Abstract
PcsB of Streptococcus pneumoniae is an essential hydrolase involved in the separation of dividing cells. In this study, it was found that PcsB localizes to the plasma membrane and is released into the growth environment, yet it is detectable on the pneumococcal surface by flow cytometry analysis. High temperature and osmolarity led to upregulation of pcsB expression.
Streptococcus pneumoniae colonizes the human nasopharynx and is a significant cause of morbidity (12). A DNA microarray based on the open reading frames of S. pneumoniae TIGR4 was used to measure differential gene regulation in S. pneumoniae D39 grown in subinhibitory concentrations of penicillin (Pen) or clarithromycin (Cln). One notable upregulated gene was pcsB (protein required for cell separation in group B streptococci [14]; The Institute for Genomic Research annotation SP2216). Based on the array results, expression of pcsB was upregulated 5.69- and 2.63-fold in response to the presence of 0.5× MIC of Pen and Cln, respectively, relative to growth in media with no antibiotic added (8).
PcsB of S. pneumoniae shows 37% identity and 50% similarity to IDG60 of Streptococcus mutans. IDG60 is not essential, and deletion mutants display pleomorphic morphology and increased sensitivity to stress conditions (2). Conditions of low pH, high salt, and high temperature upregulate IDG60 expression (3).
In contrast, PcsB was found to be essential in S. pneumoniae (15). Mutants with decreased levels of PcsB could be obtained only when the native promoter was replaced with an inducible or constitutive promoter (14). PcsB is the only essential hydrolase identified at present in S. pneumoniae (15).
Based on Signal P (1, 16) and PSORT (13) algorithms, PcsB has a signal peptidase I motif but no other motifs that would otherwise predict its localization (21). The purpose of this study was to determine cellular localization and confirm pcsB upregulation in response to stress.
Bacterial strains and growth conditions.
Pneumococcal strains D39 (serotype 2) and WU2 (serotype 3) and their respective nonencapsulated derivatives, R6 and JD908 (11), were used in this study. Pneumococci were cultured on sheep blood agar plates overnight at 37°C in 5% CO2 or grown in Todd-Hewitt broth (Difco, Detroit, MI) supplemented with 0.5% yeast extract (THY) at 37°C.
rPcsB.
PCR was used to amplify the portion of pcsB corresponding to amino acids 28 to 392 (excluding the leader sequence) with primers 565F and 565R (Table 1). The PCR product was cloned into pET100 vector (Invitrogen, Carlsbad, CA), which incorporates an N-terminal His6 tag to facilitate purification, and cloning success was confirmed by sequencing (SeqWright, Houston, TX). Recombinant PcsB (rPcsB) was purified and stored in phosphate-buffered saline (PBS) at −20°C until use. The recombinant protein migrated to the expected position as confirmed by Western analysis using Pierce INDIA His probe-horseradish peroxidase (data not shown).
TABLE 1.
Primers used in this study
| Primer | Description | Sequence (5′-3′) |
|---|---|---|
| 514F | 16S rRNA-RTPCR forward primer | CTGCGTTGTATTAGCTAGTTGGTG |
| 514R | 16S rRNA-RTPCR reverse primer | TCCGTCCATTGCCGAAGATTC |
| 559F | PcsB-RTPCR forward primer | ACCTTGGGCTGGAGACTACTG |
| 559R | PcsB-RTPCR reverse primer | TTGCTCCAACTTGAGGTGTTGAA |
| 565F | PcsB-pET100 forward primer | CACCGAAACGACTGATGACAAAATTG |
| 565R | PcsB-pET100 reverse primer | TAATCTGCATAAATATATGTAACAAAACCTTCA |
Rabbit polyclonal serum.
A New Zealand White rabbit was subcutaneously immunized with a mixture of 100 μg of rPcsB and Freund's complete adjuvant (Sigma-Aldrich, St. Louis, MO) followed by a booster with rPcsB and incomplete adjuvant 2 weeks later.
Fractionation.
Mid-exponential-phase cultures were fractionated as previously described (20) with the following modifications. To obtain proteins secreted into the culture medium, 0.5 ml of culture supernatant was precipitated with 10% trichloroacetic acid, washed twice with chilled 90% acetone, and suspended in 50 μl PBS. Protoplasts were made by incubation in protoplast buffer (20% sucrose, 5 mM Tris [pH 7.4], and 2.5 mM MgSO4) at 25°C for 24 h.
Western analysis.
Volumes of each fraction, equivalent to 100 μl of culture, were separated by 4 to 20% gradient sodium dodecyl sulfate-HEPES gel electrophoresis. After transfer onto a nitrocellulose membrane, PcsB was detected with polyclonal rabbit antiserum (1:10,000 dilution) followed by biotinylated goat anti-rabbit antibody and streptavidin-conjugated horseradish peroxidase (Southern Biotechnology Associates, Birmingham, AL). Washes were done with PBS-0.05% Tween 20. Pierce SuperSignal West Pico chemiluminescent substrate and autoradiography film were used for detection.
Flow cytometric analysis.
Flow cytometric analysis was carried out as previously described (4) with the following modifications. Exponentially growing bacteria were washed and incubated with rabbit antiserum (100 μl of a 1:100 dilution) for 30 min at room temperature. Biotinylated secondary antibody (5 μg/ml) and streptavidin-conjugated Alexa Fluor 488 (Molecular Probes, Eugene, OR) (2 μg/ml) were used to detect bound antibodies. The bacteria were fixed in 1% methanol-free paraformaldehyde and analyzed using a Becton Dickinson FACScan flow cytometer. Bacteria incubated with only the secondary antibody served as a negative control.
Real-time PCR (RTPCR).
S. pneumoniae D39 was grown to an approximate optical density at 600 nm of 0.6, and Pen or Cln was added to bacterial aliquots at a final concentration of 0.5× MIC or sterile distilled water was added. The cells were harvested after 30-min and 1-h incubations. Stress experiments under conditions of 0.5 M NaCl, pH 5.0, and 42°C in THY were conducted as previously described (3). QIAGEN RNeasy kits were used to purify RNA.
Moloney murine leukemia virus reverse transcriptase with random primers was used to make cDNA from 1 μg of RNA. Primers (0.5 μM) 514F and 514R and primers 565F and 565R (Table 1), together with cDNA at a 1:100 dilution, were used to amplify a portion of the 16S rRNA and PcsB, respectively, in experiments performed in triplicate as previously reported (18). SYBR green Supermix (Bio-Rad, Hercules, CA) was used in a MyIQ thermocycler according to the instructions of the manufacturer. Real-time data were analyzed using iQ optical system version 3.0a software (Bio-Rad) and the ΔΔCt method (10).
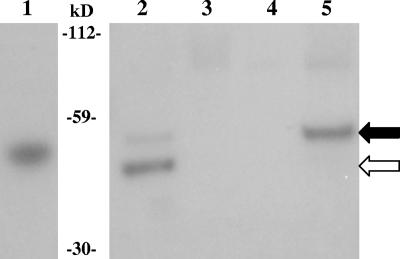
PcsB was detected in the secreted and plasma membrane fractions (Fig. 1). In both fractions, two reactive bands were present. Based on their positions relative to the rPcsB (41.4 kDa), the two proteins are possibly the preprotein (signal intact; 41.7 kDa) and the mature protein (signal cleaved; 38.9 kDa). Furthermore, the prevalent species seen in the plasma membrane and secreted fractions are those of the possible preprotein and mature protein, respectively. It was unexpected that PcsB would localize to the plasma membrane, as there is no SPaseII signal. Perhaps PcsB is lipidated by an unknown mechanism, allowing plasma membrane association. Another possibility may be that the catalytic rate of the one SPaseI of S. pneumoniae (19) is lower than the rate of PcsB synthesis, allowing detection of the uncleaved protein at the plasma membrane. Furthermore, as has been suggested previously (6), the uncleaved signal sequence may serve as the anchor in the membrane, but once the signal is cleaved, the mature protein is released into the media. There is no evidence as to whether PcsB is active while it is associated with the plasma membrane and/or after cleavage as it diffuses out from the membrane. However, adding exogenous rPcsB, which lacks the native leader sequence, to the media when growing a mutant with depressed PcsB expression does not restore the wild-type phenotype (Wai-Leung Ng and Malcolm Winkler, unpublished results), as has been shown for another cell wall hydrolase, LytB (5). This result is preliminary and may have been due to differences in posttranslational modification by S. pneumoniae and Escherichia coli, which was used to produce the recombinant protein. In addition, rPcsB may have not been able to reach the appropriate site for activity.
FIG. 1.
Western blot of D39 fractions. Lanes: 1, recombinant PcsB; 2, supernatant; 3, cell wall fraction; 4, cytoplasmic fraction; 5, plasma membrane fraction. PcsB was detected with polyclonal rabbit serum. The black arrow indicates the preprotein, with a predicted mass of 41.7 kDa, and the white arrow indicates the size of the mature protein, with a predicted mass of 38.9 kDa. The recombinant protein has a predicted mass of 41.4 kDa.
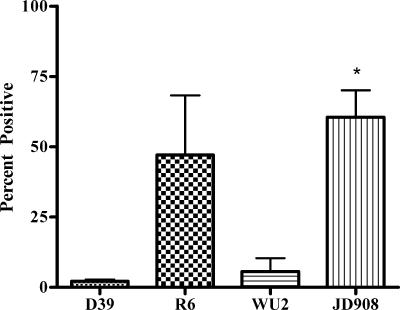
The accessibility to antibodies of PcsB on the surface of live cells was assessed by flow cytometric analysis. When S. pneumoniae D39 was analyzed by flow cytometric analysis (Fig. 2), on average 2.18% of the cells were positive, whereas when R6, a nonencapsulated derivative of strain D39, was analyzed, 47.11% were positive. Likewise, when strain WU2 was analyzed, 5.68% of cells were positive, and 59.57% of the cells of its nonencapsulated mutant, JD908, were positive. PcsB at the plasma membrane is detectable by flow cytometric analysis, although the capsule and probably to a lesser degree other structures at the surface of the cell impede antibodies from interacting with PcsB, leading to higher detection on nonencapsulated than on encapsulated cells, as has also been seen with PspA and PspC (9).
FIG. 2.
Percent positive values as determined by FACScan analysis of fresh cultures of strains D39, R6, WU2, and JD908. Error bars indicate standard errors of the means of the results of three independent experiments. *, P < 0.05 compared with WU2 results.
RTPCR experiments measuring differential expression of pcsB in late-exponential-phase strain D39 due to exposure to Pen and Cln indicated no change; therefore, the exposure to Pen and Cln, and the remaining stressors, was done in mid-exponential phase. The upregulation of pcsB in response to exposure to 0.5× MIC of Pen or Cln was below what is broadly recognized as significant (Fig. 3). However, in response to conditions of 0.5 M NaCl and 42°C, pcsB transcript levels increased by 1.9- and 2.9-fold, and no change was observed in response to low pH (Fig. 3); these results, in contrast, did not support the data from the array which initiated our investigation of PcsB.
FIG. 3.
Average fold induction of four independent experiments as determined by RTPCR of pcsB in response to 30 min of exposure to THY with one of the following stressors: 0.5 M NaCl, pH 5.0, 42°C, 0.5× MIC of clarithromycin or penicillin. Values are expressed relative to the values obtained in the presence of THY with only distilled water added.
The results of this study show that PcsB is secreted and is associated with the plasma membrane. Although the sublethal levels of antibiotics did not increase pcsB expression in these experiments, high temperature and the presence of salt did, similar to results obtained in investigations of S. mutans (3). pcsB expression increased as a result of stress; thus, PcsB may be an important protein involved in the response to changes of environment incurred in pathogenesis. The capsule by its charge and/or steric hindrance decreases the accessibility of PcsB to antibodies, as determined by flow cytometric analysis. Studies have shown that PsaA, a lipoprotein, is not readily accessible to antibodies (as determined by FACS) (7); however, immunization elicits protection against lethal pneumococcal challenge in mice (17). Immunization with PcsB may likewise provide protection against lethal pneumococcal challenge.
Footnotes
Published ahead of print on 20 April 2007.
REFERENCES
- 1.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 2.Chia, J. S., L. Y. Chang, C. T. Shun, Y. Y. Chang, and J. Y. Chen. 2001. A 60-kilodalton immunodominant glycoprotein is essential for cell wall integrity and the maintenance of cell shape in Streptococcus mutans. Infect. Immun. 69:6987-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chia, J. S., Y. Y. Lee, P. T. Huang, and J. Y. Chen. 2001. Identification of stress-responsive genes in Streptococcus mutans by differential display reverse transcription-PCR. Infect. Immun. 69:2493-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dave, S., A. Brooks-Walter, M. K. Pangburn, and L. S. McDaniel. 2001. PspC, a pneumococcal surface protein, binds human factor H. Infect. Immun. 69:3435-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Las Rivas, B., J. L. García, R. López, and P. García. 2002. Purification and polar localization of pneumococcal LytB, a putative endo-β-N-acetylglucosaminidase: the chain-dispersing murein hydrolase. J. Bacteriol. 184:4988-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desvaux, M., E. Dumas, I. Chafsey, and M. Hebraud. 2006. Protein cell surface display in Gram-positive bacteria: from single protein to macromolecular proteins structure. FEMS Microbiol. Lett. 256:1-15. [DOI] [PubMed] [Google Scholar]
- 7.Gor, D. O., X. Ding, D. E. Briles, M. R. Jacobs, and N. S. Greenspan. 2005. Relationship between surface accessibility for PpmA, PsaA, and PspA and antibody-mediated immunity to systemic infection by Streptococcus pneumoniae. Infect. Immun. 73:1304-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hooshdaran, M. Z., T. T. Liu, K. S. Barker, G. M. Hilliard, B. K. English, J. Thornton, E. Swiatlo, L. S. McDaniel, and P. D. Rogers. 2005. Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 907.
- 9.Kolberg, J., A. Aase, S. Bergmann, T. K. Herstad, G. Rødal, R. Frank, M. Rohde, and S. Hammerschmidt. 2006. Streptococcus pneumoniae enolase is important for plasminogen binding despite low abundance of enolase protein on the bacterial cell surface. Microbiology 152:1307-1317. [DOI] [PubMed] [Google Scholar]
- 10.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 11.Magee, A. D., and J. Yother. 2001. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect. Immun. 69:3755-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDaniel, L. S., and E. Swiatlo. 2004. Pneumococcal disease: pathogenesis, treatment, and prevention. Infect. Dis. Clin. Pract. 12:93-98. [Google Scholar]
- 13.Nakai, K., and M. Kanehisa. 1991. Expert system for predicting protein localization sites in Gram-negative bacteria. Proteins 11:95-110. [DOI] [PubMed] [Google Scholar]
- 14.Ng, W. L., G. T. Robertson, K. M. Kazmierczak, J. Zhao, R. Gilmour, and M. E. Winkler. 2003. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol. Microbiol. 50:1647-1663. [DOI] [PubMed] [Google Scholar]
- 15.Ng, W. L., K. M. Kazmierczak, and M. E. Winkler. 2004. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol. Microbiol. 53:1161-1175. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 17.Talkington, D. F., B. G. Brown, J. A. Tharpe, A. Koenig, and H. Russell. 1996. Protection of mice against fatal pneumococcal challenge by immunization with pneumococcal surface adhesin A (PsaA). Microb. Pathog. 21:17-22. [DOI] [PubMed] [Google Scholar]
- 18.Thornton, J., and L. S. McDaniel. 2005. Monocytes up-regulate intercellular adhesion molecule 1 in response to pneumolysin from Streptococcus pneumoniae. Infect. Immun. 73:6493-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Roosmalen, M. L., N. Geukens, J. D. H. Jongbloed, H. Tjalsma, J. Y. F. Dubois, S. Bron, J. M. van Dijl, and J. Anné. 2004. Type I signal peptidases of Gram-positive bacteria. Biochim. Biophys. Acta 1694:279-297. [DOI] [PubMed] [Google Scholar]
- 20.Vijayakumar, M. N., and D. A. Morrison. 1986. Localization of competence-induced proteins in Streptococcus pneumoniae. J. Bacteriol. 165:689-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wizemann, T. M., J. H. Heinrichs, J. E. Adamou, A. L. Erwin, C. Kunsch, G. H. Choi, S. C. Barash, C. A. Rosen, H. R. Masure, E. Tuomanen, A. Gayle, Y. A. Brewah, W. Walsh, P. Barren, R. Lathigra, M. Hanson, S. Langermann, S. Johnson, and S. Koenig. 2001. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect. Immun. 69:1593-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]