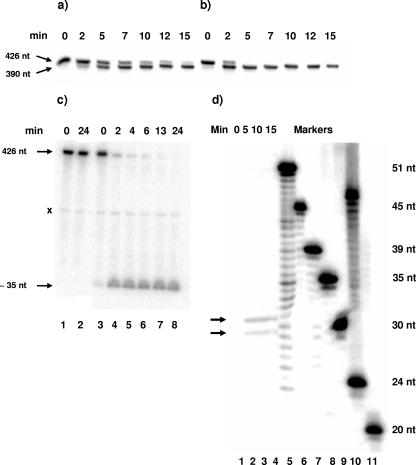
In view of the demonstrated and potential secondary structure in the cspA mRNA, we tested whether it would be a substrate for degradosomes prepared from cells grown at 37°C (Fig. 2a). We also tested whether cold shock degradosomes (32) would also cleave the cspA mRNA substrate (Fig. 2b). The two degradosome preparations were normalized to contain similar quantities of RNase E. The data in Fig. 2a and b show that in both cases the cspA mRNA substrate was cleaved readily to yield a major product of ∼390 residues which was relatively stable. This suggests that the major RNase E cleavage occurs ∼30 to 40 residues from one end of the mRNA. In this and subsequent experiments, we noted that normal degradosomes displayed a lag in attacking the substrate. We did not pursue this observation.
FIG. 2.
Time course of digestion of cspA mRNA in vitro by purified degradosomes. (a and b) Internally 32P-labeled cspA mRNA was digested with degradosomes purified from cultures grown at 30°C (a) or 15°C (b) (see Materials and Methods). The time of digestion is shown above each lane. Products of <∼75 residues would have run off the gel. The positions of the substrate and major product are shown in the left margin. (c) 3′-end-labeled cspA mRNA (see Materials and Methods) was incubated without enzyme (lanes 1 and 2) or with degradosomes purified from cultures grown at 30°C (lanes 3 to 8), and the products were separated on a denaturing 10% polyacrylamide gel. The positions of the substrate (428 residues) and product (∼35 residues) are shown in the left margin. The species labeled “x” in the left margin is found in undigested samples (cf. lanes 1 to 3), but its provenance is unknown. (d) Internally 32P-labeled cspA mRNA was digested with cold shock degradosomes for 0, 5, 10, or 15 min (lanes 1 to 4) and separated on a sequencing gel with 5′-32P-labeled oligonucleotides as markers. The sizes of the oligonucleotides are indicated in the right margin. The arrows in the left margin show the two products.
To map the cleavage site, we initially subjected the products of digestion by degradosomes to further cleavage with RNase H in the presence of specific oligonucleotides. The data showed that the initial cleavage occurred ∼40 residues from the 3′ end of the substrate (not shown). Moreover, using three primers spaced throughout the cspA sequence (CSPA-PE [Table 1]), we were unable to detect new 5′ termini in the 5′ UTR by primer extension, a finding consistent with in vivo data (12). Accordingly, we prepared 3′-end-labeled cspA mRNA and repeated the experiment of Fig. 2a, using 10% gels to retain the anticipated 3′ product. A time course of digestion is shown in Fig. 2c. The substrate disappeared within 10 min, and a product of ∼35 residues accumulated in nearly 100% yield. Additional characterization of the product was achieved by separation on a sequencing gel, using labeled DNA oligonucleotides to provide size markers. Assuming that the mobilities of the RNA products can be compared directly to those of the DNA markers, the products migrated as a doublet of 30 and 31 nt (Fig. 2d). This would correspond to cleavage immediately 5′ to the base of the terminal stem-loop (i.e., 3′ to residue 237) but possibly further 5′ between residues 233 and 234 (Fig. 1b). Heterogeneity in the products could reflect minor variations in the site of termination of transcription or in cleavage itself, analogous to the case for the major cleavage site in the rpsT mRNA (25). This cleavage site(s) lies ∼20 residues 3′ from the termination codon between two stem-loops, somewhat reminiscent of the rpsO mRNA (6). A previous inference that the site of RNase E cleavage lies in the 5′ UTR (12) is not supported by our data.
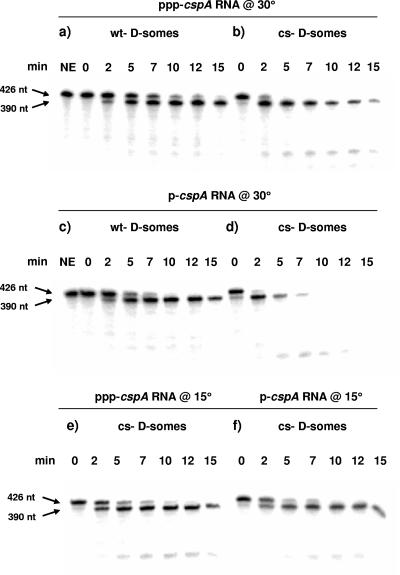
To test whether the predicted 5′ stem-loop was functional during our assays, we measured the effect of replacing the 5′-triphosphate terminus with a 5′-monophosphorylated terminus. We reasoned that if present, the predicted 5′-proximal stem-loop should negate the stimulatory effects of monophosphorylation (23). Normal or cold shock degradosomes were assayed against ppp-cspA mRNA (Fig. 3a and b) or p-cspA mRNA (Fig. 3c and d) at 30°C. In either case, the monophosphorylated substrate was digested at least 1.5- to 2.5-fold faster than ppp-cspA mRNA (compare Fig. 3a or b with Fig. 3c or d, respectively). At 15°C, however, the rate of cleavage of p-cspA mRNA by cold shock degradosomes as well as the rate of accumulation of 390-nt product was indistinguishable from that of the ppp-substrate (Fig. 3, compare panels f [p-] and e [ppp-]). Since substrates with single-stranded 5′ termini are typically stimulated up to 30-fold by monophosphorylation (23), we conclude that the 5′ terminus of cspA mRNA is sequestered in a form that is not readily recognized by RNase E, particularly at 15°C.
FIG. 3.
Effect of the status of the 5′ terminus on the rate of digestion of cspA mRNA. Internally 32P-labeled cspA mRNA containing either a 5′-triphosphate (a, b, and e) or a 5′-monophosphate (c, d, and f) was digested with degradosomes purified from cultures grown at 30°C (wt D-somes) (a and c) or 15°C (cs D-somes) (b, d, e, and f) as described in Materials and Methods. Digestions were performed at 30°C in panels a to d and at 15°C in panels e and f. The time of digestion is shown above each lane. NE, no enzyme. The positions of the substrate and major product are shown by arrows in the left margin.
DISCUSSION
The expression of the cspA gene appears to be regulated almost entirely by posttranscriptional mechanisms, including translational efficiency and mRNA stability, that are dependent on the temperature at which cultures are grown (17). We have focused on better understanding the basis for the instability of the cspA mRNA at normal temperatures. The available data show that the cspA mRNA is very labile in vivo at temperatures of >30°C, with half-lives of 47 s at 30°C (Table 2) and only 10 s at 37°C (15). This lability seems surprising in view of the 5′ stem-loop in the cspA mRNA, a feature that usually confers stability on mRNAs ostensibly by preventing a productive interaction with the 5′ sensor domain of RNase E (2, 3, 7, 11, 18, 23, 33). However, the 5′ stem-loop in cspA mRNA is only moderately stable (calculated ΔG = −3.4 kcal/mol). Nonetheless, the data in Fig. 3 suggest that its effectiveness as a stabilizing element improves at lower temperature, consistent with its requirement for the stabilization of cspA mRNA at low temperature (36). We also found that rne mutations increase the half-life of the cspA mRNA in vivo, although relatively modestly (Table 2). Taken together, these data suggest a role for RNase E in the degradation of cspA mRNA.
Our data show clearly that cspA mRNA is nearly equally sensitive to cleavage by RNase E in vitro at either 30°C or 15°C whether the RNase E resides in normal or cold shock degradosomes. Nonetheless, cspA mRNA is no more sensitive to RNase E in vitro than other substrates we have tested (e.g., rpsT mRNA and 9S pre-rRNA). However, the initial 390-nt product of cleavage is atypically stable, although prolonged digestion results in further cleavage (Fig. 3d). The secondary structure of the cspA mRNA can rationalize these observations. RNase E cleavage essentially truncates the cspA mRNA at its 3′ end, leaving the 5′ terminus intact. The preferred RNase E cleavage site is located between two stem-loops and is separated from the termination codon by 20 residues which are incorporated into an imperfect stem-loop. This situation is reminiscent of the rpsO mRNA, where the M2 cleavage site lies 10 residues from the termination codon and is adjacent to a stable 3′ terminator stem-loop (6, 10). The cleavage site in cspA mRNA is located a significant distance from the 5′ end of cspA mRNA and occurs despite the presence of a moderately protective 5′ stem-loop. Thus, this cleavage would constitute an example of the internal entry pathway by RNase E (2, 20). Such cleavages can occur on untranslated mRNAs (20). Moreover, the presence of a 5′ stem-loop and a terminal triphosphate can explain why the 390-nt 5′ fragment is relatively stable once cleaved in vitro: both features are protective, and once the preferred site is cleaved, alternative sites would display even lower affinity for RNase E. Finally, the observed cleavage would sever the body of the mRNA from the relatively stable 3′ terminator stem-loop, rendering the body of the mRNA susceptible to rapid exonucleolytic degradation.
These findings suggest a model for the behavior of cspA mRNA in exponential growth at 37°C and during cold shock. The in vitro data show that the distinctive half-lives of cspA under these different environment conditions are not a property of RNase E per se, although the 5′ stem-loop in cspA mRNA must contribute to its stabilization after cold shock (36). Rather, we believe that they reflect additional variables. In particular, at 37°C, translation of CspA is apparently very inefficient (4), although it is not known if this due to active repression. In this regard, it is likely significant that the putative Shine-Dalgarno sequence (AAGG) in cspA mRNA is weak (22) and its spacing from the AUG initiation codon is nine residues, longer than is typical in E. coli (8, 22). Moreover, both the Shine-Dalgarno sequence and the AUG codon may be sequestered into a stem-loop structure (Fig. 1b). Thus, few or no ribosomes would transit the cspA coding sequence rendering it highly vulnerable (2, 10). After cold shock, translation of the cspA mRNA is greatly enhanced (12-15, 36). This would result in frequent transit of the coding sequence by ribosomes and greater likelihood of their pausing at the termination codon prior to release. Although the distance from the termination codon to the cleavage site is greater than for the rpsO mRNA (6), the presence of an intervening stem-loop suggests that paused ribosomes could also occlude the RNase E cleavage site, enhancing stability of the cspA mRNA. In effect, the increased frequency of translation enhances the stability and abundance of the cspA mRNA following cold shock by at least two means. First, the internal entry pathway is inhibited (2), and second, paused ribosomes provide protection against RNase E cleavage at the major site in the 3′ UTR. In addition, CspA itself may chaperone its mRNA to provide further stabilization (17), while CsdA, a cold shock DEAD box RNA helicase, may promote translation and stabilization of cspA mRNA (5). Finally, the 5′ stem-loop, whose protective effect is well known (2, 3, 11, 18, 23, 33), further stabilizes cspA mRNA.
Acknowledgments
This work was supported by grant MOP-5396 to G.A.M. from the Canadian Institutes of Health Research.
We thank Sidney Kushner and Michael Cashel for providing strains.
Footnotes
Published ahead of print on 6 April 2007.
REFERENCES
- 1.Arcus, V. 2002. OB-fold domains: a snapshot of the evolution of sequence, structure and function. Curr. Opin. Struct. Biol. 12:794-801. [DOI] [PubMed] [Google Scholar]
- 2.Baker, K. E., and G. A. Mackie. 2003. Ectopic RNase E sites promote bypass of 5′-end-dependent mRNA decay in Escherichia coli. Mol. Microbiol. 47:75-88. [DOI] [PubMed] [Google Scholar]
- 3.Bouvet, P., and J. G. Belasco. 1992. Control of RNase E-mediated RNA degradation by 5′-terminal base pairing in E. coli. Nature 360:488-491. [DOI] [PubMed] [Google Scholar]
- 4.Brandi, A., P. Pietroni, C. O. Gualerzi, and C. L. Pon. 1996. Post-transcriptional regulation of CspA expression in Escherichia coli. Mol. Microbiol. 19:231-240. [DOI] [PubMed] [Google Scholar]
- 5.Brandi, A., R. Spurio, C. O. Gualerzi, and C. L. Pon. 1999. Massive presence of the Escherichia coli major cold shock protein CspA under non-stress conditions. EMBO J. 18:1653-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, F., J. Le Derout, and P. Régnier. 1998. Ribosomes inhibit an RNase E cleavage which induces the decay of the rpsO mRNA of Escherichia coli. EMBO J. 17:4790-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callaghan, A. J., M. J. Arcaida, J. A. Stead, K. J. McDowall, W. G. Scott, and B. F. Luisi. 2005. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature 437:1187-1191. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H., M. Bjerknes, R. Kumar, and E. Jay. 1994. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 22:4953-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coburn, G. A., and G. A. Mackie. 1998. Reconstitution of the degradation of the mRNA for ribosomal protein S20 with purified enzymes. J. Mol. Biol. 279:1061-1074. [DOI] [PubMed] [Google Scholar]
- 10.Deana, A., and J. G. Belasco. 2005. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 19:2526-2533. [DOI] [PubMed] [Google Scholar]
- 11.Emory, S. A., P. Bouvet, and J. G. Belasco. 1992. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 6:135-148. [DOI] [PubMed] [Google Scholar]
- 12.Fang, L., W. Jiang, W. Bae, and M. Inouye. 1997. Promoter-independent cold-shock induction of cspA and its derepression at 37°C by mRNA stabilization. Mol. Microbiol. 23:355-364. [DOI] [PubMed] [Google Scholar]
- 13.Fang, L., B. Xia, and M. Inouye. 1999. Transcription of cspA, the gene for the major cold-shock protein of Escherichia coli, is negatively regulated at 37°C by the 5′-untranslated region of its mRNA. FEMS Microbiol. Lett. 176:39-43. [DOI] [PubMed] [Google Scholar]
- 14.Goldenberg, D., I. Azar, A. B. Oppenheim, A. Brandi, C. L. Pon, and C. O. Gualerzi. 1997. Role of Escherichia coli cspA promoter sequences and adaptation of translational apparatus in the cold shock response. Mol. Gen. Genet. 256:282-290. [DOI] [PubMed] [Google Scholar]
- 15.Goldenberg, D., I. Azar, and A. B. Oppenheim. 1996. Differential mRNA stability of the cspA gene in the cold shock response of Escherichia coli. Mol. Microbiol. 19:241-248. [DOI] [PubMed] [Google Scholar]
- 16.Goldstein, J., S. Pollitt, and M. Inouye. 1990. Major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87:283-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gualerzi, C. O., A. M. Giuliodori, and C. L. Pon. 2003. Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 331:527-539. [DOI] [PubMed] [Google Scholar]
- 18.Hambraeus, G., K. Karhumaa, and B. Rutberg. 2002. A 5′ stem-loop and ribosome binding but not translation are important for the stability of B. subtilis aprE leader mRNA. Microbiology 148:1795-1803. [DOI] [PubMed] [Google Scholar]
- 19.Jiang, W., Y. Hou, and M. Inouye. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272:196-202. [DOI] [PubMed] [Google Scholar]
- 20.Joyce, S. A., and M. Dreyfus. 1998. In the absence of translation, RNase E can bypass 5′ mRNA stabilizers in Escherichia coli. J. Mol. Biol. 282:241-254. [DOI] [PubMed] [Google Scholar]
- 21.Kido, M., K. Yamanaka, T. Mitani, H. Niki, T. Ogura, and S. Hiraga. 1996. RNase E polypeptides lacking a C-terminal half suppress a mukB mutation in Escherichia coli. J. Bacteriol. 178:3917-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma, J., A. Campbell, and S. Karlin. 2002. Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bacteriol. 184:5733-5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackie, G. A. 1998. RNase E is a 5′-end-dependent endonuclease. Nature 395:720-724. [DOI] [PubMed] [Google Scholar]
- 24.Mackie, G. A. 2000. Stabilization of circulation rpsT mRNA demonstrates the 5′-end dependence of RNase E action in vivo. J. Biol. Chem. 275:25069-25072. [DOI] [PubMed] [Google Scholar]
- 25.Mackie, G. A. 1991. Specific endonucleolytic cleavage of the mRNA for ribosomal protein S20 of Escherichia coli requires the product of the ams gene in vivo and in vitro. J. Bacteriol. 173:2488-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackie, G. A., G. A. Coburn, X. Miao, D. J. Briant, and A. Prud'homme-Généreux. 2001. Preparation of the Escherichia coli Rne protein and reconstitution of the RNA degradosome. Methods Enzymol. 342:346-356. [DOI] [PubMed] [Google Scholar]
- 27.Matzura, O., and A. Wennborg. 1996. RNAdraw: an integrated program for RNA secondary structure calculation and analysis under 32-bit Microsoft Windows. Comput. Appl. Biol. Sci. 12:247-249. [DOI] [PubMed] [Google Scholar]
- 28.McDowall, K. J., R. G. Hernandez, S. Lin-Chiao, and S. N. Cohen. 1993. The ams-1 and rne-3071 temperature-sensitive mutations in the ams gene are in close proximity to each other and cause substitutions with a domain that resembles a product of the Escherichia coli mre locus. J. Bacteriol. 175:4245-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murzin, A. G. 1993. OB (oligonucleotide/oligosaccharide binding) fold: common structural and functional solution for non-homologous sequences. EMBO J. 12:861-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phadtare, S. 2004. Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 6:125-136. [PubMed] [Google Scholar]
- 31.Phadtere, S., J. Alsina, and M. Inouye. 1999. Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 2:175-180. [DOI] [PubMed] [Google Scholar]
- 32.Prud'homme-Généreux, A., R. K. Beran, I. Iost, C. S. Ramey, G. A. Mackie, and R. W. Simons. 2004. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome.’ Mol. Microbiol. 54:1409-1421. [DOI] [PubMed] [Google Scholar]
- 33.Sharp, J. S., and D. H. Bechhofer. 2005. Effect of 5′-proximal elements on decay of a model mRNA in Bacillus subtilis. Mol. Microbiol. 57:484-495. [DOI] [PubMed] [Google Scholar]
- 34.Theobald, D. L., R. M. Mitton-Fry, and D. S. Wuttke. 2003. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 32:115-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thieringer, H. A., P. G. Jones, and M. Inouye. 1998. Cold shock and adaptation. BioEssays 20:49-57. [DOI] [PubMed] [Google Scholar]
- 36.Xia, B., H. Ke, W. Jiang, and M. Inouye. 2002. The cold box stem-loop proximal to the 5′-end of the Escherichia coli cspA gene maximizes its mRNA at low temperature. J. Biol. Chem. 277:6005-6011. [DOI] [PubMed] [Google Scholar]